Abstract
The promoters of mammalian genes are commonly regulated by multiple distal enhancers, which physically interact within discrete chromatin domains. How such domains form and how the regulatory elements within them interact in single cells is not understood. To address this we developed Tri-C, a new Chromosome Conformation Capture (3C) approach to identify concurrent chromatin interactions at individual alleles. Analysis by Tri-C reveals heterogeneous patterns of single-allele interactions between CTCF boundary elements, indicating that the formation of chromatin domains likely results from a dynamic process. Within these domains, we observe specific higher-order structures involving simultaneous interactions between multiple enhancers and promoters. Such regulatory hubs provide a structural basis for understanding how multiple cis-regulatory elements act together to establish robust regulation of gene expression.
Introduction
Precise spatial and temporal patterns of gene expression during development and differentiation are controlled by cis-regulatory elements including promoters, enhancers and boundary elements. The interaction and activity of these elements depend on and influence their structural organization within the nucleus1. To date the relationship between structure and function has mainly been analyzed in populations of cells. The globin loci, which provide ideal models to elucidate the general principles of mammalian gene regulation, have been extensively studied in this way. For example, we have previously shown that the active mouse α-globin cluster and its regulatory elements are located in a decompacted ~70 kb chromatin domain that forms early in erythroid differentiation and is flanked by CTCF-binding sites2–4. Within this domain, the α-globin genes interact with five enhancer elements, which cooperate in an additive manner to upregulate gene expression5.
Although these studies have made important contributions to our understanding of chromatin architecture and gene regulation, they are based on analyses of populations of cells, which may obscure the dynamic nature of the underlying biological processes in individual cells. Therefore, it remains unknown how promoters, enhancers and boundary elements physically interact as genes are switched on in single cells. To address this important question, we developed Tri-C, a new Chromosome Conformation Capture (3C) technique, which can identify concurrent chromosomal interactions at individual alleles, and consequently provides information derived from single nuclei. By combining conventional 3C and Tri-C experiments, we performed in-depth characterization of the structural architecture of the murine globin loci.
