Abstract
MALAT1 has previously been described as a metastasis-promoting long non-coding RNA (lncRNA). Unexpectedly, we found that targeted inactivation of the Malat1 gene without altering the expression of its adjacent genes in a transgenic mouse model of breast cancer promoted lung metastasis, and importantly, this phenotype was reversed by genetic add-back of Malat1. Similarly, knockout of MALAT1 in human breast cancer cells induced their metastatic ability, which was reversed by Malat1 re-expression. Conversely, overexpression of Malat1 suppressed breast cancer metastasis in transgenic, xenograft, and syngeneic models. Mechanistically, MALAT1 binds and inactivates the pro-metastatic transcription factor TEAD, blocking TEAD from associating with its co-activator YAP and target gene promoters. Moreover, MALAT1 levels inversely correlate with breast cancer progression and metastatic ability. These findings demonstrate that MALAT1 is a metastasis-suppressing lncRNA rather than a metastasis promoter in breast cancer, calling for rectification of the model for a highly abundant and conserved lncRNA.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with no protein-coding capacity1. The nuclear lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1) is among the most conserved lncRNAs and is highly abundant in normal tissues2–4. MALAT1 localizes to nuclear speckles4 and has been shown to modulate alternative pre-mRNA splicing based on in vitro knockdown effects5. However, Malat1 knockout mice showed no phenotypic differences compared with wild-type mice, and genetic ablation of Malat1 did not affect global gene expression, nuclear speckles, splicing factors, or alternative pre-mRNA splicing in mouse tissues2,6,7.
Previous in vitro and xenograft studies demonstrated contradictory effects of MALAT1 on tumor cell growth and invasion8–12. Recently, Spector and colleagues generated mice with deletion of a 3 kb genomic region encompassing the 5′ end of Malat1 and its promoter2. After breeding these mice to a transgenic model of breast cancer, MMTV (mouse mammary tumor virus)-PyMT (polyomavirus middle T antigen)13, they observed a reduction of lung metastases14, but the mechanism underlying this observation remains unclear. Notably, this Malat1 gene deletion model exhibited significant upregulation of Malat1’s adjacent genes, including Neat1, Frmd8, Tigd3, Ehbp1l1, Ltbp3, and to a lesser extent, Map3k11, Kcnk7, Fam89b, Scyl1, Slc25a45, Dpf2, and Cdc42ep22. It is unknown whether their upregulation was due to the loss of Malat1 lncRNA or deletion of regulatory sequences for these neighboring genes.
Questions have been raised as to whether phenotypes resulting from deleting a lncRNA gene can be unequivocally attributed either to the loss of the lncRNA per se or to the loss of overlapping regulatory elements15. A recent study revealed opposite effects from the lncRNA Haunt gene deletion and insertional inactivation, and the Haunt gene deletion effect was attributed to the loss of Haunt genomic DNA16. Moreover, based on multiple examples of different or opposite phenotypes resulting from different strategies for inactivating the same lncRNA in vivo, it has been concluded that genetic rescue experiments from a separate transgene are crucial for separating lncRNA-specific effects from those arising from the manipulation of the underlying genomic DNA15. In addition to gene deletion, MALAT1 has been studied by short-hairpin RNA (shRNA) or small-interfering RNA (siRNA), which is questionable for nuclear lncRNAs, and by antisense oligonucleotides (ASOs) in a few recent studies14,17,18. However, emerging evidence revealed substantial non-specific effects of antisense RNAs and invalidated certain putative anticancer targets19,20. The MALAT1 gene deletion, ASO, or siRNA effect has never been validated to be MALAT1-specific by rescue experiments.
In this study, we observed metastasis induction by Malat1 germline insertional inactivation or somatic knockout without altering Malat1’s adjacent gene expression, and we conducted genetic rescue experiments to demonstrate that this effect was specific to Malat1 lncRNA loss. Moreover, we found that MALAT1 binds and inactivates TEAD and suppresses metastasis in a TEAD-dependent manner. These findings defy the conclusions drawn from previous MALAT1 gene deletion and antisense RNA studies lacking rescue experiments.
Results
Genetic analyses identify Malat1 as a metastasis suppressor
To study the role of Malat1 in breast cancer, instead of using the Malat1 gene deletion model showing upregulation of multiple Malat1’s adjacent genes2, we used a different Malat1 knockout mouse model in which the transcriptional terminator (lacZ and the polyadenylation sequences) was inserted 69 bp downstream of the transcriptional start site of Malat17. This targeted inactivation strategy resulted in loss of Malat1 RNA expression7.
MMTV-PyMT mice recapitulate the tumor stages, pathology, metastasis, and biomarkers of patients with metastatic breast cancer21. In this model, the breast cancer phenotypes of the FVB strain are much more aggressive than those of the C57BL/6 (B6) strain13,22, and thus we used either a B6 or an FVB background (instead of mixed background) in our studies. First, we bred Malat1 knockout mice to MMTV-PyMT mice to generate MMTV-PyMT;Malat1−/− females on a B6 background. Similar to the Spector study14, MMTV-PyMT;Malat1+/+ and MMTV-PyMT;Malat1−/− mice showed no significant difference in overall survival and mammary tumor-free survival (Supplementary Fig. 1a, b). Moreover, the weight of mammary tumors was similar between the two groups (Supplementary Fig. 1c), and histopathological analysis of mammary tissues revealed no substantial differences (Supplementary Fig. 1d). Unlike the Spector study showing that Malat1-deleted PyMT tumors were more differentiated with a dramatically increased cystic phenotype14, we found that Malat1-positive and Malat1-negative PyMT tumors exhibited similar degrees of cystic areas and high-grade carcinoma areas (Supplementary Fig. 1e). Notably, the Malat1 gene deletion model showed significant upregulation of 12 Malat1’s adjacent genes2, whereas the model used in our study had no significant changes in expression levels of these neighboring genes in normal tissues and in mammary tumors (Supplementary Fig. 2a-e).
In both MMTV-PyMT;Malat1+/+ and MMTV-PyMT;Malat1−/− groups, most females reached the endpoint due to primary mammary tumor burdens between 20 and 25 weeks of age. Unexpectedly, MMTV-PyMT;Malat1−/− mice showed an 8.3-fold increase in the number of visible metastatic nodules in the lung, compared with MMTV-PyMT;Malat1+/+ animals (4.9 vs. 40.9 nodules, P = 0.015, Fig. 1a, b). We also assessed metastatic lesions in H&E-stained lung sections (Fig. 1c), revealing that MMTV-PyMT;Malat1−/− mice had a 7.2-fold increase in the number of metastatic foci (10.0 vs. 72.2 foci, P = 0.0001, Fig. 1d) and a 31-fold increase in the percent of lung areas with metastatic lesions (1.1% vs. 34.3%, P < 0.0001, Fig. 1e).
Figure 1. Targeted inactivation and restoration of Malat1 in mice demonstrate that Malat1 is a suppressor of breast cancer lung metastasis.
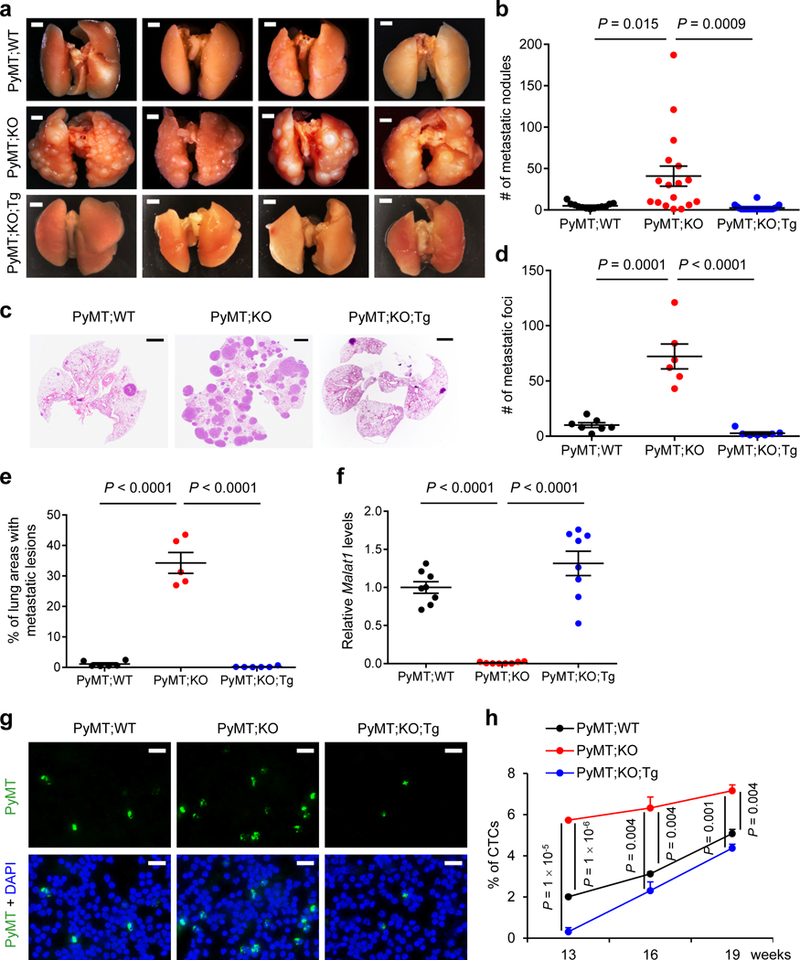
(a, b) Bright-field imaging (a) and the number of metastatic nodules (b) in the lungs of MMTV-PyMT;Malat1+/+ (PyMT;WT, n = 13 mice), MMTV-PyMT;Malat1−/− (PyMT;KO, n = 17 mice), and MMTV-PyMT;Malat1−/−;Malat1Tg (PyMT;KO;Tg, n = 22 mice) mice at the endpoint (20–25 weeks of age). Scale bars in (a), 2 mm. (c-e) H&E staining (c) and the number (d) and relative area (e) of metastatic foci in the lungs of PyMT;WT, PyMT;KO, and PyMT;KO;Tg mice at the endpoint (20–25 weeks of age). n = 7, 6, and 7 mice per group in (d); n = 6, 5, and 6 mice per group in (e). Scale bars in (c), 2 mm. (f) qPCR of Malat1 in the mammary tumors of age-matched PyMT;WT, PyMT;KO, and PyMT;KO;Tg mice. n = 8 mice per group. (g, h) Immunofluorescent staining (g) and the percentages (h) of circulating tumor cells (CTCs) in the peripheral blood from PyMT;WT, PyMT;KO, and PyMT;KO;Tg mice. CTCs from 13-, 16-, and 19-week-old mice were immunostained with a PyMT-specific antibody (green) and nuclei were stained with DAPI (blue). n = 3 mice per group. Scale bars in (g), 20 μm. Statistical significance in (b), (d), (e), (f), and (h) was determined by an unpaired t-test. Error bars are s.e.m. All mice used in this figure are females on a B6 background.
