Abstract
Background
Transcriptional profiling has been performed on biopsies from ulcerative colitis patients. Limitations in prior studies include the variability introduced by inflammation, anatomic site of biopsy, extent of disease, and medications. We sought to more globally understand the variability of gene expression from patients with ulcerative colitis to advance our understanding of its pathogenesis and to guide clinical study design.
Methods
We performed transcriptional profiling on 13 subjects, including pediatric and adult patients from 2 hospital sites. For each patient, we collected 6 biopsies from macroscopically inflamed tissue and 4 biopsies from macroscopically healthy-appearing tissue. Isolated RNA was used for microarray gene expression analysis utilizing Affymetrix Human Primeview microarrays. Ingenuity pathway analysis was used to assess over-representation of gene ontology and biological pathways. RNAseq was also performed, and differential analysis was assessed to compare affected vs unaffected samples. Finally, we modeled the minimum number of biopsies required to reliably detect gene expression across different subject numbers.
Results
Transcriptional profiles co-clustered independently of the hospital collection site, patient age, sex, and colonic location, which parallels prior gene expression findings. A small set of genes not previously described was identified. Our modeling analysis reveals the number of biopsies and patients per cohort to yield reliable results in clinical studies.
Conclusions
Key findings include concordance, including some expansion, of previously published gene expression studies and similarity among different age groups. We also established a reliable statistical model for biopsy collection for future clinical studies.
Keywords: genetics and molecular epidemiology, endoscopy, pediatrics
INTRODUCTION
Ulcerative colitis (UC) is a chronic, relapsing inflammatory bowel disease (IBD) that affects an estimated 590,000 people in the United States.1 Ulcerative colitis is characterized by superficial colonic inflammation extending proximally in a continuous fashion from the rectum and is associated with bloody diarrhea, urgency, and abdominal cramping.2, 3 Extra-intestinal manifestations can also occur, including seronegative arthritis, panniculitis, primary sclerosing cholangitis, uveitis, and episcleritis. Data from humans and animal models suggest that mucosal inflammation in UC occurs secondary to inappropriate immune responses to environmental triggers including commensal bacteria in genetically susceptible individuals.3, 4 Mucosal inflammation in UC is associated with an influx of immune cells to the lamina propria of the affected colon. Consequently, the mainstay of medical therapy involves anti-inflammatory agents such as 5-aminosalicylates (5-ASA) and corticosteroids, immunomodulators such as 6-mercaptopurine (6MP) and azathioprine (AZA), biologic agents such as anti–tumor necrosis factor alpha (anti-TNFα) antibodies, and adhesion molecule inhibitors that impair leukocyte homing to the colon such as anti-α4β7 integrin antibodies. For many patients, such available medical interventions are often unable to control disease long term.
In an effort to better understand the pathways of importance in IBD pathogenesis and guide the development of therapeutic alternatives, many have studied the genetics of IBD. A recent meta-analysis of genome-wide association studies (GWAS) identified more than 200 risk loci associated with development of IBD, including 23 that are unique to UC.4–7 Some risk loci significantly enriched in UC relate to epithelial barrier responses (HNF4A, GNA12, MUC19) and both innate (MHC, FCGR2A, MST1) and adaptive immunity (IL23R, RORC, IL12B).4 Others have looked more directly, using transcriptional profiling of intestinal mucosal biopsies of patients with IBD to shed further light on the pathogenesis of disease through exploring the RNA profiles of the affected tissue.8–16
There is a clear separation between transcriptional profiles of colonic biopsies taken from active UC patients and the healthy tissue of controls.10, 11, 17 Consistent between studies,11, 13, 15, 17 sigmoid biopsies taken from patients with UC with histologically normal rectums as compared with those of healthy controls show differential gene expression in immune-related genes, such as: defensin beta 14, SAA1, and HLA class 2.12 One of the largest studies looking at transcriptional profiling of biopsies of IBD patients taken from macroscopically inflamed and uninflamed biopsies of adults with UC, Crohn’s, and healthy controls found a predominance of increased expression of Th1-related transcription factors and antimicrobial peptide-related genes in biopsies taken from patients with active UC as compared with healthy controls.18
However, there remains significant heterogeneity across the conclusions from various groups. Specifically, although Bjerrum et al., Planel et al., and Noble et al. found that the transcriptional profiles of biopsies of uninflamed or quiescent colonic biopsies of patients with UC differed from those of healthy controls, Granlund et al.’s work showed that these were rather similar.12, 15, 17, 18 Another source of heterogeneity may be the anatomic location of colonic biopsy collection. The proximal and distal colons have distinct embryological origins. The proximal colon, including the proximal two-thirds of the transverse colon, derives from the embryologic midgut, whereas the distal colon and rectum derive from the hindgut. Some investigators identified differences in gene expression between the right and left colon in healthy controls that was lost in inflamed UC mucosa.12 In contrast, others have found no significant differences based on anatomic regional locations of colonic biopsies, whether samples were of inflamed tissue or not.10–12 To complicate matters, in a different cohort of patients, Bjerrum et al. identified that there was also differential gene expression in biopsies taken from the descending colon in patients with left-sided colitis as compared with patients with pancolitis.16 In addition to factoring in the extent of disease that may affect transcriptional profiles, one must also account for the impact of various medications. Indeed, Arijs et al. identified differential gene expression in cell adhesion molecules in inflamed biopsies of patients with UC that normalized following response to anti-TNF agents.14 Taken together, many factors can impact results and interpretations of transcriptional profiling of intestinal biopsies.
When studying transcriptional profiling, careful attention must be paid to details of sample collection. In addition to changes introduced by presence of inflammation, anatomic site of biopsy, extent of disease, and medications, one must consider the number of biopsies. The exact number of biopsies needed within a region of either inflamed or uninflamed tissue to yield statistically significant, reliable, and reproducible results remains unclear. A robust means of stringently assessing for interbiopsy differences in gene expression has not been rigorously performed to date. This is especially critical in the setting of analyses in clinical trials assessing response to various medical interventions occurring at multiple centers.
In this study, we sought to more globally understand the transcriptional profile of colonic tissue of patients with UC comparing macroscopically active inflammation with macroscopically uninflamed tissue. Unlike other studies, our cohort also includes patients with pediatric onset UC (13–20 years old) recruited from Boston Children’s Hospital and an adult cohort (31–56 years old) recruited from Brigham & Woman’s Hospital. Given the heterogeneity observed in previous studies, we sought to reproducibly assess gene expression variation in biopsy specimens. Defining such variation and establishing techniques for robust and reproducible assessments of transcriptional profiling from colonic biopsies are imperative for quality studies and clinical trials. We strived to expand on possible interbiopsy variance in expression profiling by sampling up to 6 biopsies within an area of macroscopically inflamed tissue and up to 4 biopsies from macroscopically healthy-appearing tissue within each patient.
METHODS
Sample Acquisition
Research biopsies were obtained at clinically indicated procedures. Target sample collection (based on institutional review board [IRB] approval) was 6 biopsies from “affected” mucosa with endoscopically visible evidence of mucosal inflammation and 4 biopsies from “unaffected” macroscopically normal mucosa. In most cases, “affected” and “unaffected” biopsies were collected within a single anatomic region. Biopsies were obtained in pairs within 10 cm of each other, and the anatomic area was noted by the endoscopist. A mean (range) of 9.4 (6–10) biopsies was obtained per patient (Supplementary Table 1). The target of 10 biopsies was achieved in 10/13 (85%) patients. Demographic and clinical data were noted at the time of sample acquisition. Clinical disease activity was evaluated using the Pediatric Ulcerative Colitis Activity Index (PUCAI)19 or Simple Clinical Colitis Activity Index (SCCAI)20 (Table 1).