Our analyses show that interactions between the boundary elements of the chromatin domains containing the globin loci are highly variable. The heterogeneity of these structures could reflect a dynamic process of chromatin domain formation, which is in agreement with a mechanism of loop extrusion contributing to chromatin architecture. Importantly, we observe higher-order complexes within these domains, formed by multi-way chromatin interactions between multiple enhancers and promoters at individual alleles. This shows that the globin enhancers and promoters form regulatory hubs in which they simultaneously interact to switch genes on. This is consistent with previous observations showing that fully regulated expression of the murine globin genes5,6 and many other genes7 depends on the presence of more than one enhancer element. The formation of such enhancer-promoter complexes also provides a possible function for apparently redundant enhancer elements8,9, which could play a role in the formation of robust structures required for assembly of the transcriptional machinery at gene promoters.
Results
Activation of the murine globin loci is associated with the formation of strongly compartmentalized chromatin domains in which enhancers and promoters interact
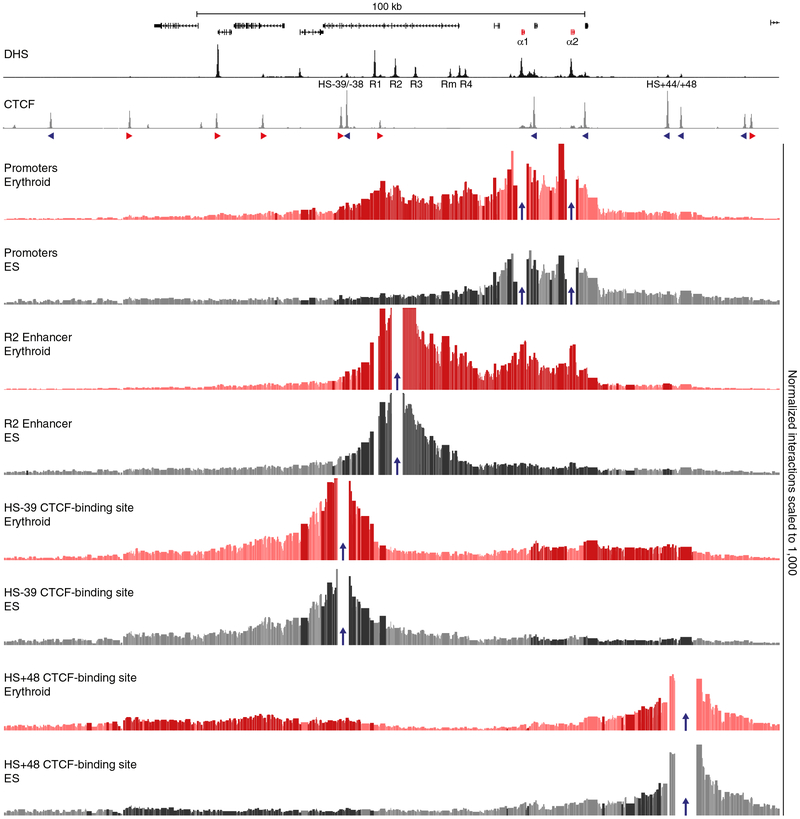
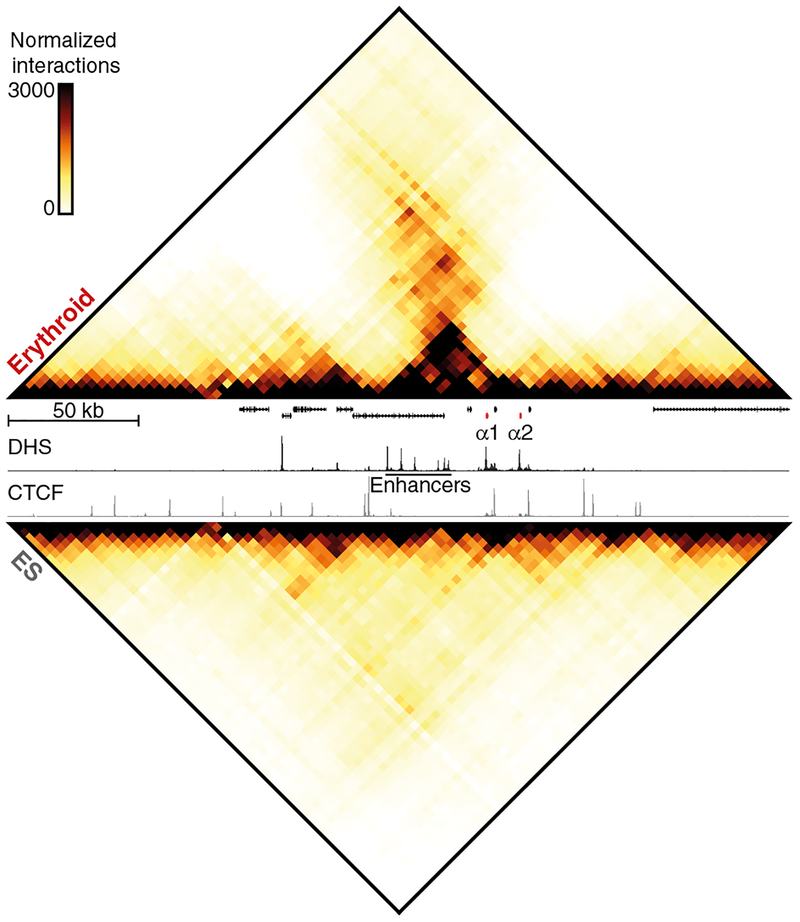
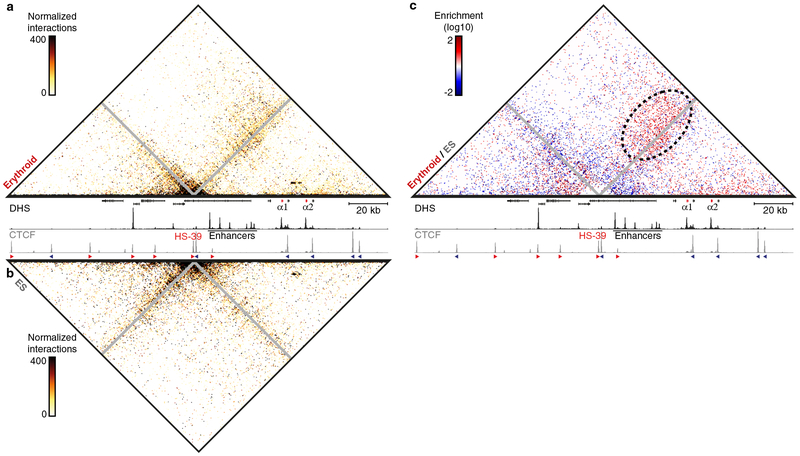
To characterize the large-scale conformations of the murine globin loci, we performed Hi-C in primary mouse erythroid cells, in which the globin genes are highly expressed, and compared this to an equivalent Hi-C dataset from mouse embryonic stem (ES) cells10, in which the globin loci are silent (Supplementary Fig. 1). This comparison shows that the chromatin regions containing the globin clusters form strong, tissue-specific, self-interacting domains in erythroid cells, but not in ES cells. To characterize the interactions within these domains at higher resolution, we performed multiplexed, high-resolution Capture-C experiments in erythroid and ES cells from ~45 viewpoints, which were closely spaced across the loci and included all known regulatory elements (Supplementary Tables 1–4). Figure 1 shows interaction profiles in the α-globin domain from the viewpoint of the promoters, the strongest enhancer element (R2)5, and the CTCF boundary elements on either side of the domain. These profiles show strong reciprocal interactions between the α-globin promoters and all its enhancer elements in erythroid cells. The flanking CTCF-binding elements do not participate in these enhancer-promoter interactions, but form diffuse interactions with the chromatin bound by CTCF on either side of the domain, spanning contiguous regions of ~50 kb. The nature of these interaction patterns is highlighted in the contact matrices derived from viewpoints tiled across a 300 kb region containing the locus (Fig. 2).
Figure 1: Characterization of the interaction landscape of the regulatory elements of the α-globin locus.