The metastasis-promoting effect of Malat1 inactivation contradicted the Malat1 gene deletion effect14. We sought to address whether the observed phenotype was specific to the loss of Malat1 lncRNA by using a genetic rescue approach. To this end, we generated mice with targeted transgenic expression of Malat1 (Malat1Tg) from the ROSA26 locus (B6 background; Supplementary Fig. 3a). The Malat1Tg mice showed normal development and growth and a 4- to 5-fold increase in Malat1 RNA levels in mammary tissues, compared with the control Malat1LSL mice (LSL: a transcriptionally inactive LoxP-STOP-LoxP allele; Supplementary Fig. 3a, b), whereas Malat1 levels showed no significant difference between Malat1LSL mice and wild-type mice (Supplementary Fig. 3c). We bred Malat1Tg mice to MMTV-PyMT;Malat1−/− mice on a B6 background, which restored Malat1 expression in mammary tumors (Fig. 1f) and reversed lung metastasis (an average of 2.4 metastatic nodules, 2.7 metastatic foci, and 0.2% metastatic area in the MMTV-PyMT;Malat1−/−;Malat1Tg triple mutant females, Fig. 1a-e). Using a PyMT-specific antibody23 to detect circulating tumor cells (CTCs), we found that the percentages of CTCs in MMTV-PyMT;Malat1−/− mice were significantly higher than those in MMTV-PyMT;Malat1+/+ mice; this increase in CTCs was also reversed by genetic add-back of Malat1 (Fig. 1g, h). Taken together, these data suggest that Malat1 suppresses dissemination and lung metastasis of mammary tumor cells.
Since the PyMT tumor and metastasis phenotypes of the FVB strain are stronger than those of the B6 strain13,22, we used the FVB strain to further determine the overexpression effect of Malat1. To this end, we backcrossed Malat1Tg mice and Malat1LSL controls to FVB mice for six generations, bred these mice to MMTV-PyMT mice on an FVB background, and confirmed that MMTV-PyMT;Malat1Tg mice had a 3.2-fold increase in Malat1 levels in their mammary tumors relative to MMTV-PyMT;Malat1LSL mice (Fig. 2a). In both groups, most females became moribund due to primary mammary tumor burdens between 12 and 13 weeks of age, and no significant difference in overall survival (Supplementary Fig. 3d), primary tumor weight (Fig. 2b), or tumor histology (Supplementary Fig. 3e) was found. By gross examination, MMTV-PyMT;Malat1Tgmice had much fewer visible metastatic nodules in the lung than MMTV-PyMT;Malat1LSL animals (P = 0.001, Fig. 2c, d). We validated this observation by H&E staining (Fig. 2e), which revealed a pronounced reduction in lung metastases in MMTV-PyMT;Malat1Tg mice, as gauged by the number of metastatic foci (P = 0.0007, Fig. 2f) and the percent of lung areas with metastatic lesions (P = 0.01, Fig. 2g). Collectively, targeted inactivation, restoration (rescue), and overexpression of Malat1 in genetic models demonstrate that Malat1 is a breast cancer lung metastasis suppressor.
Figure 2. Targeted transgenic overexpression of Malat1 in mice inhibits breast cancer metastasis.
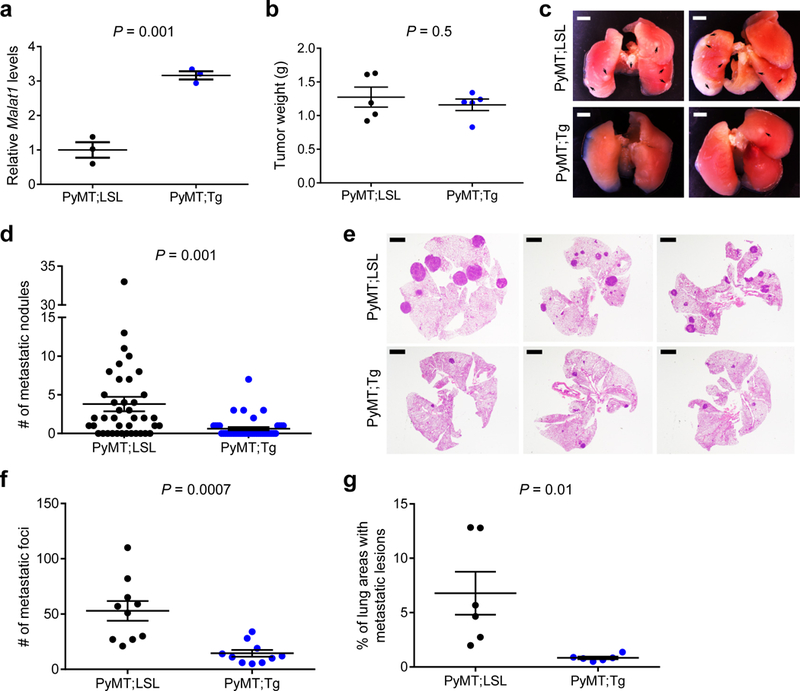
(a) qPCR of Malat1 in the mammary tumors of MMTV-PyMT;Malat1LSL (PyMT;LSL) and MMTV-PyMT;Malat1Tg (PyMT;Tg) mice. n = 3 mice per group. (b) Weight of the mammary tumors of PyMT;LSL and PyMT;Tg mice at 8 weeks of age. n = 5 mice per group. (c, d) Bright-field imaging (c, arrows indicate metastases) and the number of metastatic nodules (d) in the lungs of PyMT;LSL (n = 40 mice) and PyMT;Tg (n = 42 mice) mice at the endpoint (12–13 weeks of age). Scale bars in (c), 2 mm.(e-g) H&E staining (e) and the number (f) and relative area (g) of metastatic foci in the lungs of PyMT;LSL and PyMT;Tg mice at the endpoint (12–13 weeks of age). n = 10 mice per group in (f); n = 6 mice per group in (g). Scale bars in (e), 2 mm.Statistical significance in (a), (b), (d), (f), and (g) was determined by an unpaired t-test. Error bars are s.e.m. All mice used in this figure are females on an FVB background.
Malat1 suppresses metastatic ability of breast cancer cells
To study the relevance of MALAT1 in human breast cancer, we first examined MALAT1 expression levels in a panel of human mammary epithelial or breast cancer cell lines. The non-transformed mammary epithelial cell line MCF10A showed much higher levels of MALAT1 than all 12 breast cancer cell lines examined (Fig. 3a). Moreover, MALAT1 expression was much lower in basal-like, triple-negative breast cancer (TNBC) cells than in less aggressive/metastatic luminal-like breast cancer cells (Fig. 3a), which we further confirmed using the Cancer Cell Line Encyclopedia (CCLE)24 panel (Fig. 3b). Interestingly, a highly lung-metastatic subline of MDA-MB-231 breast cancer cells, named LM225, showed lower MALAT1 expression than the weakly metastatic parental MDA-MB-231 cells (Fig. 3a).
Figure 3. Malat1 inhibits metastatic ability of breast cancer cells.
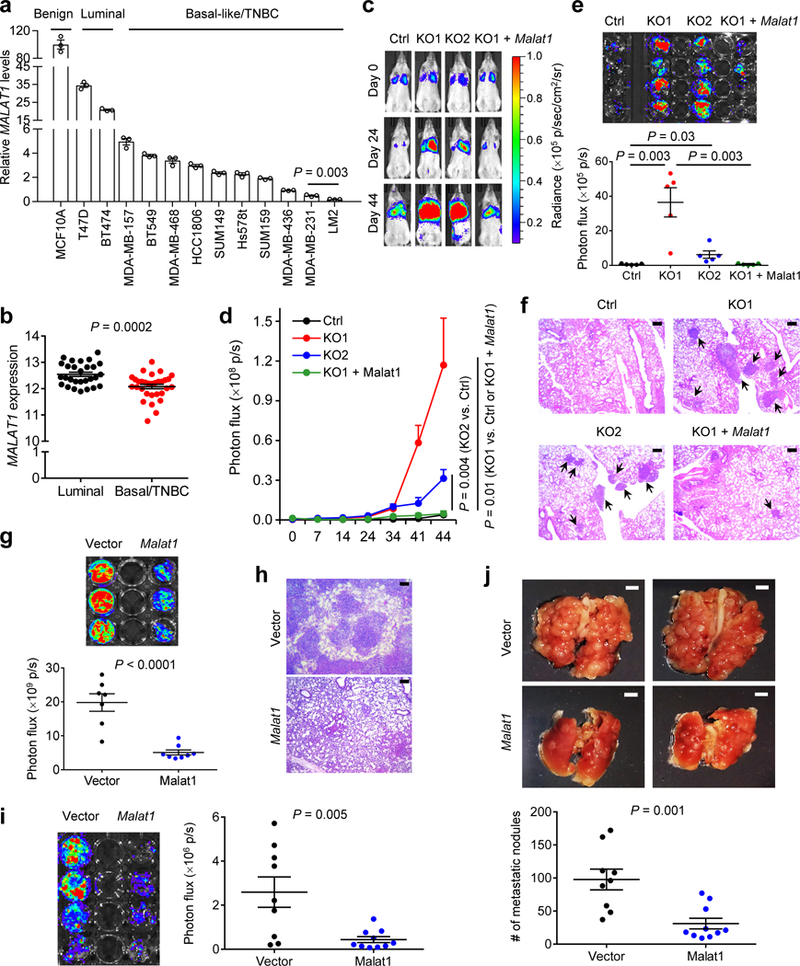
(a) qPCR of MALAT1 in a panel of cell lines. (b) MALAT1 levels in luminal (n = 28) and basal/triple-negative (n = 31) breast cancer cell lines available in CCLE. (c, d) Bioluminescent imaging (c) and quantification of photon flux (d) of NSG mice with intravenous injection of control, MALAT1 knockout, or Malat1-restored MDA-MB-231 cells. Day 0: the day of tumor cell injection. n = 5 mice per group.(e, f) Bioluminescent imaging (e, upper panel), quantification of photon flux (e, lower panel), and H&E staining (f) of the lungs from mice described in Fig. 3c, d. n = 5 mice per group in (e). Scale bars in (f), 200 μm.(g) Bioluminescent imaging (upper panel) and quantification of photon flux (lower panel) of the lungs from NSG mice with intravenous injection of control (n = 7 mice) or Malat1-overexpressing (n = 8 mice) LM2 cells. (h) H&E staining of the lungs described in Fig. 3g. Scale bars, 200 μm. (i) Bioluminescent imaging (left panel) and quantification of photon flux (right panel) of the lungs from BALB/c mice injected with control (n = 9 mice) or Malat1-overexpressing (n = 10 mice) 4T1 cells.(j) Bright-field imaging (upper panel) and the number of metastatic nodules (lower panel) in the lungs of mice described in Fig. 3i. Scale bars, 2 mm. Statistical significance in (a), (b), (d), (e), (g), (i), and (j) was determined by an unpaired t-test. Error bars are s.e.m.