Table 1:
Patient Demographics
| Subject No. | Sex | Age, y | Disease Duration, y | Extent of Disease | Clinical Score (PUCAI or SCCAI) | Concomitant Medications |
|---|---|---|---|---|---|---|
| BCH-157 | Male | 18 | 3.5 | Pancolitis | 40P | 5-ASA, corticosteroid |
| BCH-877 | Female | 13 | 8.75 | Left-sided | 40P | 5-ASA, 6-MP, corticosteroid |
| BCH-1057 | Male | 17 | 1.25 | Left-sided | 70P | Antibiotic, 6-MP, corticosteroid |
| BCH-1077 | Male | 17 | 2 | Left-sided | 35P | 5-ASA, corticosteroid |
| BCH-1120 | Female | 20 | 0 | Pancolitis | 40P | Antibiotic |
| BCH-1192 | Male | 20 | 7 | Pancolitis | 20P | Anti-TNF |
| BCH-1214 | Female | 19 | 2 | Left-sided | 0P | 6-MP, anti-TNF |
| BWH-8855 | Female | 52 | 9.86 | Left-sided | 0S | 5-ASA |
| BWH-8854 | Male | 56 | 9.86 | Left-sided | 1S | 5-ASA |
| BWH-8874 | Male | 21 | 5.92 | Left-sided | 2S | 5-ASA |
| BWH-8878 | Female | 29 | 0.95 | Pancolitis | 1S | Anti-TNF |
| BWH-8879 | Male | 45 | 2.95 | Left-sided | 0S | 5-ASA, 6-MP, anti-TNF |
| BWH-8881 | Female | 27 | 8.96 | Left-sided | 0S | 5-ASA |
Transcriptional Analysis
RNA was extracted from individual biopsies utilizing the RNeasy Lipid Tissue Kit (Qiagen, Valencia, CA, USA) and homogenized on a TissueLyser II system using steel beads. RNA concentration was assessed with a Nanodrop 8000 spectrophotometer (Wilmington, DE, USA), and quality was assessed with an Agilent 2100 BioAnalyzer (Santa Clara, CA, USA). RNA concentrations ranged from 500 ng to 12 μg. Samples used for microarray had an RNA integrity number (RIN) >7 and were normalized to the same input level for microarray (100 ng) and quantitative polymerase chain reaction (qPCR; 300 ng) (Supplementary Fig. 1). cDNA conversion was performed using the Ovation RNA Amplification System V2 (Nugens, San Carlos, CA, USA).
Microarray gene expression analysis was performed utilizing Affymetrix Human Primeview microarrays (Santa Clara, CA, USA). Arrays were processed, hybridized, washed, and scanned using GeneChip Fluidics Station 450 and GeneChip Scanner 3000 according to the manufacturer’s instructions. All Cel files were processed together utilizing the Robust Multi-array Average (RMA) algorithm, and batch control samples (Human Universal RNA, Agilent) were utilized to assess batch-to-batch variability (Supplementary Fig. 2).
Quantative polymerase chain reaction (qPCR) was performed utilizing the Applied Biosystems High-Capacity RNA-to-cDNA Kit (Carlsbad, CA, USA) for cDNA conversion, Taqman fast advanced mastermix, and Taqman real-time PCR (rtPCR) assays. Assays were run on an Applied Biosystems Viia7 rtPCR machine and processed for analysis utilizing GeneData’s Expressionist Software Suite (Basel, Switzerland).
RNAseq was performed utilizing 50 ng of RNA taken from the same aliquot used for both the microarrays and qPCR to generate the cDNA libraries using the Neoprep automated system (Illumina, San Diego, CA, USA) and the Truseq stranded mRNA kit (NP-202–1001). The resulting libraries were quantified and checked for quality using the Fragment Analyzer system (Advanced Analytical, Ankeny, IA, USA). Libraries were pooled to equimolar concentrations and sequenced on a Nextseq 500 system (Illumina, San Diego, CA, USA) targeting ~20 million 75 bp reads (40 million paired reads) per sample. Sequencing run quality was assessed using Illumina’s SAV software and demultiplexed with Illumina’s bcl2fastq algorithm (Illumina, San Diego, CA, USA). Processing of the fastq files was performed using the QuickRNAseq pipeline utilizing Hg38 for the genome and Gencode v24 for annotation.21
Statistical Analysis of Microarrays, qPCR, and RNAseq
Microarray gene expression data were analyzed within Genedata Expressionist software (Lexington, MA, USA). Probes were called as expressed with a log2 robust multi-array average (RMA) expression ≥6. For microarrays, linear models (Boston Children’s Hospital [BCH] alone, Brigham & Women’s Hospital [BWH] alone, and all data merged) with the status of the biopsies and tissue locations as fixed factors and the study ID as a random variable were utilized. Other factors, including age and sex, were checked but not found to significantly influence the analysis. Only genes with a corrected P value (Bonferroni-Hochberg [BH]) <0.05 and fold-change ≥±2 were considered for downstream analyses (Supplemental Tables 2–4). In some cases, multiple transcriptional gene probes selected the same gene. Although the quantitative value of the log2 fold change differed between probes, these all had the same directionality of fold change. In these cases, we report values of the gene for probes that reflect the highest magnitude of fold change.
Comparing Our Results With Existing Microarray Data
Differential gene expression comparisons of existing microarray data sets for UC (GSE9452, GSE6731, GSE13367, GSE38713, GSE11223, GSE47908) were prepared with the online tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) using the default settings. Samples from each study were classified into UC uninflamed and UC inflamed to yield results as comparable to this study as reasonably possible. The comparison script generated for each existing data set was downloaded and altered to return all significant results rather than the top 250 results that the online version is restricted to. These scripts were run locally to obtain a list of differentially expressed genes from each data set and compared with the list of top differentially expressed genes from this study. The analysis scripts used are available for download under the following DOI: 10.6084/m9.figshare.6163793.
For qPCR, raw cycle threshold (Ct) values were converted to delta cycle threshold (dCt; control genes used, ACTB, and GAPDH) and assessed for significance utilizing a Student t test between affected and unaffected biopsies across patients (all biopsies) and within patients (only biopsies from that individual).
For RNAseq, differential analysis was performed utilizing the EdgeR algorithm22 to compare affected vs unaffected samples. Genes with expression of less than 1 count per million (cpm) in >50% of samples in either condition were removed from analysis, and genes with a corrected P value (BH) <0.05 and fold change (FC) ≥±2 are reported as significant (Supplementary Table 5).