High-resolution Capture-C interaction profiles of the α-globin locus from the viewpoints (indicated by blue arrows) of the α-globin promoters, the R2 enhancer, and CTCF-binding sites HS-39 and HS+48 in erythroid (red) and ES (grey) cells. Profiles represent the mean number of normalized unique interactions per restriction fragment in 3 replicates. Statistically significant differential interactions (derived using DESeq2) between erythroid and ES cells are highlighted in bold colors. Gene annotation (α-globin genes highlighted in red), erythroid DNaseI hypersensitivity (DHS) and CTCF occupancy are shown at the top, with arrows indicating the orientation of the CTCF-binding motifs. Coordinates (mm9): chr11:32,050,000–32,250,000.
Figure 2: Structural conformation of the active and inactive α-globin locus.
Contact matrices (4 kb resolution) of the α-globin locus derived from Capture-C experiments with viewpoints closely spaced across the domain in erythroid (top) and ES (bottom) cells. Matrices represent the mean number of normalized unique interactions in 3 replicates. Gene annotation (α-globin genes highlighted in red), erythroid DNaseI hypersensitivity (DHS) and CTCF occupancy are shown in the middle. Coordinates (mm9): chr11:32,000,000–32,300,000.
Although the contacts between the enhancers and promoters are more discrete compared to those between the CTCF-binding sites, interaction frequencies are significantly enriched throughout the domain in erythroid cells. This suggests that interactions between enhancers and promoters might not reflect stable, distinct loops1,11. Rather, these interactions are more readily interpreted as a compartmentalized domain, in which, at some point, every region of chromatin interacts with every other, and preferred, transiently stabilized structures are formed between regulatory elements. Analysis of the β-globin locus reveals a similar interaction landscape (Supplementary Figs. 2, 3).
Our analysis by Capture-C suggests that all enhancers and promoters in the globin loci can form interactions. However, as these data are derived from pair-wise contacts in populations of cells, they reflect multiple dynamic conformations that obscure specific higher-order structures associated with productive interactions between enhancers and promoters in individual cells. In particular, it is not clear whether cis-interactions between multiple enhancers and promoters occur simultaneously in a single cell nucleus, or if these elements compete for the formation of exclusive interactions. Therefore, it remains unclear how these regulatory elements interact to ensure robust regulation of gene expression.
Tri-C detects multi-way chromatin interactions with unprecedented sensitivity
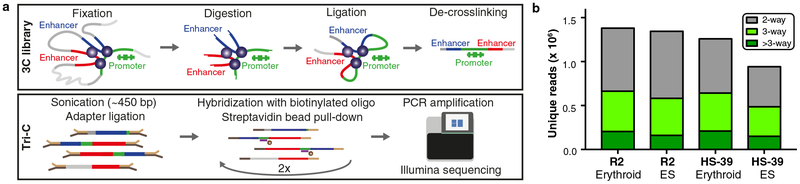
Questions regarding the structural interaction between regulatory elements can be addressed by analyzing interactions between these elements in individual cells. Though techniques such as single-cell Hi-C12–14, Genome Architecture Mapping (GAM)15 and Split-Pool Recognition of Interactions by Tag Extension (SPRITE)16 have provided insights into chromosomal structures in single cells at large scales, the resolution of these techniques is currently insufficient to be informative at the level of individual regulatory elements. To overcome these limitations, we explored a different strategy to analyze chromosomal structures in single cells. Because in situ 3C libraries contain long DNA concatemers in which neighboring fragments represent chromatin regions that were in close proximity in individual nuclei, single-allele chromatin conformations can be derived from population-based assays in which several neighboring fragments in the 3C concatemer are identified simultaneously. However, the proportion of reads containing multiple interacting fragments in conventional 3C techniques is very low (<2%)17–20 and despite advances in efficiency in a recent 3way-4C approach21, resolution and sensitivity of current techniques are insufficient for robust detection of higher-order structures formed by individual regulatory elements in single cells. To analyze such structures at high resolution, we developed Tri-C, a new 3C method that can identify multi-way interactions with viewpoints of interest with unprecedented sensitivity (Fig. 3a, Supplementary Tables 5, 6). For efficient detection of multiple ligation junctions between interacting fragments, Tri-C libraries are generated using an enzyme selected to create relatively small DNA fragments (~200 bp) for the viewpoints of interest. We chose the restriction enzyme NlaIII, which has a recognition sequence of 4 bp and produces fragments with a median size of 131 bp in the mouse genome (Supplementary Fig. 4). Sonication of these libraries to ~450 bp generates fragments of which about ~50% contain multiple ligation junctions. Using highly optimized oligonucleotide capture-mediated enrichment for viewpoints of interest, millions of multi-way contacts can be identified with Illumina sequencing, generating 3C profiles at unprecedented depth (Fig. 3b). Importantly, Tri-C allows for multiplexing both viewpoints and samples, enabling analyses of multiple genomic regions and cell types of interest in a single experiment. Because Illumina sequencing provides accurate identification of the random sonication ends of the reads, these can be used as unique molecular identifiers to filter out PCR duplicates, allowing for quantitative analysis of the detected multi-way interactions.