Next, we studied the loss- and gain-of-function effects of MALAT1 on metastatic ability of human breast cancer cells. It is difficult to target a nuclear lncRNA using shRNA or siRNA. Moreover, unlike protein-coding genes, lncRNAs cannot be depleted by single guide RNA (gRNA)-mediated frame-shift mutations. Using a pair of MALAT1 gRNAs and a Double Excision CRISPR Knockout (DECKO) approach26, we deleted ~650 bp in the 5′ end of MALAT1 in luciferase-expressing MDA-MB-231 cells and validated six MALAT1-deficient clones (Supplementary Fig. 4a, b). Whereas loss of MALAT1 did not affect MALAT1’s adjacent gene expression (Supplementary Fig. 4c), cell proliferation (Supplementary Fig. 4d), or anchorage-independent growth (Supplementary Fig. 4e), MALAT1 knockout clones showed higher migratory and invasive ability than the control cells expressing GFP gRNA (Supplementary Fig. 4f), which was reversed by ectopic expression of mouse Malat1 (Supplementary Fig. 4g, h; mouse Malat1 was used, because it is resistant to human MALAT1 gRNAs). Moreover, using time-lapse video microscopy, we observed a significant increase in the speed of movement of MALAT1 knockout cells compared with control cells, which was reversed by Malat1 re-expression (Supplementary Fig. 4i and Supplementary Videos 1–3). To determine the effect of MALAT1 loss on lung metastatic colonization, we injected control (cells expressing GFP gRNA, which had similar metastatic behavior to the parental MDA-MB-231 cells, Supplementary Fig. 4j-l), MALAT1 knockout, and Malat1-restored MDA-MB-231 cells into NSG (non-obese diabetic; severe combined immunodeficiency; interleukin-2 receptor gamma chain null) mice through the tail vein. Bioluminescent imaging of live animals (Fig. 3c, d) and whole lungs (Fig. 3e), as well as H&E staining of lung sections (Fig. 3f), revealed that knockout of MALAT1 in MDA-MB-231 cells strongly promoted lung metastasis in mice, which was fully reversed by restoration of Malat1 expression.
The lung-metastatic LM2 subline exhibited the lowest MALAT1 expression among all 13 cell lines examined (Fig. 3a). Stable transfection of luciferase-labeled LM2 cells with mouse Malat1 reduced cell movement, migration, and invasion (Supplementary Fig. 5a-c and Supplementary Videos 4–5) without affecting cell proliferation (Supplementary Fig. 5d). Similarly, overexpression of Malat1 in HCC1806 and Hs578t human breast cancer cell lines inhibited motility and invasiveness (Supplementary Fig. 5e-g). We then performed tail-vein injection of LM2 cells into NSG mice. Bioluminescent imaging of live animals revealed consistently less lung metastasis in recipients of Malat1-overexpressing LM2 cells (Supplementary Fig. 5h, i). At week 5, mice that received Malat1-overexpressing LM2 cells exhibited a 74% reduction in lung metastases relative to the control group (Fig. 3g), which was confirmed by histopathological analysis (Fig. 3h). Similarly, stable transfection of 4T1 mouse mammary tumor cells with Malat1 (Supplementary Fig. 5j) markedly reduced their colonization of the lungs of syngeneic BALB/c mice, as gauged by live animal imaging (Supplementary Fig. 5k, l), ex vivo lung imaging (Fig. 3i), and the number of visible metastatic nodules (Fig. 3j). These data provide additional in vivo proof that MALAT1 suppresses metastatic ability of human and mouse mammary tumor cells.
We next analyzed the RNA-Seq data from The Cancer Genome Atlas (TCGA)27 and found that MALAT1 was significantly underexpressed in human breast tumors compared with normal mammary tissues (Supplementary Fig. 6a, b). Using an Oncomine data-mining platform, we found that MALAT1 was significantly underexpressed in higher-grade breast tumors (Supplementary Fig. 6c), and that breast cancer metastases had lower MALAT1 expression than primary mammary tumors (Supplementary Fig. 6d). In addition, Kaplan-Meier (KM) plotter28 analysis showed that lower MALAT1 levels correlated with shorter distant metastasis-free survival both in total breast cancers as well as in luminal A and basal subtypes (Supplementary Fig. 6e).
To corroborate the observed correlation, we orthotopically implanted G418-resistant, luciferase-expressing 4T1 cells into syngeneic BALB/c mice, and isolated G418-resistant cells from mammary tumors and lungs. Interestingly, Malat1 levels were significantly lower in metastasized tumor cells than in paired primary tumor cells (Supplementary Fig. 6f). In addition, compared with 4T1 cells, the non-metastatic 67NR cell line and the weakly metastatic 168FARN and 4TO7 cell lines29 showed higher Malat1 expression (Supplementary Fig. 6g). Taken together, these data demonstrate that MALAT1 levels are negatively associated with breast cancer progression and metastasis.
MALAT1 interacts with TEAD family members
To elucidate the mechanism by which MALAT1 regulates metastasis, we attempted to identify its endogenous binding proteins by performing chromatin isolation by RNA purification coupled to mass spectrometry (ChIRP-MS)30. We collected the tumors from MMTV-PyMT mice and pulled down endogenous Malat1 lncRNA using mouse Malat1-specific, biotinylated DNA probes and streptavidin beads. DNA probes for U1 nuclear RNA and probe-free conditions were included as negative controls to validate the specificity of Malat1 pulldown (Fig. 4a). Our ChIRP-MS analysis identified 970 Malat1-interacting proteins, including previously reported Malat1 interactors, such as splicing factors and RNA-binding proteins5,8,9. Most of them, however, interacted with both Malat1 and U1. Therefore, we screened for proteins specifically bound to Malat1 by excluding bound proteins in the two negative controls (U1 and probe-free beads). Only 23 out of 970 proteins met this criterion; among them, the Tead family stood out because all four Tead proteins were identified as Malat1’s binding partners (Supplementary Fig. 7a).
Figure 4. MALAT1 interacts with TEAD family members.
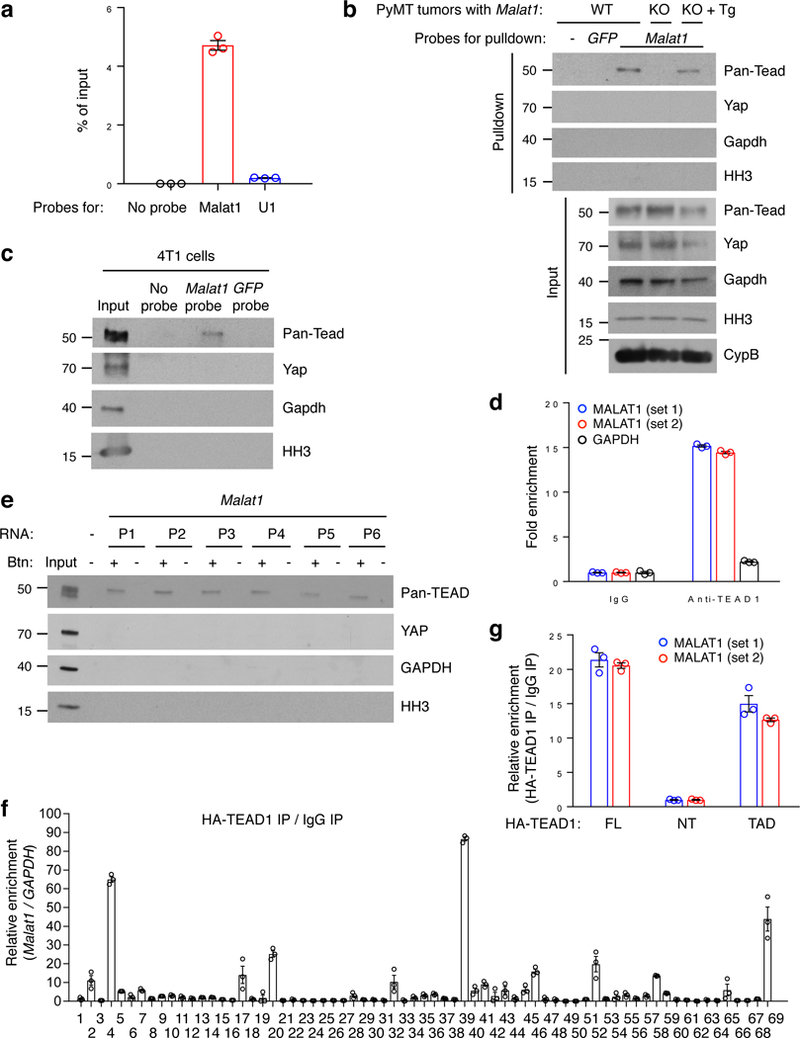
(a) qPCR of Malat1 in ChIRP samples. Probes for mouse Malat1 or U1 nuclear RNA were used to pull down endogenous Malat1 or U1 from PyMT mammary tumor samples. (b, c) Western blot analysis of ChIRP samples. Mouse Malat1-specific probes were used to pull down endogenous Malat1 from the mammary tumors of MMTV-PyMT;Malat1+/+ (WT), MMTV-PyMT;Malat1−/− (KO), and MMTV-PyMT;Malat1−/−;Malat1Tg (KO + Tg) mice (b), or from 4T1 cells (c), followed by immunoblotting with antibodies against pan-Tead, Yap, Gapdh, and histone H3 (HH3).(d) RNA immunoprecipitation assay. Endogenous TEAD1 was immunoprecipitated from crosslinked MDA-MB-231 cells. TEAD1-bound MALAT1 was quantitated by qPCR with two primer sets. GAPDH was used as a negative control. (e) RNA pulldown assay. Unlabeled and biotinylated Malat1 fragments (P1-P6) were synthesized by in vitro transcription, incubated with HEK293FT cell lysate, and pulled down with streptavidin beads. The bound proteins were eluted by boiling in Laemmli sample buffer and immunoblotted with antibodies against pan-TEAD, YAP, GAPDH, and histone H3. Btn: biotinylation. (f) CLIP-qPCR assay of Hela cells overexpressing HA-TEAD1 and mouse Malat1. The protected Malat1 RNA segments bound by TEAD1 were detected by qPCR using 69 pairs of primers. (g) RNA immunoprecipitation assay. Hela cells were transfected with HA-tagged full-length TEAD1 (FL), N-terminal region (NT), or transactivation domain (TAD), crosslinked, and subjected to immunoprecipitation with a HA-specific antibody. TEAD1-bound MALAT1 was quantitated by qPCR with two primer sets. All error bars are s.e.m. Uncropped blots are shown in Supplementary Fig. 10.
