To the Editor:
Chronic rhinosinusitis (CRS) is a chronic inflammatory disease involving the mucosal tissue of the upper airways, including the nose and paranasal sinuses.(1) Asthma is a related chronic inflammatory disease of the lower airways that is often comorbid with CRS. Uncontrolled upper airway inflammation in the context of CRS is associated with lower airway T-helper-2 mediated inflammation and recalcitrant asthma, however; the underlying mechanism of this link is rather complicated and currently under investigation.(2) Rhinosinusitis is also linked to increased asthma severity and exacerbation rate.(3) Nevertheless, fundamental questions regarding the mechanisms of chronic mucosal inflammation in CRS, and how chronic sinonasal inflammation may affect the lower airways remain unanswered. Considering the likely possibility that microorganisms extant in the upper airways will be aspirated into the lungs, studies of the sinonasal microbiome in the context of lung health appear to be worthwhile. Microbiome studies provide important knowledge about both commensal and pathogenic microbes residing in the airways.(1) Prior studies have shown that CRS patients have significant differences in nasal microbiomes compared to healthy individuals(1), and one study reported further differentiation between CRS patients with or without asthma.(4) In the present study, we investigated whether the composition of the nasal microbiome is associated with asthma control and severity in patients with comorbid CRS.
A previously reported cohort of 111 CRS patients visiting a tertiary care center were included in this prospective study.(5) Diagnosis of CRS was confirmed with at least 12 weeks of rhinosinusitis symptoms and evidence of sinusitis in computer tomography scans based on European position paper on rhinosinusitis (EPOS) 2012 criteria.(6) Exclusion criteria included use of antibiotics within 3 months or undergoing FESS 6 months prior to sampling. Samples for microbiome analysis were collected by rhinologists experienced in nasal sample collection after complete nasal endoscopy; using endoscopy guided small size nasal swabs from the middle meatus. All cases underwent thorough assessment for allergic conditions and asthma by an allergist. Individuals with asthma, confirmed per GINA (Global Initiative for Asthma) criteria, completed an Asthma Control Test (ACT) on the day of nasal swabbing and underwent an office spirometry (performed within 2 months of sample collection). Enrolled asthmatics were interviewed using a questionnaire that captures information about current asthma control, asthma emergency room(ER) visits and hospitalization during the two-year period prior to the study. Additionally, all participating patients were followed up for 12 months for asthma-related events, including ER visits or hospitalizations. Asthmatic CRS patients were grouped based on ACT scores (ACT ≥20, ACT<20) and FEV1 performance which was done in our office (Group 1: FEV1≤ 60% predicted, Group 2: FEV1 61–75% predicted, Group 3: FEV1≥76% predicted). Furthermore, patients were divided using the National Institutes of Health Expert Panel Report 3 (EPR-3) asthma guidelines into intermittent, mild, moderate and severe persistent asthmatic groups. Mild and moderate persistent asthmatics were merged due to their small numbers (7 mild, 9 moderate). Microbiome composition was analyzed using 16S-ribosomal-RNA sequencing of the V4 region as described previously(5) and data was clustered into operational taxonomic units (OTUs) at 97% similarity. Alpha diversity indices were calculated within the software package Primer7. The relative abundance of individual taxa from the taxonomic levels of phylum to genus were compared independently from Alpha indices results and reported using the Kruskal-Wallis non-parametric analysis of variance tests; corrected for false discovery rate and accepted at a significance of (FDR-P˂0.05). This study was approved by Rush University Institutional review board and all participants signed consent forms.
Among 111 CRS cases, 46 (41.4%) had concurrent physician-diagnosed asthma per GINA criteria which included clinical investigations and spirometry measurements. Of note, there was no difference between asthmatics and non-asthmatics in terms of demographic factors, nasal polyps, number of past functional endoscopic sinus surgeries, duration of CRS, sinonasal outcome test score (SNOT-22) and Lund Mackay score (LMS). Asthmatic patients had higher rates of allergic rhinitis(AR) diagnosed based on rhinitis symptoms plus positive skin prick test results to aeroallergens; (60.9% asthmatic vs. 26.2% non-asthmatic, p<0.05).