Figure 3: Overview of the experimental procedure and data output of Tri-C.
(a) Overview of Tri-C. 3C libraries are generated by fixation, digestion and proximity ligation of chromatin. De-crosslinked 3C concatemers are sonicated to ~450 bp fragments and ligated to sequencing adapters, after which fragments containing viewpoints of interest are enriched by oligonucleotide capture. Small viewpoint restriction fragments are targeted to allow for efficient detection of multiple ligation junctions within single sonicated 3C fragments using Illumina sequencing platforms. (b) Number of unique reads containing pair-wise and multi-way interactions generated by Tri-C for viewpoints in the α-globin locus.
To validate that Tri-C detects reliable multi-way interactions in individual cells, we performed several additional experiments and analyses. First, to confirm that capturing multiple ligation junctions simultaneously did not introduce a bias in the detected interactions, we compared the pair-wise interactions derived from Tri-C to Capture-C interaction profiles from the same viewpoint (Supplementary Fig. 5). To validate the detected multi-way interactions, we developed an additional method to analyze multiple ligation junctions within a single 3C concatemer based on long-read Nanopore sequencing. This method, named C-Trap, uses long-range PCR amplification with primers targeting two interacting fragments of interest, to ‘trap’ and enrich for the intervening fragments in the concatemer that were interacting simultaneously (Supplementary Fig. 6a and Supplementary Table 7). Though the sensitivity of C-Trap is limited (Supplementary Fig. 6b and Supplementary Table 8), as it relies on the relatively low-throughput Nanopore sequencing platform, the detected patterns of multi-way interactions in the α-globin locus are very similar to the multi-way interactions identified by Tri-C (Supplementary Fig. 6c). Finally, to confirm that Tri-C interactions represent single-cell chromosome conformations, we show that the detected multi-way interactions are allele-specific (Supplementary Fig. 7).
Multiple enhancers and promoters form higher-order chromatin structures in which they interact simultaneously
We used Tri-C to analyze the higher-order structures around cis-regulatory elements in the globin loci in erythroid and ES cells. To visualize these 3D structures, we represented the multi-way interactions in contact matrices in which we excluded the viewpoint of interest and plotted the frequencies with which two elements were captured simultaneously with this viewpoint. Note that although these matrices contain data derived from populations of cells, they only show interactions occurring simultaneously at single alleles, and therefore provide insight into the patterns of interactions and 3D relationships between multiple cis-regulatory elements in individual nuclei. Mutually exclusive contacts between elements appear as depletions at the intersections between these elements in the contact matrix, whereas preferential simultaneous interactions in higher-order structures are visible as enrichments at these foci.
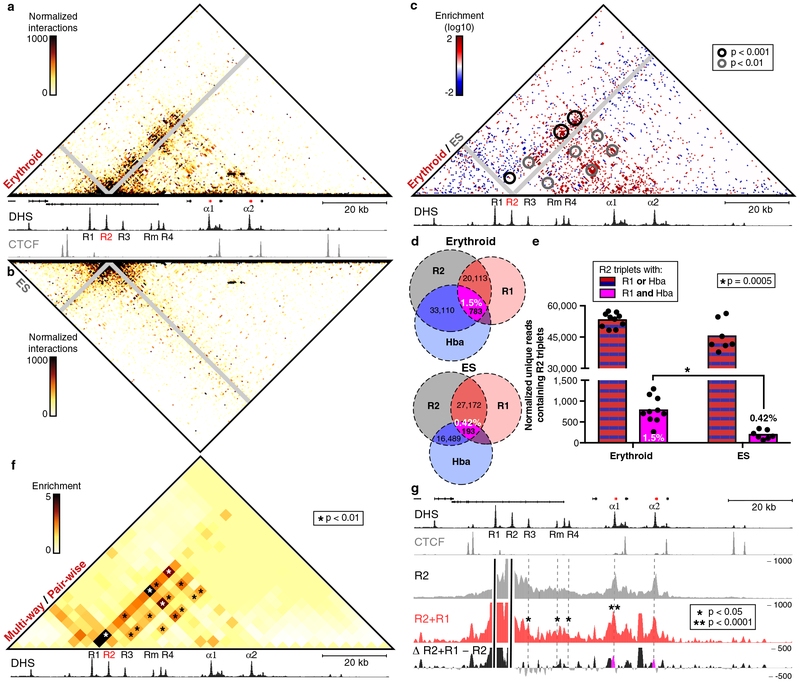
Analysis from the R2 viewpoint, the strongest α-globin enhancer5, shows that specific multi-way interactions with the other enhancers (R1, R3, Rm and R4) and promoters are formed in erythroid cells (Fig. 4a), which are highly reproducible between replicates (Supplementary Fig. 8). These interactions are absent in ES cells (Fig. 4b) and direct comparison shows strong, significant enrichments for these multi-way enhancer-promoter interactions in erythroid cells (Fig. 4c). For example, multi-way interactions between R2, R1 (the second strongest α-globin enhancer5), and either of the α-globin promoters (‘R2-R1-Hba triplets’) are over ~4-fold enriched (Fig. 4d). Importantly, these interactions are still highly significantly enriched in erythroid cells compared to ES cells after correcting for the increased pair-wise interactions between the individual enhancers and the α-globin promoters (~3.5-fold; p = 0.0005; Fig. 4e). This shows that these multi-way enhancer-promoter interactions represent erythroid-specific higher-order structures, and do not simply reflect increased pair-wise interactions between the participating elements in erythroid cells.