Next, we performed Western blot analysis of ChIRP samples, which validated the interaction between endogenous Malat1 and Tead proteins in both PyMT tumors (Fig. 4b) and in 4T1 cells (Fig. 4c). Importantly, the interaction was abolished in Malat1-null PyMT tumors, but was restored in tumors from the MMTV-PyMT;Malat1−/−;Malat1Tg mutants (Fig. 4b), suggesting that this interaction is Malat1 RNA-specific. In both PyMT tumors and 4T1 cells, Malat1 did not interact with the cytoplasmic marker Gapdh, the nuclear marker histone H3, or the Tead coactivator Yap (Fig. 4b, c). To further corroborate our result, we pulled down TEAD1 protein from crosslinked MDA-MB-231, Hela, BT549, or MDA-MB-468 cells and isolated its associated RNAs. RT-qPCR analysis revealed that MALAT1 lncRNA was highly enriched in TEAD1 immunoprecipitates (Fig. 4d and Supplementary Fig. 7b).
To identify the TEAD-binding region(s), we generated six non-overlapping biotinylated Malat1 fragments (P1-P6, 1.1–1.2 kb each) spanning full-length mouse Malat1 by in vitro transcription. All six fragments, but not U1, bound to TEAD proteins (Fig. 4e and Supplementary Fig. 7c, d), suggesting that the TEAD-binding sites may be distributed diffusely on Malat1 lncRNA. In contrast, GAPDH, YAP, and histone H3 did not interact with any region of Malat1 (Fig. 4e), validating the specificity of the MALAT1-TEAD binding. To further map the TEAD-binding sites on Malat1, we performed a UV crosslinking-immunoprecipitation and qPCR (CLIP-qPCR) assay31,32 using 69 pairs of primers with overlapping 200 bp amplicons, which allowed detection of the protected Malat1 RNA segments bound by TEAD1 and the mapping of TEAD1-binding sites on Malat1 at 200 nt intervals (Supplementary Fig. 7e). At a threshold enrichment value of 2, all six fragments (P1-P6) showed multiple peaks; at a threshold enrichment value of 10, each of the six fragments showed at least one major peak and a total of 10 major peaks were detected (Fig. 4f), suggesting that Malat1 contains multiple TEAD-binding sites.
We sought to identify the Malat1-binding domain on TEAD1. TEAD1 consists of two functional regions: the N-terminal region (NT) containing the TEA domain responsible for DNA binding and the C-terminal transactivation domain (TAD) responsible for YAP binding33 (Supplementary Fig. 7f). Accordingly, we generated two TEAD1 truncation mutants (Supplementary Fig. 7f, g) and performed RNA immunoprecipitation. Interestingly, Malat1 was enriched in the immunoprecipitates of full-length TEAD1 or the transactivation domain, but not the N-terminal region (Fig. 4g), suggesting that Malat1 interacts with TEAD1’s transactivation domain, the same domain that mediates the YAP-TEAD1 interaction33.
MALAT1 inhibits the transcriptional activity of TEAD
The TEAD transcription factors and their coactivators YAP and TAZ promote tumor progression and metastasis through the transcriptional activity34. In the nucleus, TEAD proteins interact with YAP or TAZ to activate the expression of target genes, including classical TEAD target genes CTGF, CYR61, ANKRD1, AMOTL2, AJUBA, AXL, and WTIP35–38. We asked whether MALAT1 regulates TEAD’s transcriptional activity. Indeed, ectopic expression of Malat1 reduced, while knockout of MALAT1 increased the activity of a TEAD luciferase reporter containing tandem TEAD-binding sites39 (Fig. 5a, b). On the other hand, fractionation assays and immunofluorescent staining demonstrated that TEAD proteins were localized exclusively in the nucleus of both control and MALAT1 knockout MDA-MB-231 cells (Supplementary Fig. 7h, i), suggesting that MALAT1 does not affect TEADs’ nuclear localization.
Figure 5. MALAT1 inactivates TEAD.
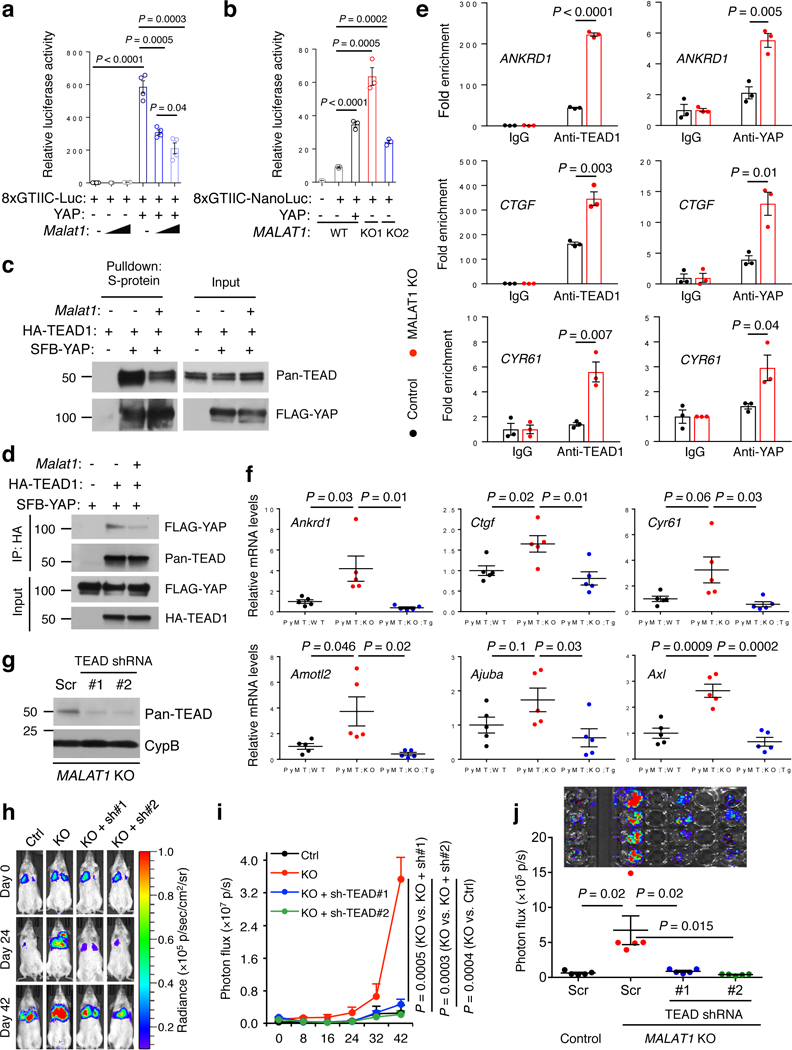
(a) Luciferase activity in HEK293FT cells co-transfected with indicated plasmids. n = 4 cell culture replicates per group. (b) Luciferase activity in control and MALAT1 knockout MDA-MB-231 cells co-transfected with indicated plasmids. n = 3 cell culture replicates per group. (c, d) HEK293FT cells were co-transfected with Malat1, SFB-YAP, and HA-TEAD1, and were subjected to pulldown with S-protein beads (c) or an HA-specific antibody (d), followed by immunoblotting with antibodies against pan-TEAD and FLAG. (e) ChIP-qPCR analysis showing the occupancy of ANKRD1, CTGF, and CYR61 promoters by TEAD1 or YAP immunoprecipitated from control or MALAT1 knockout MDA-MB-231 cells. (f) qPCR of YAP-TEAD target genes in the tumors of MMTV-PyMT;Malat1+/+ (PyMT;WT), MMTV-PyMT;Malat1−/− (PyMT;KO), and MMTV-PyMT;Malat1−/−;Malat1Tg (PyMT;KO;Tg) mice. n = 5 mice per group. (g) Immunoblotting of pan-TEAD and cyclophilin B (CypB) in MALAT1 knockout MDA-MB-231 cells with or without transduction of TEAD shRNA. Scr: scramble control. (h, i) Bioluminescent imaging (h) and photon flux quantification (i) of NSG mice with intravenous injection of control and MALAT1 knockout MDA-MB-231 cells with or without transduction of TEAD shRNA. Day 0: the day of tumor cell injection. n = 5 mice per group. (j) Bioluminescent imaging (upper panel) and photon flux quantification (lower panel) of the lungs from mice described in Fig. 5h, i. n = 5 mice per group. Statistical significance in (a), (b), (e), (f), (i), and (j) was determined by an unpaired t-test. Error bars are s.e.m. Uncropped blots are shown in Supplementary Fig. 10.
Because MALAT1 RNA is highly abundant2, and because the TEAD-binding sites are distributed throughout MALAT1 (Fig. 4e, f), we speculated that MALAT1 may sequester TEAD, thereby blocking TEAD’s ability to bind YAP and/or the target genes. To test this hypothesis, we first performed co-immunoprecipitation of TEAD1 and YAP. Upon Malat1 overexpression, we observed a clear reduction in YAP-TEAD1 interaction (Fig. 5c, d). Next, we analyzed YAP-TEAD target gene promoters by chromatin immunoprecipitation (ChIP) assays. Ectopic expression of Malat1 in LM2 cells significantly decreased the occupancy of three classical target gene (ANKRD1, CTGF, and CYR61) promoters by endogenous TEAD1 or YAP (Supplementary Fig. 8a); conversely, in MALAT1 knockout MDA-MB-231 cells, the occupancy of these three gene promoters by endogenous TEAD1 or YAP was prominently increased (Fig. 5e).