Pathway Analysis
Gene ontology (GO) and pathway analysis were performed on differential gene expression results using Ingenuity Pathway Analysis (IPA; Qiagen). The core analysis function (default settings except added causal network option) was utilized on the 3 different model results (BCH alone, BWH alone, and all data merged). Canonical pathways, diseases and functions, and upstream regulators were assessed for significant enrichment and directionality of effect utilizing a z score >±2 and P value <0.05 (right-tailed Fisher’s exact test).23
Modeling the Number of Biopsies Required for Reliable Transcriptomic Analysis
To guide future clinical trial design, gene expression data were used to model the minimum number of biopsies required to reliably detect gene expression from patient samples, based on the number of subjects per group. One to 12 samples were randomly selected with replacement. For each selected sample, 1–4 pairs of inflamed and uninflamed biopsies were selected with replacement. Consequently, a total of 48 data sets were generated containing expression data from different numbers of samples and pairs of biopsies. For each data set, differentially expressed genes between affected and unaffected biopsies were identified using mixed effect or linear models with a false discovery rate (FDR) <0.01. Analysis included 100 permutations of each paired comparison, and differentially expressed genes were recorded for each.
ETHICAL CONSIDERATIONS
IRB Approval
This study was conducted at Boston Children’s Hospital and the Brigham & Women’s Hospital, Boston, Massachusetts, between January 2014 and December 2015, following IRB approval (BCH IRB P00000529, Partners Healthcare IRB 2010P002317). Adult patients and legal caregivers provided written informed consent, and pediatric patients provided their assent to participate in this study.
RESULTS
Patient Characteristics
Thirteen patients with an established diagnosis of UC were recruited, 7 at BCH and 6 at BWH; 6/13 (46%) were female. Four had pancolitis, and 9 had left-sided disease. Patients recruited at BCH were younger than patients recruited at BWH (median [range], 18 [13–20] vs 37 [21–56] years; P = 0.004) but with a similar disease duration (2 [1.25–7] vs 7.44 [2.45–9.86] years; P = 0.17). Patients recruited at BCH were more likely to have clinically active disease (6/7 vs 0/6; P = 0.005; PUCAI ≥ 10 or SCCAI > 2). Demographic details, including medications, are summarized in Table 1.
Transcriptional Profiling by Microarray
We performed transcriptomic profiling using Affymetrix on biopsies taken from affected and unaffected intestinal biopsies. Please refer to Supplementary Table 1 for anatomic location of collected biopsies. The number of probes called as expressed (log2 RMA) with expression ≥6 across either status or tissue location in any subject on these arrays was 38,237/49,395, which translates to 16,532 unique genes (with a known gene symbol) (Supplementary Fig. 3).
There were 1563 unique genes differentially expressed between affected vs unaffected sites, with a corrected P value <0.05 and fold-change ≥±2 (Supplementary Fig. 3). Within our cohort, differential expression was mainly influenced by macroscopic evidence of disease activity and less by anatomic location of the samples, as 1448 genes of the original 1563 (~93%) were still considered significant when differential analysis was limited to only the left-sided samples. Similarly, when we limited our analysis to biopsies collected only on the right colon (cecum, ascending and transverse colon), we captured 67.4% of significantly differentially expressed genes. In general, the microarray profiles co-clustered, independently of hospital site of collection (Fig. 1A) and patient age (Supplementary Fig. 4A) and sex (Supplementary Fig. 4B).
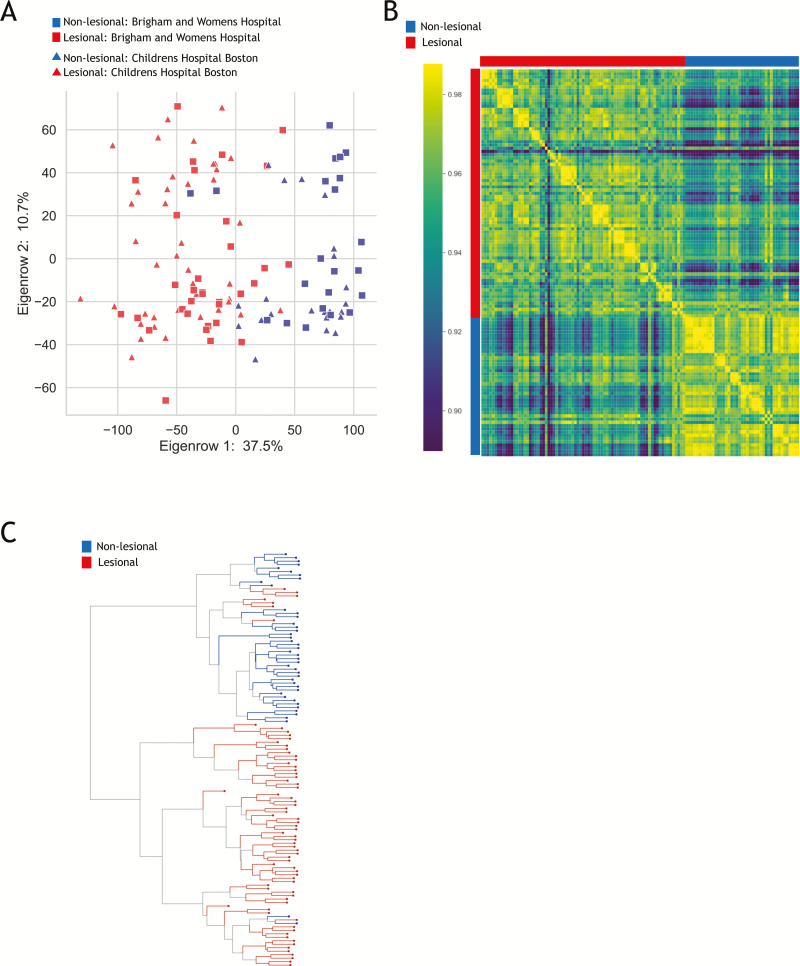
FIGURE 1.
Microarray results. A, PCA plot of transcriptional profiles comparing biopsies collected from areas of macroscopically active inflammation (red) with those collected from macroscopically noninflamed tissue (blue). Samples collected at BWH are in squares whereas those collected at BCH are depicted as triangles. Note that there is no strong difference between biopsies taken at different centers. B, Heatmap also reflecting high correlation among the set of affected biopsies and within the set of unaffected biopsies. C, Hierarchical clustering also illustrating clear distinction between the majority of affected and unaffected biopsies.
The most significant differences were appreciated in transcriptional profiles between biopsies collected from areas of macroscopically active inflammation, as compared to those collected from macroscopically noninflamed tissue (Fig. 1). There was high correlation within the set of affected biopsies, similarly high correlation within the set of unaffected biopsies, and significantly reduced correlation when comparing affected with unaffected biopsies (Fig. 1B). Principle component analysis (PCA) (Fig. 1A) and hierarchical clustering (Fig. 1C) also depict the generally clear distinction between affected and unaffected. There did not appear to be strong differences between biopsies taken at different centers independent of all analyzed metrics (Fig. 1A–C).