There were no significant differences in alpha diversity between CRS patients with and without asthma. However, compared to non-asthmatic CRS patients, asthmatic CRS had significantly higher relative abundance (RA) of the Streptococcus genus, with a mean of 1037.2 vs. 318.7; p=0.001 (Figure 1A). Asthmatic CRS also trended towards increased Burkholderia genera abundance, with a mean of 685.8 vs. 285.8; p=0.083. Overall, 8 (17%) asthmatics had at least one ER visit due to asthma exacerbation in the three-year period (two years prior and one year follow-up). Asthma-related ER visits were associated with significant nasal microbiome changes. RA of Proteobacteria phylum was significantly higher in asthmatics with ER visits vs. asthmatics without ER visits (mean±SD of 4287.1±3047.4 vs. 1835.1±2170.8, p=0.02; Figure 1B). Furthermore, Burkholderia within this phylum was significantly increased in asthmatics with at least one ER visit (mean±SD of 685.8±894.4 vs. 319.3±324.2, p=0.02; Figure 1B). Patients in Group 1(FEV1≤ 60%) trended towards increased Burkholderia compared to Group 2 and 3 (those with FEV1 61–75% and FEV1≥76% respectively); with mean±SD of 705.35±701.38, 411.62±402.05 and 234.6±262.5 in Groups 1,2 and 3, respectively, p=0.067; Figure 1C. No significant variation existed in nasal microbiomes associated with NIH EPR-3 categories (Figure 1E) or ACT scores (Figure 1D). There were no significant differences in terms of alpha-diversity indices of nasal bacterial communities associated with asthma ER visits, FEV1 groups or ACT groups. All results remained unchanged after adjusting for age, gender and allergic rhinitis by logistic regression analysis.
FIG 1:
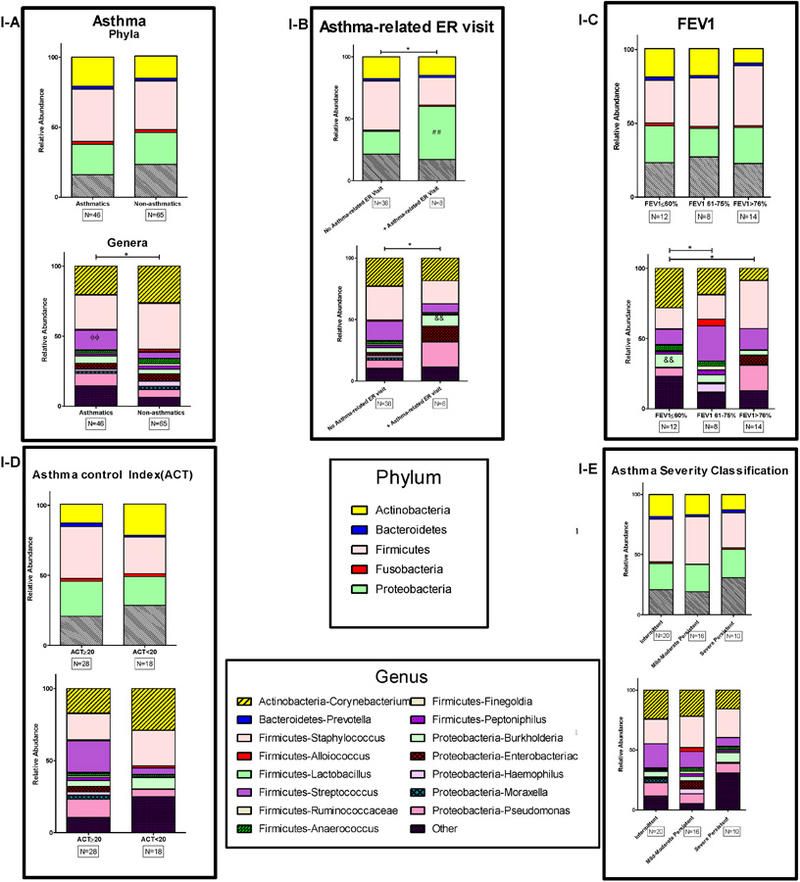
1-I. Nasal microbiome of study groups based on relative abundance of bacteria at phylum and genus levels. The relative abundance of bacterial compositions were compared in asthmatic and non-asthmatic CRS (A), asthma related ER visits (B), Forced Expiratory Volume in 1 Second (FEV1); which was measured in 34 asthmatic subjects (C), Asthma Control Test scores (D), and asthma severity classification based on persistence determined by NIH Expert Panel Report 3 (EPR-3) guidelines (E). * p< 0.05.
We have recently shown that CRS is associated with changes in the nasal microbiome(5). The findings of this current prospective study provide additional evidence that nasal microbial populations vary between patients with both asthma and CRS and those with just CRS alone regardless of the presence of AR. It is tempting to speculate that alterations in upper airway microbial flora in CRS may facilitate the onset or severity of asthma in the lower airways. In agreement with one prior study(4), we found CRS patients with asthma have significantly different nasal microbiota compared to those without asthma. Specifically, the RA of Streptococcus was elevated in asthmatic CRS versus non-asthmatic CRS. Streptococcus is one of the most frequently captured genera in both culture-based and sequencing CRS studies.(1) Importantly, early asymptomatic colonization of upper airways with Streptococcus in infancy is linked to early childhood wheezing(7) and strongly predicts asthma.(7, 8) Our results reinforce the link between this genus and asthma in adult CRS patients, directing future studies to evaluate the underlying mechanism of this observed association.