YAP is a transcriptional co-factor lacking the DNA-binding domain, and TEAD proteins mediate YAP’s association with chromatin40. Importantly, MALAT1 does not bind YAP (Fig. 4b, c, e). To further exclude the possibility that MALAT1 directly regulates YAP, we generated GAL4 DNA binding domain (DBD)-fused YAP constructs (i.e., TEAD-independent YAP mutants capable of binding to DNA without TEAD) and gauged their transcriptional activity using a GAL4 DBD-responsive luciferase reporter. When fused to the GAL4 DBD, both full-length YAP and its transactivation domain (TAD) exhibited significant transcriptional activity, which was not altered by overexpression of Malat1 (Supplementary Fig. 8b). This suggests that repression of YAP-TEAD’s transcriptional activity by Malat1 is TEAD-dependent.
We examined whether YAP-TEAD target gene expression is regulated by MALAT1. Indeed, in Malat1-overexpressing LM2 cells, the expression of four of seven classical target genes examined was significantly repressed (Supplementary Fig. 8c). Conversely, these target genes were upregulated in MALAT1 knockout clones of MDA-MB-231 cells (Supplementary Fig. 8d). Importantly, compared with control PyMT mouse mammary tumors, Malat1-deficient PyMT tumors showed an increase in expression levels of these classical YAP-TEAD target genes, which was reversed by genetic add-back of Malat1 (Fig. 5f).
To determine the functional relevance to metastasis, we used shRNAs to knock down multiple TEAD family members41 in MALAT1 knockout MDA-MB-231 cells (Fig. 5g). Notably, depletion of TEAD proteins reversed migration, invasion, and in vivo metastasis (Fig. 5h-j and Supplementary Fig. 8e-g) induced by the loss of MALAT1, with only a marginal inhibitory effect on migration and invasion of control MDA-MB-231 cells (Supplementary Fig. 8f, g), suggesting that the metastasis-promoting effect of MALAT1 depletion is TEAD-dependent. Conversely, overexpression of Malat1 in LM2, BT549, and SUM149 cells decreased migration and invasion, which was reversed by TEAD1 overexpression (Supplementary Fig. 8h, i), suggesting that Malat1 inhibits cell motility and invasiveness through TEAD.
ITGB4 and VEGFA are TEAD target genes regulated by MALAT1
In addition to validating that known TEAD target genes are downregulated by MALAT1, we sought to identify novel MALAT1-regulated genes. To this end, we performed RNA-Seq analysis and identified nine genes that were most significantly upregulated in MMTV-PyMT;Malat1−/− tumors, compared with both MMTV-PyMT;Malat1+/+ tumors and MMTV-PyMT;Malat1−/−;Malat1Tg tumors (Fig. 6a). We also performed metastasis gene-specific qPCR array analysis and identified three genes that were most significantly downregulated in Malat1-overexpressing LM2 cells (Supplementary Table 1). Two of these 12 Malat1-downregulated genes, Itgb4 and Vegfa, are well-established metastasis promoters and were shown to be bound by YAP-TEAD42. In addition, from the paired-end RNA-Seq analysis, only 51 out of 16,034 cassette exons (0.3%) exhibited significant changes in the splicing pattern in Malat1 knockout PyMT tumors compared with Malat1 wild-type PyMT tumors (Supplementary Fig. 9a). Thus, Malat1 has little effect on global pre-mRNA splicing.
Figure 6. ITGB4 and VEGFA are TEAD target genes and are regulated by MALAT1.
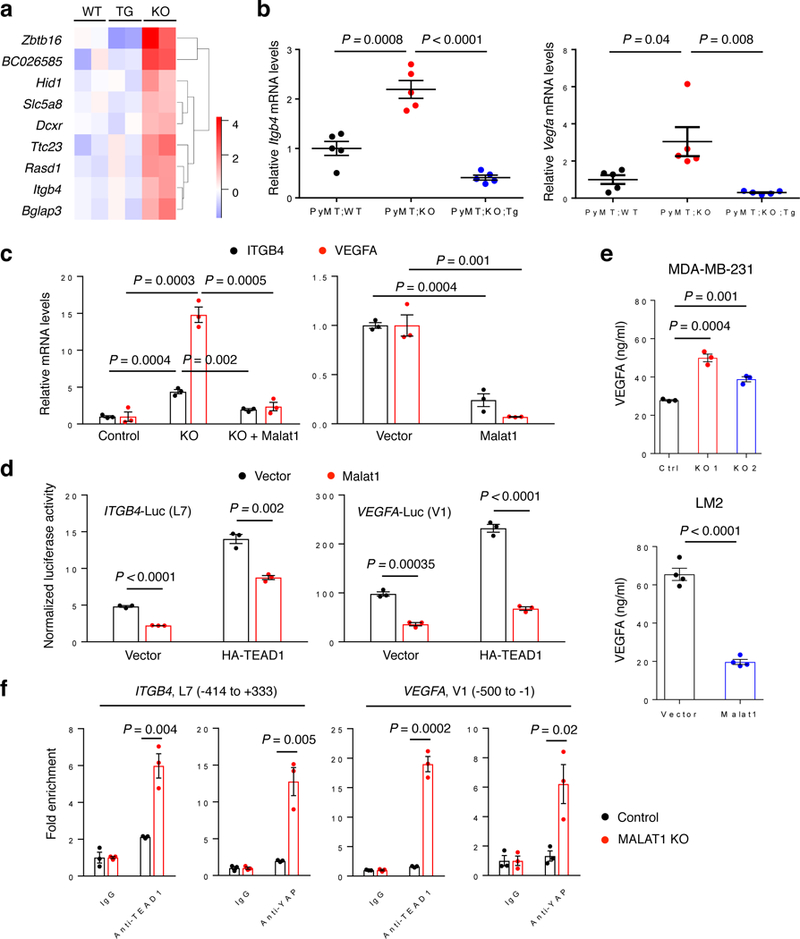
(a) Heat map of nine genes that were identified by RNA-Seq analysis to be commonly upregulated in MMTV-PyMT;Malat1−/− tumors (KO), compared with both MMTV-PyMT;Malat1+/+ tumors (WT) and MMTV-PyMT;Malat1−/−;Malat1Tg tumors (TG). n = 2 mice per group. (b) qPCR of Itgb4 (left panel) and Vegfa (right panel) in the mammary tumors of MMTV-PyMT;Malat1+/+ (PyMT;WT), MMTV-PyMT;Malat1−/− (PyMT;KO), and MMTV-PyMT;Malat1−/−;Malat1Tg (PyMT;KO;Tg) mice. n = 5 mice per group. (c) qPCR of ITGB4 and VEGFA in control, MALAT1 knockout, and Malat1-restored MDA-MB-231 cells (left panel), and in control and Malat1-overexpressing LM2 cells (right panel). (d) Luciferase activity in HEK293FT cells co-transfected with Malat1, HA-TEAD1, an ITGB4 (left panel) or VEGFA (right panel) luciferase reporter, and a Renilla luciferase reporter. n = 3 cell culture replicates per group. (e) ELISA of VEGFA secreted by MALAT1 knockout MDA-MB-231 cells (upper panel, n = 3 cell culture replicates per group) and by Malat1-overexpressing LM2 cells (lower panel, n = 4 cell culture replicates per group).(f) ChIP-qPCR analysis showing the occupancy of ITGB4 (two left panels) and VEGFA (two right panels) promoters by TEAD1 or YAP. Endogenous TEAD1 and YAP were immunoprecipitated from control or MALAT1 knockout MDA-MB-231 cells. Statistical significance in (b) – (f) was determined by an unpaired t-test. Error bars are s.e.m.
ITGB4 encodes integrin β4, which forms a heterodimer with integrin α6 to promote tumor progression and to direct lung-tropic metastasis43–46. VEGFA encodes vascular endothelial growth factor, a promoter of angiogenesis and metastasis47. By RT-qPCR analysis, we confirmed that ITGB4 and VEGFA mRNA levels were significantly upregulated by the loss of Malat1 both in PyMT tumors (Fig. 6b) and in MDA-MB-231 cells (Fig. 6c), while re-expression of Malat1 in MMTV-PyMT;Malat1−/− mice (Fig. 6b) and in MALAT1 knockout MDA-MB-231 cells (Fig. 6c) reversed the induction of ITGB4 and VEGFA expression. Moreover, ectopic expression of Malat1 in LM2 cells reduced ITGB4 and VEGFA levels (Fig. 6c).
We asked whether the expression of ITGB4 and VEGFA is activated by TEAD and whether Malat1 opposes it. By testing a series of upstream regulatory regions of the human ITGB4 or VEGFA gene cloned into a luciferase reporter vector48,49, we identified two regions, named L7 and V1, respectively, as the minimal promoter/enhancer regions of ITGB4 and VEGFA that are responsive to TEAD (Supplementary Fig. 9b-d). Next, using the luciferase construct containing the L7 or V1 region, we found that overexpression of Malat1 suppressed the transcriptional activity of ITGB4 and VEGFA promoters both at the basal level and upon TEAD1 overexpression (Fig. 6d). VEGFA is a secreted protein, and ELISA assays showed that secreted VEGFA was upregulated by MALAT1 depletion in MDA-MB-231 cells and was downregulated by Malat1 overexpression in LM2 cells (Fig. 6e). Furthermore, ChIP assays revealed that knockout of MALAT1 increased (Fig. 6f), while overexpression of Malat1 reduced (Supplementary Fig. 9e) the occupancy of the ITGB4 and VEGFA promoters by TEAD1 and YAP. Taken together, these data demonstrate that ITGB4 and VEGFA are TEAD target genes and are negatively regulated by MALAT1.
VEGFA is known for its function in angiogenesis50. Moreover, tumor cells respond to autocrine and paracrine VEGFA signals through their VEGF receptor tyrosine kinases and neuropilins47,51–54, and autocrine VEGFA signaling stimulates cancer cell migration and invasion47,52,54. Indeed, we found that recombinant human VEGFA165 (the most abundant isoform)53 promoted MDA-MB-231 cell invasion (Supplementary Fig. 9f). Furthermore, knockdown of VEGFA in MALAT1 knockout MDA-MB-231 cells reversed the induction of cell invasiveness (Supplementary Fig. 9g-i). Thus, VEGFA may be a functional YAP-TEAD target that is upregulated by MALAT1 depletion.