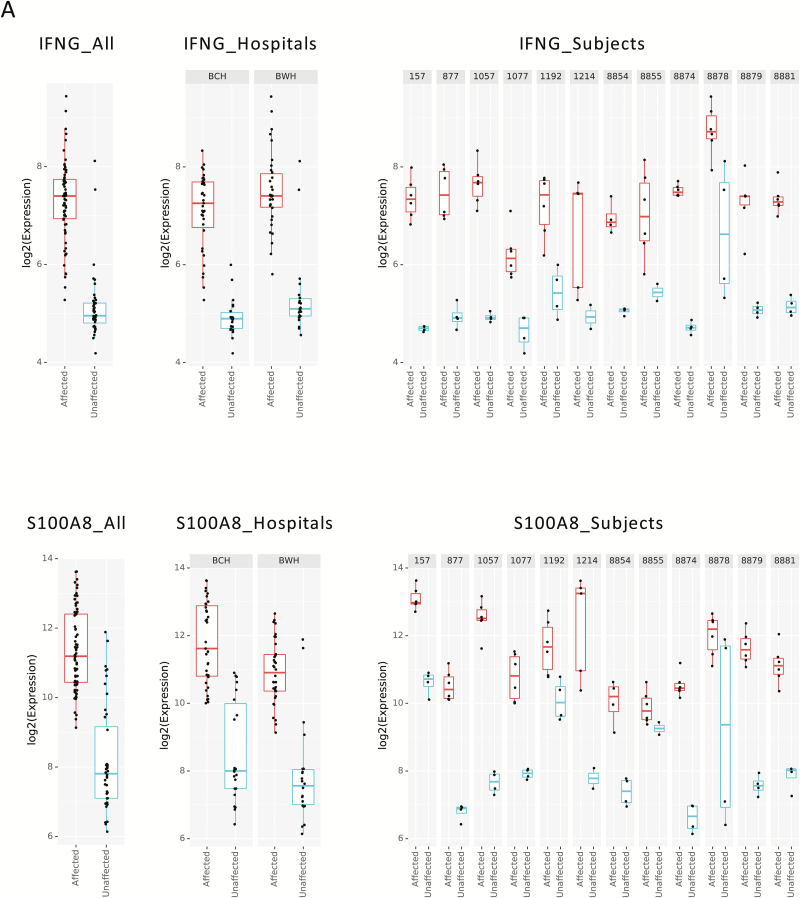
When visualizing the differences in single genes, the differences between affected and unaffected biopsies replicated across the cohort as a whole, between the 2 hospital collections sites, and even down to the level of each patient’s individual biopsy samples. Two examples of genes important in UC pathogenesis already established as upregulated UC in prior transcriptomic studies12, 18 include IFNG, a type 2 interferon important for response to viral and microbial infections, and S100A8, related to antimicrobial peptides. Differential expression patterns of IFNG and S100A8 are therefore presented as illustrative examples of the very clear and reproducible patterns observed in our data set (Fig. 2).
FIGURE 2.
Illustrative examples of gene expression. A, IFNG transcriptional level reflected in combined affected vs unaffected samples in both hospitals combined (left panel), center-specific data (middle panel), and among individual subjects (right panel), reflecting consistency in the data. B, S100A8 transcriptional level reflected in combined data from affected vs unaffected samples in both hospitals combined (left panel), center-specific data (middle panel), and among individual subjects (right panel), reflecting consistency in the data.
We further investigated the top 20 upregulated and downregulated genes in affected tissue as compared to unaffected tissues, expressed in our combined data set. Tables 2 and 3 reflect the top 20 up- and downregulated genes, respectively, with corrected P value (BH) <0.05 and fold-change ≥±2, after removing all duplicate probes and sorting by most significant fold-change. Of these 40 top differentially regulated genes, 36 have been reported in at least one other UC gene expression study (ether between affected and unaffected tissues or between UC and healthy controls) as being significantly up/downregulated in the same direction as we report.11, 12, 15–17 Of the 4 remaining, partial matches of the gene probes for SAA1:SAA2:SAA2-SAA4, LOC100509620:AQP7P1:AQP7, and CELA3B:CELA3A were similarly reported as being significantly up/down regulated in these studies, whereas MS4A10 was not reported in them. This confirms the quality and similarity of our data with prior work.
Table 2:
Top 20 Upregulated Genes in Affected Biopsies of Patients With UC
| Gene | P (BH-Q) | Log2 Fold-Change in Affected vs Unaffected |
|---|---|---|
| SLC6A14 | 1.69E-21 | 5.04 |
| REG3A | 7.09E-12 | 4.90 |
| SAA1; SAA2; SAA2-SAA4 | 2.01E-16 | 4.87 |
| MMP7 | 1.06E-24 | 4.84 |
| DEFB4B; DEFB4A | 1.01E-15 | 4.83 |
| MMP3 | 3.43E-13 | 4.79 |
| DEFA6 | 3.20E-09 | 4.58 |
| CHI3L1 | 3.70E-17 | 4.53 |
| REG1A | 5.19E-14 | 4.39 |
| MMP10 | 1.23E-13 | 4.30 |
| TNIP3 | 3.22E-15 | 4.30 |
| VNN1 | 5.31E-18 | 4.21 |
| DUOXA2 | 6.66E-20 | 4.18 |
| REG1B | 1.03E-10 | 4.15 |
| DUOX2 | 4.24E-15 | 4.10 |
| DEFA5 | 2.49E-08 | 4.08 |
| IL8 | 1.64E-15 | 3.87 |
| CXCL1 | 9.18E-19 | 3.80 |
| CXCL5 | 1.27E-09 | 3.75 |
| CXCL6 | 8.66E-18 | 3.73 |
Abbreviation: BH-Q, Bonferroni-Hochberg corrected P value.
Table 3:
Top 20 Downregulated Genes in Affected Biopsies of Patients With UC
| Gene | P (BH-Q) | Log2 Fold-Change in Affected vs Unaffected |
|---|---|---|
| AQ8 | 1.21E-19 | –4.83 |
| SLC38A4 | 4.51E-16 | –4.27 |
| SLC51A | 5.54E-15 | –4.01 |
| PITX2 | 2.41E-05 | –3.73 |
| MS4A10 | 6.41E-14 | –3.49 |
| SLC26A2 | 8.49E-15 | –3.48 |
| ABCG2 | 1.49E-22 | –3.45 |
| BMP3 | 3.32E-14 | –3.39 |
| MEP1B | 1.26E-11 | –3.22 |
| HSD3B2 | 2.36E-04 | –3.17 |
| LOC100509620; AQP7P1; AQP7 | 3.38E-17 | –3.16 |
| PCK1 | 2.07E-13 | –3.14 |
| CYP2B7P1 | 2.52E-08 | –3.01 |
| HSPB3 | 7.96E-10 | –2.95 |
| CELA3B; CELA3A | 2.27.E-11 | –2.87 |
| PDE6A | 2.24E-14 | –2.87 |
| SLC3A1 | 2.53E-12 | –2.786 |
| ANPEP | 2.63E-15 | –2.82 |
| OTOP2 | 2.87E-18 | –2.81 |
| G6PC | 2.00E-08 | –2.80 |
Abbreviation: BH-Q, Bonferroni-Hochberg corrected P value.
There were no discrepant data between top genes found to be up- or downregulated between the 2 hospital collection sites (Supplementary Tables 6 and 7). Although the overall concordance was high, there were a small number of site-specific differentially expressed genes (Supplementary Figs. 5 and 6). Given the strong overlap between the data collected from both hospital sites, we present a combined data set of all significant genes comparing affected with unaffected biopsies among all patients with UC (Supplementary Table 2), and further analysis was performed on this joined data set. Please see Supplementary Tables 3 (BCH) and 4 (BWH) for hospital site-specific differential analysis of all significantly differentially expressed genes.