Figure 4: Analysis of multi-way interactions between enhancers and promoters in the α-globin locus.
Tri-C matrices represent the mean number of normalized, unique interaction counts in 10 erythroid or 7 ES replicates, displayed at 500 bp resolution, with proximity contacts around the R2 viewpoint excluded (grey diagonal). Matrices are annotated with gene positions (α-globin genes highlighted in red), erythroid DNaseI hypersensitivity (DHS) and/or CTCF occupancy. Coordinates (mm9): chr11:32,120,000–32,240,000. (a) Contact matrix showing multi-way interactions with R2 in erythroid cells. (b) Contact matrix showing multi-way interactions with R2 in ES cells. (c) Contact matrix showing differential multi-way interactions with R2 that are enriched in erythroid (red) or ES (blue) cells. Statistically significant enriched interactions in erythroid cells (derived using DESeq2) are highlighted. (d) Venn diagram illustrating the proportion of R2-R1-Hba triplets relative to all triplets containing pair-wise R2-R1 and R2-Hba interactions in erythroid and ES cells. Numbers represent normalized counts of unique reads containing the specified three-way or pair-wise interactions. (e) Comparison of the absolute numbers of R2-R1-Hba triplets (shown on the y-axis) and the proportion of these relative to all triplets containing pairwise R2-R1 and R2-Hba interactions (represented as percentages in the bar graphs) in erythroid and ES cells. Bar graphs represent average normalized unique read counts in 10 erythroid and 7 ES replicates, with individual data points overlaid as dot plots. Both absolute and relative R2-R1-Hba triplet counts are significantly enriched in erythroid cells (~4-fold, p = 0.0002 and ~3.5-fold, p = 0.0005, respectively; statistics derived using unpaired, two-tailed t-tests). (f) Contact matrix (4 kb resolution) showing enrichment of multi-way interactions with R2 in erythroid cells over pair-wise contact frequencies derived from multiplexed Capture-C data in 3 replicates. Statistically significant enriched multi-way interactions (derived using unpaired, two-tailed t-tests, corrected for multiple comparisons) are highlighted. (g) Comparison of a double-anchored interaction profile containing fragments interacting with both the R2 and R1 enhancers (red) to the conventional R2 interaction profile (grey) in erythroid cells. Enrichment of multi-way R2-R1 interactions over pair-wise R2 interactions are shown in the differential profile at the bottom (black), with the R2-R1-Hba interactions highlighted (magenta). Profiles represent windowed, normalized, unique interaction counts in 10 replicates. Statistically significant enriched multi-way interactions with cis-regulatory elements of α-globin (derived using DESeq2) are indicated. Note that the double-anchored interaction profile has a relatively high signal-to-noise ratio and therefore contains some spurious peaks, including a strong, yet insignificant, enrichment of an unusually large (> 1.5 kb) NlaIII fragment containing many repeats in between the two α-globin promoters.
To further investigate how the multi-way R2 interactions detected in erythroid cells relate to the underlying pair-wise contact distribution, we compared the R2 Tri-C matrices to the contact matrices derived from the multiplexed Capture-C experiments at 4 kb resolution (Fig. 4f). This shows that multi-way interactions between R2, the other α-globin enhancers and promoters are significantly enriched, beyond what would be expected based on the pair-wise interactions between these elements in erythroid cells. To examine these enrichments at higher resolution, we analyzed ‘double-anchored’ interaction profiles generated from reads containing both the R2 and R1 enhancer. To investigate whether reads containing both major α-globin enhancers were more likely to interact with the other α-globin enhancers and promoters, we compared these profiles to conventional interaction profiles containing all interactions with the R2 viewpoint. Though the double-anchored profiles have a relatively high signal-to-noise ratio due to the many possible triplets that can be formed, interactions with all the other enhancers and the Hba-a1 promoter are significantly enriched. Together, these analyses demonstrate that the α-globin enhancers and promoters form regulatory hubs in which they simultaneously interact together to switch genes on. Importantly, Tri-C analysis of HS2, one of the strongest β-globin enhancers6, also shows the formation of such erythroid-specific higher-order structures (Supplementary Fig. 9).
CTCF-binding sites form dynamic interactions supportive of a loop extrusion mechanism underlying boundary formation
We have previously shown that CTCF-binding sites flanking the α-globin locus contribute to the formation of a domain that delimits the region of chromatin within which the observed complexes between enhancers and promoters can be formed2. However, the processes underlying the formation of such chromatin domains and their contribution to enhancer-promoter specificity remain unclear. Based on Hi-C data, it has been suggested that CTCF-binding sites located at domain boundaries form specific loops and that multiple CTCF-binding sites might form multi-anchored and/or nested structures22. Our Capture-C data show that the functionally validated CTCF boundary element HS-392 located upstream of the α-globin cluster predominantly interacts with a region downstream of the domain, which contains many CTCF-binding sites (Figs. 1, 2). However, the pattern of interactions is very broad compared to the interactions between enhancers and promoters and it is unclear whether these interactions represent stable loops between multiple CTCF-binding sites or represent a more dynamic mechanism by which domain boundaries are formed.