Discussion
In both genetically engineered mouse models and xenograft models, we found that MALAT1 overexpression inhibited, while MALAT1 deficiency induced breast cancer metastasis, which was reversed by add-back of MALAT1. We found that MALAT1 sequesters the transcription factor TEAD, leading to inhibition of TEAD’s transcriptional activity. Whereas our finding is a big departure from the literature, our approaches are highly rigorous. There is no evidence that the previously reported Malat1 gene deletion or ASO/siRNA phenotype was specific to Malat1 lncRNA loss. In contrast, several critical considerations have been taken into account in our study: first, we used a transcriptional terminator insertion strategy that inactivates the Malat1 gene without altering the expression of its neighboring genes, instead of deleting a several kb genomic region which led to upregulation of multiple Malat1’s adjacent genes; second, we conducted genetic rescue experiments to demonstrate that the metastasis induction by MALAT1 germline insertional inactivation or somatic knockout was specific to MALAT1 lncRNA loss; third, we found that overexpression of Malat1 suppressed breast cancer metastasis in transgenic, xenograft, and syngeneic models; fourth, we used either a B6 or an FVB background (instead of mixed background) for all compound mouse mutants, which is crucial for breast cancer models. Mechanistically, we captured endogenous MALAT1-TEAD interaction from primary mammary tumors, and discovered that MALAT1 binds and inactivates the pro-metastatic transcription factor TEAD. Taken together, our study reveals the unexpected function of MALAT1 through comprehensive targeted inactivation, restoration (rescue), and overexpression approaches in multiple in vivo models, calls for the need to reassess the ongoing efforts to target MALAT1 as an anti-metastatic therapeutic strategy, and provides a general framework for rigorous characterization of lncRNAs.
Methods
Mouse models.
The 7 kb full-length mouse Malat1 gene (NR_002847), including 47 bp upstream genomic sequence and 19 bp downstream genomic sequence, was cloned into the pGEM-T vector (Promega, #A362A) and then subcloned into the RMCE (Recombinase-Mediated Cassette Exchange) vector (Supplementary Fig. 3a). The subsequent generation of targeted Malat1 transgenic mice was performed at Taconic (see the Supplementary Note for details).
Malat1 knockout mice with targeted disruption of Malat1 (Malat1−/−) were from Shinichi Nakagawa’s lab stock. We bred MMTV-PyMT males (on C57BL/6, provided by William Muller, McGill University, Canada) to Malat1−/− females, and then further bred MMTV-PyMT;Malat1+/− males to Malat1+/− females to obtain MMTV-PyMT;Malat1−/− mice. To restore Malat1 expression in MMTV-PyMT;Malat1−/− mice, we bred Malat1−/− mice to Malat1Tg mice and further mated their offsprings to produce Malat1−/−;Malat1Tg mice. MMTV-PyMT;Malat1−/− males were then bred to Malat1−/−;Malat1Tg females to obtain MMTV-PyMT;Malat1−/−;Malat1Tg triple mutants. All mice described here were on a C57BL/6 background.
To generate Malat1Tg animals on an FVB/N background, we backcrossed Malat1Tg mice on C57BL/6 to FVB/N mice for 6 generations. Then Malat1Tg females on FVB/N were bred to MMTV-PyMT males on FVB/N (The Jackson Laboratory, stock #002374) to produce MMTV-PyMT;Malat1Tg mice. MMTV-PyMT;Malat1LSL mice were generated and used as the control.
Genotyping of MMTV-PyMT transgenic mice and Malat1 knockout mice was performed as described previously7,23. Primer sequences for PCR genotyping are listed in Supplementary Table 2. The purity of all mouse strains used in this study is greater than 98%.
Cell culture.
The HEK293FT cell line was from ThermoFisher Scientific. Hela, MCF10A, and a panel of breast cancer cell lines (Fig. 3a, except SUM149 and SUM159) were from American Type Culture Collection (ATCC) and were cultured under conditions specified by the manufacturer. SUM149 and SUM159 cell lines were from Li Ma’s lab stock (originally from Stephen P. Ethier, Medical University of South Carolina, Charleston) and were cultured in Ham’s F-12 medium supplemented with 5% FBS, 10 mM HEPES, 1 μg/ml hydrocortisone, and 5 μg/ml insulin. 67NR, 168FARN, 4TO7, and 4T1 cell lines were from Li Ma’s lab stock (originally from Fred R. Miller, Wayne State University School of Medicine, Detroit) and were cultured in DMEM medium supplemented with 10% FBS. The luciferase-expressing LM2 cell line was from Xiang Zhang (Baylor College of Medicine, Houston) and the G418-resistant, luciferase-expressing 4T1 cell line was from Mien-Chie Hung’s lab stock; both were cultured in DMEM medium supplemented with 10% FBS. Short tandem repeat (STR) profiling and mycoplasma tests were done by ATCC and MD Anderson’s Characterized Cell Line Core Facility.
Tumor and metastasis studies in GEM models.
All animal studies were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. Mammary tumor-free survival was determined by palpation. Mice were euthanized when they met the institutional euthanasia criteria for tumor size (2 cm in diameter) or overall health condition. MMTV-PyMT;Malat1+/+, MMTV-PyMT;Malat1−/−, and MMTV-PyMT;Malat1−/−;Malat1Tg female mice on a C57BL/6 background were euthanized at 13, 16, and 19 weeks of age and at the endpoint (20–25 weeks of age, upon euthanasia notice). MMTV-PyMT;Malat1LSL and MMTV-PyMT;Malat1Tg female mice on an FVB/N background were euthanized at 8 weeks of age and at the endpoint (12–13 weeks of age, upon euthanasia notice). Whole mammary glands or tumors and lung tissues were collected, weighed, and processed for histopathological analysis. Lung metastases were analyzed by gross examination of freshly dissected lungs and histopathological review of hematoxylin and eosin (H&E)-stained lung sections.
Circulating tumor cell isolation and staining.
~150 μl peripheral blood was collected from live animals via retro-orbital bleeding and red blood cells were lysed with RBC lysing buffer (Gibco, #A10492–01). Nucleated cells were spun onto glass slides using Cytospin and fixed in 10% formalin. For immunofluorescent staining of the PyMT protein, fixed cells were permeabilized with 0.25% Triton X-100 in phosphate-buffered saline (PBS). Endogenous peroxidase was blocked with 1.5% H2O2 in 0.05% Tween-20 in PBS (PBST). The cells were then incubated with a PyMT-specific primary antibody (Abcam, #ab15085, 1:200) and horseradish peroxidase-conjugated anti-rat secondary antibody (Vector laboratories, #PI-9401, 1:500). The signal was amplified using a Tyramide Signal Amplification Kit (Perkin Elmer, #NEL741001KT). Stained slides were mounted with VECTASHIELD Antifade Mounting Medium with DAPI (Vector laboratories, #H-1200). For CTC quantification, the ratio of PyMT+;DAPI+ cells to total DAPI+ cells was calculated.
Experimental metastasis assays.
Tumor cells were injected into the tail vein of 6- to 8-week-old female mice: NSG mice were injected with 2 × 105 MDA-MB-231 cells or 1 × 105 LM2 cells, and BALB/c mice were injected with 5 × 105 4T1 cells. Metastasis was monitored by luciferase imaging of live animals using an IVIS-200 bioluminescence imaging system (Perkin Elmer) after intraperitoneal injection of 100 μl D-luciferin substrate (25 mg/ml in PBS, Perkin Elmer). Mice were euthanized when they met the institutional euthanasia criteria for overall health condition. The lungs were collected, imaged with D-luciferin substrate (150 μg/ml in PBS), and then processed for histopathological analysis.
Immunoblotting.
Cells were lysed in RIPA lysis buffer (Millipore) containing protease inhibitors and phosphatase inhibitors (GenDEPOT). Proteins were resolved on 4–20% precast gradient gels (Bio-Rad) and transferred to a PVDF membrane. After blocking with 5% non-fat milk in Tris-buffered saline with 0.05% Tween-20 (TBST), membranes were incubated with the primary antibody followed by the secondary antibody conjugated with horseradish peroxidase. After washing, the bands were visualized with enhanced chemiluminescence substrate (Denville). Primary antibodies used are as follows: antibodies against pan-TEAD (1:1,000, Cell Signaling Technology, #13295), FLAG (1:5,000, Sigma, #F7425), HA (1:2,000, Santa Cruz Biotechnology, #sc-7392), cyclophilin B (1:5,000, ThermoFisher Scientific, #PA1–027A), YAP (1:1,000, Cell Signaling Technology, #14074), histone H3 (1:1,000, Cell Signaling Technology, #9715), Lamin B1 (1:1,000, Cell Signaling Technology, #12586), α-tubulin (1:1,000, Sigma, #T5168), HSP90 (1:5,000, BD Biosciences, #610419), and GAPDH (1:1,000, ThermoFisher Scientific, #MA5–15738).
Lentiviral vectors and lentivirus production.
Lentiviral vectors containing a pair of gRNAs targeting human MALAT1 (pDECKO_MALAT1_C, Addgene #72622)26 and Cas9 (lentiCas9-Blast, Addgene #52962)55 were from Addgene. Two shRNAs targeting TEAD1/3/441 were cloned by restriction enzymes AgeI and EcoRI into the pLKO.1-neo vector (Addgene #13425). The FU-luciferase-CRW/RFP vector was from Li Xin (Baylor College of Medicine, Houston). HEK293FT cells were co-transfected with the lentiviral vector, an envelope plasmid (pCMV-VSV-G, Addgene #8454), and a packaging plasmid (pCMV-dR8.2 dvpr, Addgene #8455)56. 2 days post transfection, viral supernatant was harvested, filtered through a 0.45 μm filter, and added to target cells.
Malat1 overexpression and CRISPR-Cas9-based MALAT1 knockout.
MDA-MB-231 and 4T1 cells were infected with the FU-luciferase-CRW/RFP lentivirus and sorted by red fluorescent protein (RFP). Luciferase-labeled MDA-MB-231 cells were then infected with the lentiCas9-Blast lentivirus and selected with blasticidin (10 μg/ml). Surviving cells were infected with the pDECKO_MALAT1_C lentivirus and selected with puromycin (1.5 μg/ml). After selection, single cells were plated in 96-well plates using a flow cytometer and grown for 1–2 weeks. The isolated single clones were subjected to qPCR, PCR, and DNA sequencing for knockout validation. DNA sequencing results revealed that nt 871–1539 and nt 857–1539 of MALAT1 were deleted in KO1 and KO2 (the two knockout clones used for functional assays), respectively. For qPCR of MALAT1, we used the MALAT1 TaqMan probe (ThermoFisher Scientific, Hs00273907_s1) and 5 sets of qPCR primers including 4 previously described sets26 (primer sequences are listed in Supplementary Table 2). We used gRNAs targeting GFP (pDECKO_GFP, Addgene, #72619) as control gRNAs and the control cells were bulk population. To restore Malat1 in MALAT1 knockout MDA-MB-231 cells and to overexpress Malat1 in LM2 and 4T1 cells, we subcloned full-length mouse Malat1 from the pGEM-T vector to the pcDNA3.1(−)-hygro vector, and transfected it into cells using Lipofectamine 2000 (Invitrogen). 3 days post transfection, hygromycin (300 μg/ml for LM2 and 800 μg/ml for 4T1) was added to select for stable cell lines.