Pathway and Gene Ontology Analysis of Microarray Results
Ingenuity pathway analysis was used to explore gene ontology in the combined gene expression data set. The top 10 highly enriched canonical pathways, diseases, and functions and the predicted upstream regulators are shown in Tables 4–6. In keeping with the overall similarities observed in gene expression between the BCH, BWH, and combined data sets, IPA comparison analysis confirmed that the GO enrichment profiles were similar across these 3 data sets (Supplementary Tables 8–10).
Table 4:
IPA Analysis: Top Enriched Canonical Pathways
| Pathway | P | Z |
|---|---|---|
| Dendritic cell maturation | 1.58 × 10-13 | 4.84 |
| Acute phase response signaling | 4.79 × 10-10 | 2.56 |
| TREM1 signaling | 2.04 × 10-9 | 4.58 |
| Th2 pathway | 8.71 × 10-8 | 2.83 |
| Th1 pathway | 4.90 × 10-7 | 2.99 |
| Role of IL-17F in allergic inflammatory airway diseases | 1.23 × 10-6 | 2.89 |
| Role of NFAT in regulation of the immune response | 2.29 × 10-6 | 3.78 |
| CD28 signaling in T-helper cells | 1.23 × 10-5 | 2.18 |
| iCOS-iCOSL signaling in T-helper cells | 1.29 × 10-5 | 3.13 |
| HMGB1 signaling | 1.55 × 10-5 | 2.84 |
Interrogating top canonical pathways within the differentially regulated gene list from the combined data set reveals dendritic cell maturation and acute phase response signaling as the top most enriched pathways (Table 4). These highlight the strong pro-inflammatory signal observed in active UC.
The top diseases and functions enriched in the combined data set were cell movement of blood cells and leukocyte migration (Table 5). Similarly to the top pathways observed, this highlights the importance of immune cell movement and trafficking within affected tissue of patients with UC.
Table 5:
IPA Analysis: Top Enriched Diseases and Functions
| Diseases and Functions | P | Activation Z | Predicted Activation State |
|---|---|---|---|
| Cell movement of blood cells | 5.35 × 10-71 | 5.56 | Increased |
| Leukocyte migration | 4.79 × 10-70 | 5.74 | Increased |
| Cancer | 3.13 × 10-68 | 4.29 | Increased |
| Malignant solid tumor | 4.53 × 10-68 | 3.36 | Increased |
| Cell movement of leukocytes | 1.13 × 10-65 | 5.59 | Increased |
| Cell movement | 1.23 × 10-65 | 6.09 | Increased |
| Nonmelanoma solid tumor | 2.83 × 10-65 | 3.82 | Increased |
| Migration of cells | 4.02 × 10-65 | 6.30 | Increased |
| Abdominal neoplasm | 5.69 × 10-60 | 2.68 | Increased |
| Activation of cells | 1.72 × 10-57 | 5.67 | Increased |
A large number of predicted upstream regulators were identified in the combined data set, including lipopolysaccharide (LPS) and TNF, as depicted in Table 6. Both LPS and TNF were predicted to be activated in affected biopsies, in addition to many other potentially interesting regulatory molecules most likely reflecting the chronic inflammatory state of affected intestinal tissues in patients with UC. Of note, dexamethasone-dependent signaling is predicted to be inhibited. Additionally, although it did not meet our z score cutoff for activity, the anti-inflammatory cytokine IL-10, which is believed to play an important role in immune homeostasis, was the fourth most significant upstream regulator by P value and trended toward inhibition of activity in the affected UC biopsies (data not shown).
Table 6:
IPA Analysis: Top Enriched Upstream Regulators
| Upstream Regulator | P of Overlap | Activation Z | Predicted Activation State |
|---|---|---|---|
| Lipopolysaccharide | 1.73 × 10-76 | 11.80 | Activation |
| TNF | 4.03 × 10-69 | 10.53 | Activation |
| TGFB1 | 1.36 × 10-49 | 4.61 | Activation |
| IL1B | 5.68 × 10-46 | 10.31 | Activation |
| IL13 | 7.74 × 10-46 | 2.32 | Activation |
| IL4 | 1.26 × 10-44 | 3.11 | Activation |
| IFNG | 7.27 × 10-44 | 8.76 | Activation |
| IL6 | 1.61 × 10-43 | 7.36 | Activation |
| STAT3 | 2.15 × 10-42 | 6.41 | Activation |
| Dexamethasone | 4.44 × 10-42 | –3.18 | Inhibition |
Modeling the Number of Biopsies Required for Reliable Transcriptomic Analysis
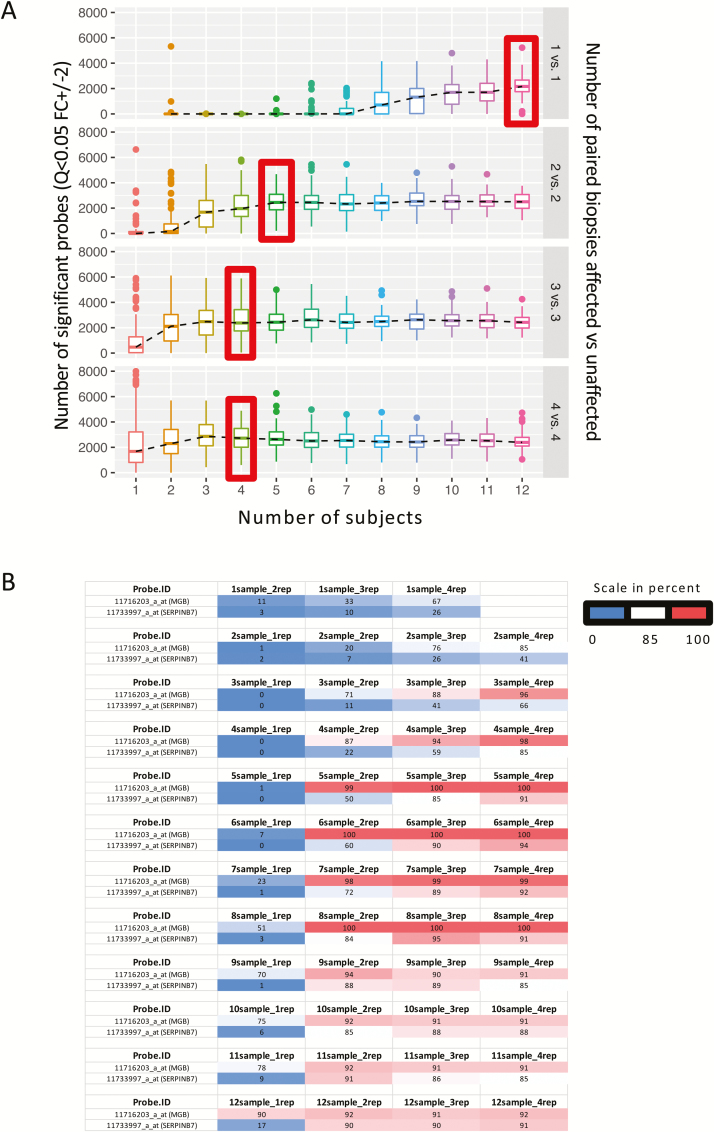
Given the nature of our biopsy collections and the importance of reliable transcriptomic data in clinical studies, we were able to utilize the full microarray data set to test the reliability of observing differential gene expression changes in relationship to the number of subjects and the number of biopsies per subject (Fig. 3A). Increasing either subject numbers or the number of biopsies from a given subject improved the power to detect differential expression between affected and unaffected biopsies in patients with UC. Two representative genes shown in Fig. 3B show the value of both increased sample subject number and increased replicate biopsies from within a subject. MGB is significantly regulated in single biopsies in the majority of simulations when including at least 8 sample subjects (51%), and as few as 2 sample subjects when triplicate biopsies were utilized (76%). Conversely, SERPINB7 was not significantly regulated in the majority of simulations from a single sample subject all the way up to 12 subjects (17%) but was called significantly when at least 2 biopsies were utilized from 6 sample subjects (60%).