To resolve these structures at individual alleles, we performed Tri-C analysis from the viewpoint of the HS-39 CTCF-binding site in the α-globin locus. Consistent with the pair-wise interaction data, multi-way interactions with HS-39 are preferentially formed with the region located downstream of the α-globin domain containing many CTCF-binding sites. A model in which CTCF boundaries are formed by stable, multi-anchored loops would result in specific enrichments of multi-way interactions between these CTCF-binding sites. However, the HS-39 Tri-C contact matrix shows a diffuse enrichment with the entire region on the other side of the α-globin domain in erythroid cells (Fig. 5a). Comparison of these interactions to ES cells (Fig. 5b) shows a trend of general enrichment of this diffuse interaction pattern in erythroid cells, though none of these interactions are significantly enriched (Fig. 5c). Tri-C analysis from the viewpoint of the 3’HS1 CTCF-binding site in the β-globin locus shows similar interaction patterns in erythroid and ES cells (Supplementary Fig. 10).
Figure 5: Analysis of multi-way interactions between CTCF-binding sites in the α-globin locus.
Tri-C matrices represent the mean number of normalized, unique interaction counts in 10 erythroid or 7 ES replicates, displayed at 500 bp resolution, with proximity contacts around the HS-39 viewpoint excluded (grey diagonal). Matrices are annotated with gene positions (α-globin genes highlighted in red), erythroid DNaseI hypersensitivity (DHS) and CTCF occupancy, with arrows indicating the orientation of CTCF-binding motifs. Coordinates (mm9): chr11:32,040,000–32,240,000. (a) Contact matrix showing multi-way interactions with HS-39 in erythroid cells. (b) Contact matrix showing multi-way interactions with HS-39 in ES cells. (c) Contact matrix showing differential multi-way interactions with HS-39 that are enriched in erythroid (red) or ES (blue) cells. Although the diffuse interactions with the region spanning the other side of the α-globin domain are enriched in erythroid cells (highlighted), there are no significant enrichments of specific interactions between multiple CTCF-binding sites (p > 0.5; statistics derived using DESeq2).
Our data therefore indicate that CTCF-binding elements do not form higher-order hub-like structures in which multiple CTCF-binding sites interact simultaneously. The observed diffuse interaction patterns of the HS-39 and 3’HS1 CTCF-binding sites could be explained by the formation of dynamic interactions with the entire region flanking the opposite side of the globin domains, with these heterogeneous interactions representing single-cell snapshots of the dynamic interactions throughout this chromatin region. These Tri-C interaction patterns are therefore consistent with a loop extrusion mechanism, in which the formation of chromatin domains is mediated by protein complexes, likely involving Cohesin, that translocate along chromosomes, bringing every region into contact with each other23–25. Continuous scanning across chromatin regions and transient stalling of these protein complexes at CTCF boundary elements could explain the enrichment over a broad region of chromatin containing many CTCF-binding sites.
Discussion
Formation of chromatin domains by the proposed loop extrusion mechanism could explain many features of chromosome organization23–25. However, the process of loop extrusion and the resulting dynamic chromatin structures have not been observed directly in vivo, and current evidence is derived from polymer model predictions24,25 and perturbations of specific components of the loop extrusion machinery26–28. Here, we show that high-resolution single-allele chromatin structures do not support a model of stable loops forming between individual CTCF-binding sites, but indicate a dynamic mechanism such as loop extrusion underlying the formation of chromatin domains. Importantly, the diffuse enrichment patterns we observe are indicative of transient CTCF-binding site interactions, which is in agreement with the kinetics of CTCF and Cohesin binding to the genome29.
The preferential interactions between CTCF-binding sites at opposite ends of the globin domains in erythroid cells explain the formation of tissue-specific domains in erythroid cells (Fig. 5 and Supplementary Fig. 10). We have previously shown that CTCF occupancy is similar in both cell types2. These different structures therefore likely reflect differences in the processivity of loop extruding factors such as Cohesin, which could result from tissue-specific recruitment locations and/or extrusion rates. It has been shown that Mediator, a transcriptional coactivator bound at active enhancers and promoters, forms complexes with Cohesin and the Cohesin-loading factor Nipbl30, and we have previously observed binding of Cohesin and Mediator at the α-globin enhancers in erythroid cells2,5. The erythroid-specific formation of the globin domains could therefore be explained by a loop extrusion model in which Cohesin is recruited to the genome at active enhancers and/or promoters. Recent findings have suggested that Cohesin translocation is stimulated by Nipbl, suggesting that Nipbl abundance determines extrusion rates31. Differences in Nipbl distribution could therefore also contribute to tissue-specific interaction patterns, though this remains to be further explored.