Chromatin isolation by RNA purification (ChIRP).
The procedure was adapted and modified from a previous publication57. Buffers (lysis buffer, hybridization buffer, wash buffer, and RNA proteinase K buffer) were used as previously described57. Mammary tumors from MMTV-PyMT female mice were collected and frozen in liquid nitrogen. ~300 mg frozen tumor tissues were pulverized using a sample pulverizer (Covaris). Cells or pulverized tissues were crosslinked in 4% formaldehyde in PBS by inverting at room temperature for 30 min. The crosslinking reaction was quenched with 1/10 volume (0.125 M) of 1.25 M glycine at room temperature for 5 min. After centrifugation and removal of the supernatant, the pellet was washed with chilled PBS, resuspended in lysis buffer containing protease inhibitors (GenDEPOT), PMSF (1 mM), and RNase inhibitor (Ambion), and sonicated. After centrifugation of sonicated samples, the supernatant was pre-cleared twice with streptavidin beads (Invitrogen) by shaking at 37 °C for 30 min. 1% of pre-cleared lysate was saved for RNA and protein input. 1 μl 3′-biotinylated DNA probes (100 μM of 32 Malat1 probes or a probe for U1 or GFP; see probe sequences in Supplementary Table 3) was added to 1 ml lysate, and then 2× lysate volume of hybridization buffer containing protease inhibitors, PMSF (1 mM), and RNase inhibitor was added to the lysate. Hybridization was performed at 37 °C with shaking overnight. Next day, streptavidin beads were added to the hybridization reaction and incubated at 37 °C with shaking for 30 min (100 μl beads per 100 pmole probes). After five washes, the beads were resuspended in wash buffer. 1/10 volume was transferred to a new tube for RNA isolation and 9/10 volume was used for protein elution. Wash buffer was removed from the tube containing 9/10 bead volume.
For RNA isolation from the input and streptavidin-bound samples, RNA proteinase K buffer (Ambion) was added to the input and streptavidin-bound samples (total 95 μl and 195 μl, respectively). Then 5 μl proteinase K (Ambion) was added and incubated at 50 °C with shaking for 45 min. After brief spin-down and boiling at 95 °C for 10 min, the samples were chilled on ice and 500 μl TRIzol reagent was added. Tubes were vortexed for 10 sec and incubated at room temperature for 10 min. RNA was isolated using the miRNeasy Mini Kit and DNase I (Qiagen). One-step RT-qPCR (Bio-Rad, #1725150) was performed on the isolated RNA to examine Malat1 levels.
For protein elution from the streptavidin-bound samples, wash buffer was removed from the beads, and the bound proteins were eluted by boiling in Laemmli buffer and subjected to Western blot analysis or mass spectrometric analysis (see the Supplementary Note for details).
Chromatin immunoprecipitation (ChIP) assay.
A ChIP assay kit from Millipore (#17–371) was used according to the manufacturer’s protocol. Briefly, Hela cells were transfected with pPGS-3HA-TEAD1 (Addgene #33055)33 and/or pcDNA3.1(−)-Malat1. After crosslinking, 5 μg of the antibody against HA (Abcam, #ab9110), YAP (Cell Signaling Technology, #14074), or normal rabbit IgG (Santa Cruz Biotechnology, #sc-2027) was added to immunoprecipitate HA-tagged TEAD1 or endogenous YAP. For MALAT1 knockout MDA-MB-231 and Malat1-overexpressing LM2 cells, 5 μg of the antibody against TEAD1 (BD Biosciences, #610922), YAP (Cell Signaling Technology, #14074), normal mouse IgG (Santa Cruz Biotechnology, #sc-2025), or normal rabbit IgG (Santa Cruz Biotechnology, #sc-2027) was added to immunoprecipitate endogenous TEAD1 or endogenous YAP. After immunoprecipitation, protein-DNA crosslinks were reversed and DNA was purified to remove the chromatin proteins and used for qPCR. Primers specific for known YAP-TEAD target gene promoters (ANKRD1, CTGF, and CYR61) were from a previous study40. Primers specific for ITGB4 and VEGFA promoters were designed in this study. Primer sequences are listed in Supplementary Table 2. The results are presented as fold enrichment (normalized to IgG).
RNA pulldown assay.
Full-length mouse Malat1 (NR_002847) was divided into 6 non-overlapping pieces (P1-P6, 1.1–1.2 kb each) and each piece was cloned into the pGEM-T vector (Promega, #A362A). At the 3′ end of each piece in the vector, NotI was used to linearize the vector and produce 5′ overhang. The linearized vectors were gel-purified and used as templates for T7 RNA polymerase-mediated in vitro transcription (ThermoFisher Scientific, #K0441). The genomic sequence of full-length U1 nuclear RNA was amplified from mouse genomic DNA by PCR using the primer pair containing the T7 promoter at the 5′ end, and the PCR product was gel-purified and used as a template for T7 RNA polymerase-mediated in vitro transcription. Biotin-16-UTP (Roche, #11388908910) was used to biotinylate the RNAs. Non-biotinylated RNAs and biotinylated U1 were synthesized as negative controls. After in vitro transcription, synthesized RNA was isolated using the RNeasy Mini Kit (Qiagen) with DNase I treatment to remove the template DNA. The subsequent RNA pulldown procedure was adapted from Howard Chang’s laboratory protocol (see URLs). Briefly, 3 μg of biotin-labeled or biotin-free RNA was heated at 90 °C for 2 min and chilled on ice for 2 min. After RNA structure buffer (2×; 20 mM Tris-HCl at pH 7.4, 0.2 M KCl, 20 mM MgCl2, 2 mM DTT) containing RNase inhibitor was added, RNA samples were placed at room temperature for 20 min for proper secondary structure formation. Subconfluent HEK293FT cells were harvested, washed, lysed in RIPA buffer (150 mM KCl, 25 mM Tris-HCl at pH 7.4, 0.5 mM DTT, 0.5% NP-40) containing 1 mM PMSF, protease inhibitors (GenDEPOT), and RNase inhibitor (Ambion), and sonicated. Cell lysate was pre-cleared twice with streptavidin beads (Invitrogen) at room temperature. 3 mg of pre-cleared cell lysate was added to each folded RNA sample and incubated at room temperature overnight. Streptavidin beads were added and incubated at room temperature for 1 hour. The bound proteins were eluted by boiling in Laemmli buffer and subjected to Western blot analysis.
RNA immunoprecipitation (RIP) assay.
The RIP procedure was adapted from a previous publication58. Briefly, sub-confluent cells in two 15-cm dishes were harvested, washed in PBS, and crosslinked with 1% formaldehyde. Glycine (0.125 M) was added to quench the formaldehyde. Cells were pelleted by centrifugation and washed with PBS. IP lysis buffer (50 mM HEPES at pH 7.5, 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, 10% glycerol) containing 1 mM PMSF, protease inhibitors (GenDEPOT), and RNase inhibitor (Ambion) was added to the cell pellet. After sonication, cell lysate was pre-cleared with washed protein G agarose (Millipore). 5 μg of the antibody against HA (Abcam, #ab9110; control: normal rabbit IgG, Santa Cruz Biotechnology, #sc-2027) or the antibody against TEAD1 (BD Biosciences, #610922; control: normal mouse IgG, Santa Cruz Biotechnology, #sc-2025) was added to pre-cleared cell lysate and incubated at 4 °C overnight. Washed protein G agarose was added and incubated at 4 °C for 1 hour. Agarose beads were washed with IP lysis buffer and pelleted by centrifugation. RIP buffer (50 mM HEPES at pH 7.5, 0.1 M NaCl, 5 mM EDTA, 10 mM DTT, 0.5% Triton X-100, 10% glycerol, 1% SDS) containing RNase inhibitor was added to the pellet and incubated at 70 °C to reverse the crosslinks. After centrifugation, the supernatant was used for RNA extraction using the RNeasy Mini Kit (Qiagen) and DNase I treatment. One-step RT-qPCR (Bio-Rad, #1725150) was performed with primers listed in Supplementary Table 2. The results are presented as fold enrichment (normalized to IgG).
UV crosslinking-immunoprecipitation (CLIP) assay.
The CLIP procedure was adapted from previous publications31,32. Hela cells overexpressing both HA-TEAD1 and mouse Malat1 were plated in eight 15 cm dishes. 16 hours before UV crosslinking, 4-thiouridine (4-SU) was added to the cells to a final concentration of 100 μM. Next day, cells were washed in ice-cold PBS and PBS was removed completely. Plates were placed in a UV crosslinker and irradiated with 150 mJ/cm2 of UVA (365 nm). Cells were harvested in PBS and lysed in NP-40 lysis buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5 % NP-40) supplemented with protease inhibitor and 1 mM DTT. Clear lysate was collected by centrifugation and incubated with RNase T1 (Life Technologies, #EN0541) at 1 U/μl at 22 °C for 5 min to digest RNAs that were not protected from bound proteins. Protein A/G agarose beads (Life Technologies, #26159) were incubated with 10 μg of the HA-specific antibody (Abcam, #ab9110) or normal rabbit IgG in NT2 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05 % NP-40) at 4 °C. RNase T1-treated cell lysate was incubated with the washed antibody-protein A/G agarose complex at 4 °C overnight, and then the beads were pelleted and washed in NP-40 lysis buffer. Supernatant was completely removed from the beads and proteinase K buffer (100 mM NaCl, 10 mM Tris-Cl at pH 7.0, 1 mM EDTA, 0.5% SDS) was added to the pelleted beads. Proteinase K (Ambion, #AM2546) was added at 0.5 mg/ml and incubated at 55 °C for 30 min. 500 μl TRIzol was added and vortexed. Total RNA was isolated using the PureLink RNA Mini Kit (Life Technologies, #12183018A) with DNase treatment. After RNA isolation, One-step RT-qPCR (Bio-Rad, #1725150) was performed using 69 primer pairs covering the full-length mouse Malat1. Data are normalized to IgG (HA-TEAD1 IP/IgG IP) and to GAPDH as described previously31.
Quantitative PCR (qPCR).