FIGURE 3.
Modeling of multiple biopsies. A, Graph depicting that with increased number of subjects (x axis) or increased number of paired biopsies (1 in affected vs 1 in unaffected, as compared with 2 in each group, 3 in each group, or 4 in each group, right y axis), the number of significant probes increases (left y axis). B, Table exemplifying the chance (in percent) for 2 illustrative genes (MGB and SERPINB7) to be called as significantly different between affected and unaffected biopsies when increasing either the number of subjects (referred to as samples) or number of biopsies (replicates referred to as rep).
Additional Transcriptional Profiling by mRNAseq
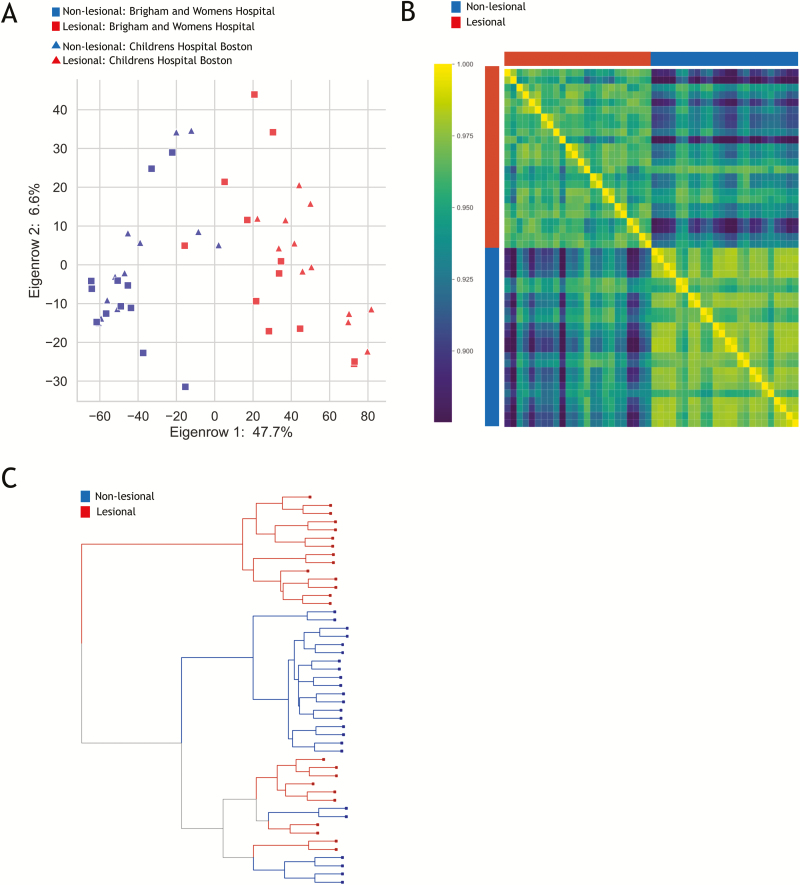
Four biopsies (2 affected and 2 unaffected with clear separation by microarray analysis) from each UC subject (except BWH subject 1120, who did not have matched unaffected colon) were selected for additional interrogation by RNA sequencing. As expected, the gene expression profiles obtained by mRNAseq replicated the results of the microarrays with clear separation by disease status (affected vs unaffected biopsies), as illustrated by PCA (Fig. 4A), correlation heatmap (Fig. 4B), and hierarchical clustering (Fig. 4C). Differential analysis of RNAseq revealed 1557 genes with Q <0.05 and FC ≥±2, and the top 20 differentially up- and downregulated genes by fold-change are presented in Tables 7 and 8.
FIGURE 4.
RNAseq of selected biopsies. A, PCA plot of gene expression profiles of comparing areas of macroscopically active inflammation (red) with those collected from macroscopically noninflamed tissue (blue). Samples collected at BWH are in squares, whereas those collected at BCH are depicted as triangles. Note that there is no strong difference between biopsies taken at different centers. B, Heatmap reflecting high correlation among the set of affected biopsies and within the set of unaffected biopsies. C, Hierarchical clustering also illustrating clear distinction between majority of affected and unaffected biopsies.
Table 7:
Top 20 Upregulated Genes RNAseq
| Gene | P (BH-Q) | Log2 Fold-Change in Affected vs Unaffected |
|---|---|---|
| IGHG3 | 2.19E-24 | 5.625495 |
| DUOXA2 | 1.48E-15 | 5.561099 |
| PI3 | 2.05E-16 | 5.558909 |
| IGHG1 | 7.52E-29 | 5.490249 |
| DUOX2 | 3.30E-21 | 5.457258 |
| REG1A | 2.72E-14 | 5.357473 |
| CXCL1 | 9.13E-25 | 4.995123 |
| DEFA5 | 6.39E-10 | 4.828204 |
| REG3A | 7.23E-14 | 4.642116 |
| DEFA6 | 2.93E-12 | 4.545738 |
| CHI3L1 | 6.49E-29 | 4.526236 |
| LCN2 | 1.89E-21 | 4.438648 |
| IGHG4 | 6.96E-23 | 4.357734 |
| IGHG2 | 5.75E-29 | 4.25801 |
| MMP3 | 2.41E-12 | 4.232053 |
| SLC6A14 | 5.84E-10 | 4.048162 |
| NOS2 | 2.74E-27 | 3.948762 |
| IGHGP | 6.33E-31 | 3.824646 |
| SAA1 | 1.63E-20 | 3.805473 |
| S100A9 | 1.37E-24 | 3.673356 |
Abbreviation: BH-Q, Bonferroni-Hochberg corrected P value.
Table 8:
Top 20 Downregulated Genes RNAseq
| Gene | P (BH-Q) | Log2 Fold-Change in Affected vs Unaffected |
|---|---|---|
| AQ8 | 1.09E-16 | –7.00592 |
| HMGCS2 | 1.48E-24 | –4.97338 |
| SLC26A2 | 7.70E-27 | –4.21223 |
| CA1 | 3.10E-20 | –4.20152 |
| ANPEP | 6.52E-20 | –3.60182 |
| GUCA2A | 4.68E-23 | –3.47234 |
| ABCG2 | 3.17E-30 | –3.28788 |
| TMIGD1 | 1.17E-17 | –3.28017 |
| PCK1 | 1.97E-17 | –3.21961 |
| PRAP1 | 5.06E-20 | –3.20506 |
| CHP2 | 3.65E-24 | –3.17849 |
| GUCA2B | 4.57E-20 | –3.12947 |
| RP11-396O20.2 | 7.73E-24 | –3.11336 |
| OTOP2 | 1.75E-13 | –3.03967 |
| PADI2 | 1.59E-26 | –2.93305 |
| SLC51A | 5.94E-16 | –2.91663 |
| UGT2A3 | 2.65E-08 | –2.90774 |
| ADH1C | 9.97E-20 | –2.8775 |
| FABP1 | 2.38E-31 | –2.83198 |
| SLC38A4 | 1.57E-28 | –2.81218 |
Abbreviation: BH-Q, Bonferroni-Hochberg corrected P value.