The increase in Proteobacteria in asthmatics with ER visits during the study reflects prior findings in lower respiratory tracts of asthmatics, in which genera within this phylum are found to be elevated.(9) The asthmatic group also had increasing trend of Burkholderia, a genus within Proteobacteria. Further, the same trend was magnified in the asthmatic CRS group with ER visits (Figure 1B) and trended towards significance in association with low FEV1 (Figure 1C). Interestingly, colonization with the species Burkholderia Cepacia is associated with decreased FEV1 in patients with cystic fibrosis(CF).(10) Other Burkholderia species are notorious for their virulence and can result in significant inflammatory response in lower airways.(11) Sinuses have been shown to act as a reservoir for these organisms, potentially linked to lower airways infection and inflammation.(12) Our data may point out to a link between upper airways colonization of these bacteria and inflammation in lower airways, echoing observations made in bacteria that colonize both upper and lower airways of CF patients.(13) In reverse, initial colonization of lower airways may act as a reservoir and source for upper airways bacteria.
Limitations of this study include absence of healthy controls and subsequently a reference sinonasal microbiome for comparison. As the sampling was performed by passing a swab through nasal passage, despite all efforts and use of very small swabs, there is possibility of contamination of the sample by the nasal vestibular flora. In addition, we have excluded 12 cases in the data point regarding FEV1 measurement, as spirometry measurements were taken over one week apart from sampling due to logistic issues, which, however, did not affect final results.
Whether microbiome changes are a cause or an effect of allergic and inflammatory diseases remains to be studied. Microbiota imbalances could be the initial trigger of immune reaction and inflammation. A dysfunctional immune barrier along with an inflamed mucosal epithelium can promote suitable conditions for certain microorganisms and dysbiosis. In addition, multiple topical and systemic medications that patients with airways diseases are often treated with may affect the local microbiome. The observed trend towards an association of increased Burkholderia RA with asthma severity and poor outcome (increased ER visits) in CRS indicates a possible role for this bacterium and a mechanism by which CRS may affect asthma, calling for further investigations.
Acknowledgments
Funding: This study is partly supported by a research grant from Medtronic. MM is also supported by research grants from Brinson Foundation and Iranian Association of Gastroenterology and Hepatology. RPS is supported in part by the Ernest S. Bazley Foundation, U19 AI106683 from the National Institute of Allergy and Infectious Diseases, and R37 HL068546 from the National Heart, Lung, and Blood Institute. AK is supported by R01 AT007143-05, R01 AA023417-02 and R01 AA020216-05 from NIH.
Footnotes
Conflict of Interest:
The authors declare no conflict of interest for this research.
References:
- 1.Mahdavinia M, Keshavarzian A, Tobin MC, Landay AL, Schleimer RP. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin Exp Allergy 2016;46(1):21–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee TJ, Fu CH, Wang CH, Huang CC, Huang CC, Chang PH, et al. Impact of chronic rhinosinusitis on severe asthma patients. PLoS One 2017;12(2):e0171047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon AE, Kaminsky DA, Holbrook JT, Wise RA, Shade DM, Irvin CG. Allergic rhinitis and sinusitis in asthma: differential effects on symptoms and pulmonary function. Chest 2006;130(2):429–35. [DOI] [PubMed] [Google Scholar]
- 4.Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol 2015;136(2):334–42 e1. [DOI] [PubMed] [Google Scholar]
- 5.Mahdavinia M, Engen PA, LoSavio PS, Naqib A, Khan RJ, Tobin MC, et al. The nasal microbiome in patients with chronic rhinosinusitis: Analyzing the effects of atopy and bacterial functional pathways in 111 patients. J Allergy Clin Immunol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology 2012;50(1):1–12. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas PA, Cooper PJ, Cox MJ, Chico M, Arias C, Moffatt MF, et al. Upper airways microbiota in antibiotic-naive wheezing and healthy infants from the tropics of rural Ecuador. PLoS One 2012;7(10):e46803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015;17(5):704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YJ, Marsland BJ, Bunyavanich S, O’Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol 2017;139(4):1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro J, Rainisio M, Harms HK, Hodson ME, Koch C, Mastella G, et al. Factors associated with poor pulmonary function: cross-sectional analysis of data from the ERCF. European Epidemiologic Registry of Cystic Fibrosis. Eur Respir J 2001;18(2):298–305. [DOI] [PubMed] [Google Scholar]
- 11.David J, Bell RE, Clark GC. Mechanisms of Disease: Host-Pathogen Interactions between Burkholderia Species and Lung Epithelial Cells. Front Cell Infect Microbiol 2015;5:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aanaes K Bacterial sinusitis can be a focus for initial lung colonisation and chronic lung infection in patients with cystic fibrosis. J Cyst Fibros 2013;12 Suppl 2:S1–20. [DOI] [PubMed] [Google Scholar]
- 13.Lucas SK, Yang R, Dunitz JM, Boyer HC, Hunter RC. 16S rRNA gene sequencing reveals site-specific signatures of the upper and lower airways of cystic fibrosis patients. J Cyst Fibros 2018;17(2):204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]