For gene expression analysis, total RNA from human cells or mouse tissues were isolated using the RNeasy Mini Kit (Qiagen) and DNase I treatment according to the manufacturer’s protocol. For ChIP-qPCR assays, chromatin samples were obtained from chromatin immunoprecipitation as described above. Real-time PCR and data collection were performed with SYBR Green reagent (Bio-Rad) or TaqMan reagent (ThermoFisher Scientific) on a CFX96 instrument (Bio-Rad). Primer sequences are listed in Supplementary Table 2. For all qPCR assays of cell lines, we used n = 3 technical replicates per sample, and a representative set from 2–3 independent experiments is shown.
RNA-Seq analysis.
Malat1 wild-type (WT), Malat1 knockout (KO), and Malat1-restored (TG) PyMT mammary tumor samples (duplicates per group) were subjected to mRNA sequencing at MD Anderson’s Sequencing and Microarray Core Facility. The sequencing platform was HiSeq4000 and the paired end reads were in 2×76 bp. We mapped FASTQ raw reads and performed differential gene expression analysis using Tophat2 alignment with default parameters, HTSeq-count with mode “union”, followed by EdgeR. We identified differentially expressed genes by comparing each pair (WT vs. KO, WT vs. TG, and KO vs. TG) using the EdgeR likelihood ratio test. Genes that were commonly upregulated (by 2-fold or more) in MMTV-PyMT;Malat1−/− tumors (KO1 and KO2), compared with both MMTV-PyMT;Malat1+/+ tumors (WT1 and WT2) and MMTV-PyMT;Malat1−/−;Malat1Tg tumors (TG1 and TG2), were selected for further analysis.
Luciferase reporter assay.
Two days post transfection, firefly and Renilla luciferase activities were measured using a Dual-Luciferase Reporter Assay system (Promega, #E1910) on a Gen5 Microplate Reader (BioTek). For NanoLuc luciferase assays with firefly luciferase-labeled cells, NanoLuc luciferase activity and β-galactosidase enzyme activity were measured using the Nano-Glo Dual-Luciferase Reporter Assay System (Promega, #N1610) and the β-Galactosidase Enzyme Assay System (Promega, #E2000), respectively (see the Supplementary Note for details).
TCGA and computational data analysis.
To compare MALAT1 RNA expression levels between normal and tumor tissues, we used TCGA breast cancer RNA-Seq data (generated by the Illumina HiSeq 2000 RNA Sequencing Version 2 analysis platform) and performed the Wilcoxon test on the log2-transformed expression values (i.e., RNA-Seq by Expectation Maximization, RSEM). From CCLE (Cancer Cell Line Encyclopedia, see URLs), 66 human breast cancer cell lines are available and 59 of them have MALAT1 expression data. We grouped these 59 cell lines into two subtypes, luminal (n = 28) and basal/TNBC (n = 31), according to previous reports59–61, and performed an unpaired t-test to compare MALAT1 expression levels between the two subtypes. To compare MALAT1 levels in human breast tumors by tumor grades and tumor sites (primary vs. metastatic), we performed Oncomine data analysis (see URLs). For comparing tumors of different grades, a threshold P value of 0.005 was applied to screen datasets associated with published papers. For comparing tumors of different sites, a threshold P value of 0.05 was applied to screen datasets associated with published papers. Original datasets were downloaded and an unpaired t-test was performed on the relative expression level (log2 median-centered intensity). To assess the correlation of MALAT1 expression with clinical outcomes, we used the KM plotter28 and performed a log-rank test to compare high and low expression groups. To examine Malat1’s adjacent gene expression in the Malat1 knockout mouse model used in this study, we used the microarray data (downloaded from the NCBI GEO: GSE37707) from a previous study7.
Statistical analysis.
The experiments were repeated 2–3 times. Unless otherwise noted, data are presented as mean ± s.e.m., and a two-tailed t-test (unpaired or paired, as indicated) was used to compare two groups of independent samples. The log-rank test was used to compare Kaplan-Meier survival curves. Statistical methods used for RNA-Seq analysis and TCGA data analysis were described above. P < 0.05 was considered statistically significant.
Data availability.
The data that support the findings of this study are available from the corresponding author upon request. The RNA-Seq data have been deposited at the Gene Expression Omnibus (see URLs) under the accession number GSE110239.
URLs.
Howard Chang’s laboratory protocol, http://changlab.stanford.edu/RNA_pull-down_assay.pdf; Quantas Documentation, https://zhanglab.c2b2.columbia.edu/index.php/Quantas_Documentation; Cancer Cell Line Encyclopedia, https://portals.broadinstitute.org/ccle; Oncomine data-mining platform, https://www.oncomine.org; Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/; Kaplan Meier Plotter, http://kmplot.com/analysis/.
Supplementary Material
Acknowledgements
We thank W. Muller for providing MMTV-PyMT mice (C57BL/6 background), X. Zhang for providing luciferase-expressing LM2 cells, and J. Jacobson and L. Xin for providing ITGB4-luciferase and FU-luciferase-CRW/RFP constructs, respectively. We thank J. Zhang and MD Anderson’s shRNA and ORFeome Core, Small Animal Imaging Facility, Flow Cytometry and Cellular Imaging Core Facility, Sequencing and Microarray Facility, and Characterized Cell Line Core Facility for technical assistance. We are grateful to all members of the Ma Lab for discussion, to J. Chen for critical reading of the manuscript, and to J.-H. Yoon and M. Gorospe for advice on the CLIP assay. L.M. is supported by US National Institutes of Health (NIH) grants R01CA166051 and R01CA181029, a Cancer Prevention and Research Institute of Texas (CPRIT) grant RP150319, and a Stand Up To Cancer Innovative Research Grant (award number: 403235). M.J.Y. was supported in part by NIH R01CA164346, R01CA200703, and CPRIT RP140402. M.-C.H. is supported by National Breast Cancer Foundation, Inc. and The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund. H.L. (Han Liang) is supported by NIH R01CA175486 and U24CA209851. M.J.E is supported by CPRIT RR140033. B.G. is supported by NIH R01CA181196 and R01CA190370.
Footnotes
Competing interests
The authors declare no completing interests.
Additional information
Supplementary information contains ten figures, three tables, five videos, and the Supplementary Note.
References
- 1.Evans JR, Feng FY & Chinnaiyan AM The bright side of dark matter: lncRNAs in cancer. J Clin Invest 126, 2775–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2, 111–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilusz JE, Freier SM & Spector DL 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson JN et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tripathi V et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39, 925–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eissmann M et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9, 1076–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagawa S et al. Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L et al. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A 106, 12956–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji Q et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer 111, 736–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao S et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res 6, 2561–2574 (2016). [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis 7, e2123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latorre E et al. The Ribonucleic Complex HuR-MALAT1 Represses CD133 Expression and Suppresses Epithelial-Mesenchymal Transition in Breast Cancer. Cancer Res 76, 2626–36 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Guy CT, Cardiff RD & Muller WJ Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12, 954–61 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arun G et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30, 34–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassett AR et al. Considerations when investigating lncRNA function in vivo. Elife 3, e03058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Y et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell 16, 504–16 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Gutschner T et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73, 1180–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jadaliha M et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 7, 40418–40436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin A, Giuliano CJ, Sayles NM & Sheltzer JM CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang HT et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin EY et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163, 2113–26 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davie SA et al. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res 16, 193–201 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J et al. Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways. Cancer Res 76, 6424–6435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minn AJ et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–24 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aparicio-Prat E et al. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics 16, 846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyorffy B et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123, 725–31 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Aslakson CJ & Miller FR Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res 52, 1399–405 (1992). [PubMed] [Google Scholar]
- 30.Chu C et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon JH et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 4, 2939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon JH & Gorospe M Cross-Linking Immunoprecipitation and qPCR (CLIP-qPCR) Analysis to Map Interactions Between Long Noncoding RNAs and RNA-Binding Proteins. Methods Mol Biol 1402, 11–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z et al. Structural insights into the YAP and TEAD complex. Genes Dev 24, 235–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pobbati AV & Hong W Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther 14, 390–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan D The hippo signaling pathway in development and cancer. Dev Cell 19, 491–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Li L, Lei Q & Guan KL The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24, 862–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moroishi T, Hansen CG & Guan KL The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 15, 73–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanconato F et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17, 1218–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupont S et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–83 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Stein C et al. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet 11, e1005465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao B et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22, 1962–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lian I et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24, 1106–18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoshino A et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao C, Lotz MM, Clarke AC & Mercurio AM A function for the integrin alpha6beta4 in the invasive properties of colorectal carcinoma cells. Cancer Res 56, 4811–9 (1996). [PubMed] [Google Scholar]
- 45.Guo W et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell 126, 489–502 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Leng C et al. An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK-AKT pathway. Cancer Lett 376, 188–96 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Goel HL & Mercurio AM VEGF targets the tumour cell. Nat Rev Cancer 13, 871–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takaoka AS et al. Cloning and characterization of the human beta4-integrin gene promoter and enhancers. J Biol Chem 273, 33848–55 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Wood LW et al. Thyroid Transcription Factor 1 Reprograms Angiogenic Activities of Secretome. Sci Rep 6, 19857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung DW, Cachianes G, Kuang WJ, Goeddel DV & Ferrara N Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246, 1306–9 (1989). [DOI] [PubMed] [Google Scholar]
- 51.Bhattacharya R et al. Intracrine VEGF signalling mediates colorectal cancer cell migration and invasion. Br J Cancer 117, 848–855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo M et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-kappaB and beta-catenin. Cancer Lett 373, 1–11 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Oommen S, Gupta SK & Vlahakis NE Vascular endothelial growth factor A (VEGF-A) induces endothelial and cancer cell migration through direct binding to integrin {alpha}9{beta}1: identification of a specific {alpha}9{beta}1 binding site. J Biol Chem 286, 1083–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perrot-Applanat M & Di Benedetto M Autocrine functions of VEGF in breast tumor cells: adhesion, survival, migration and invasion. Cell Adh Migr 6, 547–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart SA et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu C, Quinn J & Chang HY Chromatin isolation by RNA purification (ChIRP). J Vis Exp (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt M & Taylor V Cross-linked RNA Immunoprecipitation. Bio-protocol 3(2013). [Google Scholar]
- 59.Neve RM et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai X, Cheng H, Bai Z & Li J Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J Cancer 8, 3131–3141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang G et al. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genomics 17 Suppl 7, 525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The RNA-Seq data have been deposited at the Gene Expression Omnibus (see URLs) under the accession number GSE110239.