Comparison of mRNAseq and Microarray Results
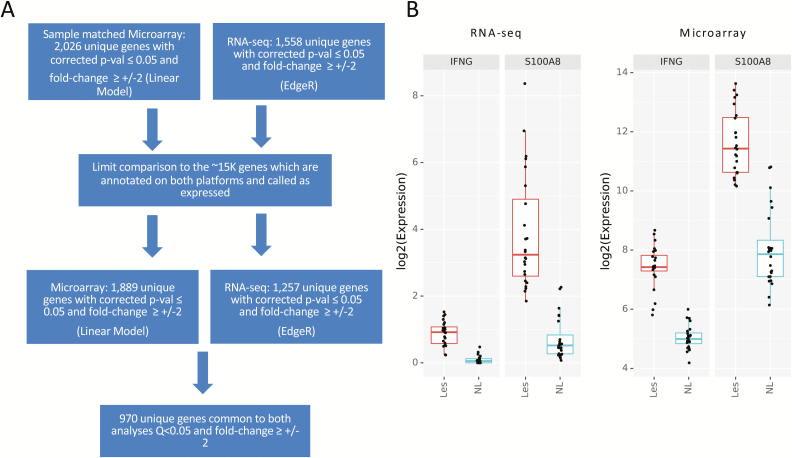
Approximately 16K gene annotations were expressed in both data types and were used for comparison analyses. Differential analysis for the microarrays were re-run, limiting to the matching samples used in mRNAseq (Fig. 5A). Analysis of genes annotated and called as expressed by both platforms with Q <0.05 and an FC cutoff of ≥±2 revealed 970 differentially expressed genes in common between these analyses. The directionality changes observed in this common gene set were concordant, and the corrected P values of the top upregulated and downregulated genes were highly significant by both methods. Visualization of the example genes IFNG and S100A8 shows the same differential gene expression patterns and relative gene abundance between affected and unaffected biopsies (Fig. 5B).
FIGURE 5.
RNAseq compared with matched microarray. A, Flow chart depicting differential analysis for microarray limited to both the matching samples in mRNAseq and matching genes called as expressed, identifying a total of 970 differentially expressed genes in common between the platforms. B, Illustrative example of the differential gene expression of 2 genes, IFNG and S100A8, using RNAseq (left) and microarry (right) among affected (Les) and unaffected (NL) tissue.
DISCUSSION
Several groups have studied the gene expression of colonic biopsies from patients with UC. However, limitations to date include numerous confounding factors that affect results of such transcriptional analysis, highlighted by the incongruences in conclusions reached between different groups. We sought to address this variability in transcriptomic analyses from biopsies of patients with UC by collecting multiple biopsies of both macroscopically inflamed and uninflamed colon. This work confirms much of the current knowledge of the pathogenesis of UC and supports underlying gene expression patterns already identified in the diseased tissue. Moreover, we identify additional novel genes that are significantly upregulated and downregulated that have not been reported by other groups. More importantly, we provide a robust model enabling researchers to reliably identify the number of biopsies and subjects required to guide reproducible expression profiling for future clinical studies.
In congruence with other studies, the majority of our top differentially regulated genes 1475/1564 (94%) have been identified as significant in previous UC studies.12, 15, 18 Consistent with results from Granlund et al., we found that expression of IL23A was significantly increased in affected samples as compared with unaffected ones, and this was observed among samples collected either at BCH or BWH. Similarly, we were also able to confirm that genes related to antimicrobial peptides such as DEFA5, DEFA6, DEFB1, DEFB4A, LYZ, and GNLY, alongside SAA1, DEFA5, DEFA6, S100A8, S100A9, MMP3, MMP7, IL8, and TNIP3, were all differentially regulated in the expected direction.12, 18 Genes in the IBD2 locus12 and cell adhesion molecules, chemokines, and chemokine receptors previously reported10, 11, 14, 16, 18 were all significantly different between affected and unaffected tissues. One limitation to our study is that we did not have the capability to routinely collect additional biopsies from each site for clinical confirmation of pathologic inflammation. Indeed, in the 4 patients with pancolitis, the samples collected at similar sites for clinical histology did reveal microscopic colitis in the macroscopically labeled uninflamed areas in 3 subjects, and no histologic inflammation in the fourth subject. This could have had some implicatons in the differentially expressed genes identified. It is also important to consider the effect that medications can have on the genetic profiling of patients, as previously shown by Arijs et al.14 Although medication use may have an effect on our results, and microscopic inflammation in biopsies labeled as macroscopically uninflammed, we consistently found that the greatest indicator of differences of up/downregulated genes was presence of macroscopic inflammation; thus, we presume that many of the conclusions described, which are also in congruence with other papers, hold true.
As may be expected, although novel identification of up- and downregulated genes did not appear within our top 20 gene lists, our robust collection of multiple biopsies within macroscopically affected and unaffected sites revealed a novel set of 89 genes with FC ≥2 and Q <0.05 that had not been previously identified in other large studies to date (Supplementary Table 11). Interestingly only 49 of these genes are associated with known gene ontology categories, 3 of which have known genetic hits for IBD (FAM92B, IDO2, and TNFRSF1A). Another novel gene we identified as differentially expressed, LYPD8, prevents invasion of flagellated microbiota in murine colonic epithelium and has been recently associated with IBD as LYPD8-deficient mice had exacerbated colitis in a dextran sulfate sodium colitis model.24 Additionally, a new gene we identified as having increased expression in inflamed intestinal biopsies of patients with UC is thrombopoietin (TPO), previously shown to be elevated in serum of patients with IBD.25 An example of a novel downregulated gene is FAM213A, which serves to maintain bone mass. This may be related to and inform the pathogenesis of metabolic bone disease in both pediatric and adult patients with IBD.26 Interestingly, many of the novel genes are expressed in infiltrating immune cells and/or colonic epithelia and are in pathways of clear relevance to IBD pathogenesis (eg, VIP, CD300E, CLDN14, CRTAM, and GRPR).
This paper also reveals strong similarity between data sets from 2 different hospital sites, BCH and BWH, reflecting similar expression profiling among older and younger patients with UC. As the age range of patients recruited at the pediatric center (BCH) did not include subjects younger than 10 years of age, this data set would not be expected to identify gene expression differences potentially unique to patients with very early-onset IBD.27 We appreciate that the severity of disease at the time of biopsy collection was minimal in our adult population, and their SCCAI scores were of smaller magnitude than the PUCAI scores of our pediatric population, which could have affected the number and magnitude of significantly differential genes captured in each of these populations. Additionally, we found that the highly significant differences observed comparing affected with unaffected biopsies were found across our samples regardless of which anatomic segment of the colon they came from, in line with previous work showing loss of segment-specific gene expression in active UC.12 Indeed, we found that differential expression was mainly influenced by macroscopic evidence of disease activity and less so by anatomic location of the samples; for example, 1448 genes of the original 1563 (~93%) remained significant when differential analysis was limited to only the left-sided samples.