Within the dynamic chromatin domains containing the globin gene clusters, we identify regulatory hubs in which multiple enhancers and promoters interact simultaneously at individual alleles. These findings contrast with a study based on a 3-way 4C approach, in which such higher-order structures could not be reliably detected, and which therefore suggested that active chromatin structures are primarily driven by pair-wise interactions21. However, as the sensitivity of 3-way 4C is relatively low compared to Tri-C (Supplementary Table 6), it is possible that this study did not have sufficient power for robust identification of hub-like structures. In a recently published study, a novel method named multi-contact 4C was developed to analyze multi-way DNA interactions. This method uses long-read Nanopore sequencing to read out multiple ligation junctions, somewhat similar to C-Trap (Supplementary Figure 6). Though the use of Nanopore sequencing has limitations regarding read accuracy, data depth and throughput, multi-contact 4C was able to detect interaction patterns indicative of the formation of ‘enhancer hubs’32. This provides additional support for the regulatory hubs we describe, in which multiple enhancers and promoters interact to switch genes on. Importantly, we also show that contacts between the cis-regulatory elements within these hubs do not represent stable, exclusive enhancer-promoter loops11, but preferred interactions within dynamic compartmentalized domains, in agreement with polymer model predictions33–35 and single-cell super-resolution analysis36.
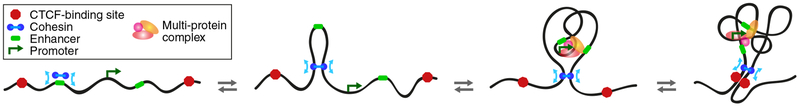
A recent study has suggested that interactions between cis-regulatory elements could be directly mediated by a ‘reeling in’ mechanism. In this model, cohesin is loaded near a CTCF-binding site close to a strong cis-regulatory element, which blocks extrusion at one end, resulting in direct tethering of promoters and enhancers by extensive one-sided loop extrusion at the other end37. Though it is likely that loop extrusion contributes to the formation of the observed regulatory hubs38, the complexity of these higher-order enhancer-promoter structures might not be explained by CTCF/Cohesin-mediated loop extrusion alone and reflect a combination of several distinct mechanisms contributing to chromatin architecture. This is consistent with polymer models39 and recent studies in which Cohesin activity was perturbed, but interactions between cis-regulatory elements still occurred, though more promiscuous27,37. Our data could be explained by a loop extrusion mechanism that brings regulatory elements into close proximity and enables subsequent formation of more complex, stabilized structures (Fig. 6). It is likely that interactions between these elements are mediated by multi-protein complexes bound at promoters and enhancers. Such interactions could contribute to or result from the formation of phase-separated assemblies of components of the transcriptional machinery40.
Figure 6: Graphical summary.
Our data are supportive of a loop extrusion mechanism contributing to the formation of compartmentalized chromatin domains. Complex higher-order structures, in which multiple enhancers and promoters interact, are formed within these domains by tissue-specific mechanisms, likely involving interactions between multi-protein complexes bound at these cis-regulatory elements.
The formation of regulatory hubs in which multiple enhancers simultaneously interact with the genes they regulate indicates cooperation rather than competition between enhancer elements. This is highlighted by the observation that the promoter of the housekeeping gene Nprl3, which is six-fold upregulated in erythroid cells2, is included in the complexes formed with the α-globin enhancers and promoters (Fig. 4). Such cooperative hubs are consistent with the observed additive effects of individual enhancers at the globin5,6 and many other genes7. Importantly, we have previously shown that no single enhancer element in the α-globin locus is critical for the formation of the chromatin structures associated with active α-globin transcription3,5. The formation of complexes in which multiple enhancers and promoters interact simultaneously therefore provides a structural basis for the observed functional cooperativity and also suggests a role for apparently redundant enhancer elements8,9. Such ‘shadow’ enhancers could have a structural function in forming and maintaining effective platforms for assembly of the transcriptional machinery and ensure the formation of robust complexes, even in the context of mutations or deletions in other enhancer elements.
This highlights that Tri-C analyses not only contribute to our fundamental understanding of the relationship between genome structure and function, but are also a valuable tool to interpret how genetic variations can disrupt complex chromatin structures and cause misregulation of gene expression and disease.
Online Methods
Cells
Primary murine ter119+ erythroid cells were obtained from spleens of female C57BL/6 mice treated with phenylhydrazine as previously described41. Mouse embryonic stem (ES) cells (129/Ola) were derived from mice at embryonic day 14 and cultured and harvested as previously described41. Protocols were approved through the Oxford University Local Ethical Review process. Experimental procedures were performed in accordance with European Union Directive 2010/63/EU and/or the UK Animals (Scientific Procedures) Act, 1986.
Hi-C – Experimental procedure
Hi-C in primary murine ter119+ erythroid cells was performed as previously described42. An equivalent dataset, generated following the same protocol, in mouse ES cells (Cast/129) was used for comparative analysis10.