Our IPA analyses are congruent with what is already known about pathways of importance in UC, including targets already approved or being actively investigated for therapeutic intervention. Specifically, the important role of leukocyte and cellular migration overall supports the robust development of antitrafficking compounds for IBD. Similarly, appreciating the importance of TNF as a main upstream regulator driving inflammation is in line with use of various anti-TNF antibodies for the management of UC.
This paper also provides the first guide for systematically and reliably identifying the numbers of biopsies and subjects to enable dependable expression profiling to guide future work in IBD in randomized controlled clinical studies. Depending on the projected size of the patient arms in a particular study, this model can be used as a baseline to help guide the minimum number of biopsies needed to be collected for gene expression analysis for adequate power. For example, our model predicts that in a small phase 1b study where subject number may be limited per arm to as few as 4–5 subjects, taking at least 2 biopsies per region of the colon sampled would yield reliable changes in gene expression (fold-change ≥ ±2) for 2000 probes, whereas in larger phase 2 studies where the number of subjects per arm is higher than 12, only single biopsies for gene expression are required to detect the same level of significant differences. It is noted that our model inherently holds wide confidence intervals, and although additional biopsies will always improve one’s discriminatory power, using this model, one can ascertain the appropriate level of sampling to shift additional samples toward other clinical or exploratory end points.
In comparing the Affymetrix microarray results with the Illumina mRNAseq, like many others,28 we observed a high concordance between the differential calls by both methods. Specifically, 12 of the genes were found as top 20 upregulated genes regardless of the method employed, and the majority of the rest were also found upregulated, but to a smaller magnitude of change as compared with the other method. Moreover, only 3 were not represented in both methods, and 2 of these are explained by the probes for these genes not being included in the microarray set employed in this study (IGHG4 and IGHGP). We feel that either method may be employed in future studies, depending on the site’s ability to perform transcriptional profiling in a reliable and consistent manner.
In summary, in this manuscript, we report an extension of previous studies focused on understanding the transcriptional profile of biopsies from subjects with ulcerative colitis utilizing microarray technology, in an effort to continue to elucidate the key genes and pathways dysregulated in the active inflammatory state. We identified both known and novel differentially regulated genes, and we were able to show concordance in data collected from 2 different clinical sites. We also re-profiled a subset of the samples by a second newer methodology, mRNAseq, and showed that the majority of genes defined by both methods are reliably called significant by the other, yet each also provides additional unique information of potential value. Additionally, and perhaps most importantly, through the collection of multiple biological samples from each subject in both an affected segment and an unaffected segment, we have provided a model for determining the appropriate sample size for small-scale clinical studies to generate reliable transcriptomic data.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients and families who contributed to this work. This work was performed jointly with Pfizer Worldwide Research & Development.
Conflicts of interest: None declared.
Supported by: This work was supported in part by a grant from Pfizer Worldwide Research & Development.
Author contributions: Study concept: S.B.S. and H.L.W. Study design: J.D.O., J.B.C., W.G., J.R.K., K.H.C., and S.B.S. Data analysis and interpretation: J.B.C., J.D.O., W.G., K.H.C., J.R.K., and S.B.S. Manuscript drafting and critical revision for important intellectual content: J.D.O., J.B.C., W.G., J.R.K., and S.B.S. All authors contributed to the critical revision of the manuscript and have read and approved the final version of this manuscript. J.D.O., J.B.C., and W.G. contributed equally.
Guarantor of the manuscript: Scott Snapper, MD, PhD.
REFERENCES
- 1. Kappelman MD, Moore KR, Allen JK, et al. . Recent trends in the prevalence of Crohn’s disease and ulcerative colitis in a commercially insured US population. Dig Dis Sci. 2013;58:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. [DOI] [PubMed] [Google Scholar]
- 3. Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cutler DJ, Zwick ME, Okou DT, et al. ; PRO-KIIDS Research Group Dissecting allele architecture of early onset IBD using high-density genotyping. PLoS One. 2015;10:e0128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haberman Y, Tickle TL, Dexheimer PJ, et al. . Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Lange KM, Moutsianas L, Lee JC, et al. . Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dieckgraefe BK, Stenson WF, Korzenik JR, et al. . Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics. 2000;4:1–11. [DOI] [PubMed] [Google Scholar]
- 9. Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445–456. [DOI] [PubMed] [Google Scholar]
- 10. Costello CM, Mah N, Häsler R, et al. . Dissection of the inflammatory bowel disease transcriptome using genome-wide CDNA microarrays. PLoS Med. 2005;2:e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu F, Dassopoulos T, Cope L, et al. . Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. [DOI] [PubMed] [Google Scholar]
- 12. Noble CL, Abbas AR, Cornelius J, et al. . Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut. 2008;57:1398–1405. [DOI] [PubMed] [Google Scholar]
- 13. Olsen J, Gerds TA, Seidelin JB, et al. . Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis. 2009;15:1032–1038. [DOI] [PubMed] [Google Scholar]
- 14. Arijs I, De Hertogh G, Machiels K, et al. . Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol. 2011;106:748–761. [DOI] [PubMed] [Google Scholar]
- 15. Planell N, Lozano JJ, Mora-Buch R, et al. . Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62:967–976. [DOI] [PubMed] [Google Scholar]
- 16. Bjerrum JT, Nielsen OH, Riis LB, et al. . Transcriptional analysis of left-sided colitis, pancolitis, and ulcerative colitis-associated dysplasia. Inflamm Bowel Dis. 2014;20:2340–2352. [DOI] [PubMed] [Google Scholar]
- 17. Bjerrum JT, Hansen M, Olsen J, et al. . Genome-wide gene expression analysis of mucosal colonic biopsies and isolated colonocytes suggests a continuous inflammatory state in the lamina propria of patients with quiescent ulcerative colitis. Inflamm Bowel Dis. 2010;16:999–1007. [DOI] [PubMed] [Google Scholar]
- 18. Granlund Av, Flatberg A, Østvik AE, et al. . Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One. 2013;8:e56818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner D, Otley AR, Mack D, et al. . Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology. 2007;133:423–432. [DOI] [PubMed] [Google Scholar]
- 20. Walmsley RS, Ayres RC, Pounder RE, et al. . A simple clinical colitis activity index. Gut. 1998;43:29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao S, Xi L, Quan J, et al. . Quickrnaseq lifts large-scale RNA-seq data analyses to the next level of automation and interactive visualization. BMC Genomics. 2016;17:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson MD, McCarthy DJ, Smyth GK. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krämer A, Green J, Pollard J Jr, et al. . Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okumura R, Kurakawa T, Nakano T, et al. . Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117–121. [DOI] [PubMed] [Google Scholar]
- 25. Heits F, Stahl M, Ludwig D, et al. . Elevated serum thrombopoietin and interleukin-6 concentrations in thrombocytosis associated with inflammatory bowel disease. J Interferon Cytokine Res. 1999;19:757–760. [DOI] [PubMed] [Google Scholar]
- 26. Bjarnason I, Macpherson A, Mackintosh C, et al. . Reduced bone density in patients with inflammatory bowel disease. Gut. 1997;40:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uhlig HH, Schwerd T, Koletzko S, et al. ; COLORS in IBD Study Group and NEOPICS The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. 2014;147:990–1007.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kogenaru S, Qing Y, Guo Y, et al. . RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics. 2012;13:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.