Hi-C – Data analysis
Both Hi-C datasets were analyzed using the HiC-Pro pipeline43. Reads were aligned to the mm9 reference genome using Bowtie2, with minor modifications to the recommended options (erythroid: -k 3 --score-min L,−0.6,−0.2; ES: -k 3 --score-min L,−0.6,−0.6) to allow for multi-mapping in the duplicated regions in the globin loci and better alignment of the ES data in the β-globin region, which contains many SNPs in the Cast/129 strain compared to the mm9 reference. Despite these adjustments, alignment of the ES Hi-C data in the β-globin region remained somewhat compromised.
The erythroid Hi-C library was sequenced to a depth of 1,256,434,953 read pairs, which resulted in 714,421,216 valid interactions, with 63.3% of the interactions occurring in cis. These data are therefore suitable to display at 10–20 kb resolution. However, for comparative analysis with the ES Hi-C dataset (445,453,769 reads; 161,276,943 valid interactions; 51.3% of interactions in cis), we down-sampled the erythroid data to 156,921,018 valid interactions. The displayed contact matrices show interactions in both cell types at 40 kb resolution after ICE normalization implemented in the HiC-Pro pipeline. TADs were identified based on insulation indices using TADtool44.
Note that due to compromised mapping of the ES data at the β-globin locus, noise levels in the ES contact matrix of this region are relatively high. Though this complicates direct quantitative comparison of interactions between erythroid and ES cells, differences in domain structure are apparent.
Capture-C – Experimental procedure
Capture-C data were generated using the Next-Generation Capture-C protocol41,45. The DpnII restriction enzyme was used for digestion during 3C library preparation.
Because exclusion zones around all viewpoints analyzed in a multiplexed experiment are removed from analysis, we performed several Capture-C experiments to characterize the complete interaction landscapes of all cis-regulatory elements of interest in the globin loci. We used the following combinations of viewpoints in 6 independent experiments:
α-globin locus: R1 and R2 (enhancers); β-globin locus: HS1 and HS2 (enhancers);
α-globin locus: R3, Rm and R4 (enhancers); β-globin locus: HS3 and HS4 (enhancers);
α-globin locus: Hbq-1 and Hbq-2 (θ-globin promoters/CTCF-binding sites); β-globin locus: HS5 and HS6 (enhancers/CTCF-binding sites);
α-globin locus: HS-38 and HS-39 (upstream CTCF-binding sites); β-globin locus: HS-57 (enhancer), HS-60 and HS-90 (upstream CTCF-binding sites);
α-globin locus: HS+44 and HS+48 (downstream CTCF-binding sites); β-globin locus: 3’HS1 and 3’HS2 (downstream CTCF-binding sites);
α-globin locus: Il9r, Snrnp25, Rhbdf1, Mpg and Nprl3 (promoters);
To cover the remaining regions of the α- and β-globin loci for the generation of a high-resolution all vs all contact matrix, we performed an additional multiplexed experiment with viewpoints tiled across a 300 kb window around both globin clusters.
Capture oligonucleotides were designed using CapSequm41. Overviews of the viewpoint DpnII fragments in the α- and β-globin loci are shown in Supplementary Tables 1 and 2, respectively.
We used 3 independent biological replicates of primary murine ter119+ erythroid and ES cells in every experiment (derived from separate spleens and cultures, respectively), which were pooled after ligation of indexed sequencing adapters.
The generated Capture-C libraries were sequenced using Illumina sequencing platforms (V2 chemistry; 150 bp paired-end reads).
Capture-C – Data analysis
Capture-C data were analyzed as previously described41.
Briefly, reads were aligned to the mm9 reference genome using Bowtie1 with the following options: -p 1 -m 2 -v 3 --best --strata. The -m 2 option was used to allow for multi-mapping in the duplicated regions in the globin loci. The -v 3 option was used to allow for up to 3 mismatches to improve alignment of the ES data in the β-globin region, which contains many SNPs in the 129/Ola strain compared to the mm9 reference. However, as some SNPs are located in close proximity to or even in DpnII cut sites, alignment of reads with viewpoints in these DpnII fragments remained sub-optimal. The quality of some Capture-C profiles in the β-globin region in ES cells is therefore somewhat compromised, though still highly interpretable (Supplementary Tables 3, 4)41,46,47.
As PCR duplicates are removed during data analysis, Capture-C accurately quantifies chromatin interactions46,48. The Capture-C profiles in the figures represent the mean number of unique interactions per restriction fragment from 3 replicates, normalized for a total of 100,000 interactions on the chromosome analyzed, and scaled to 1,000.
Statistical analysis of differential Capture-C interactions in 3 replicates of erythroid and ES cells was performed using the Bioconductor package DESeq2, as previously described41. Statistically significant interactions indicated in Figure 1 and Supplementary Figure 2 represent restriction fragments with significantly different interaction frequencies in erythroid or ES cells, with a p-value < 0.01, adjusted for multiple testing.
Tri-C – Experimental procedure
The R2 enhancer and HS-39 CTCF-binding site in the α-globin locus, and the HS2 enhancer and 3’HS1 CTCF-binding site in the β-globin locus are located on small NlaIII restriction fragments (Supplementary Table 5). We therefore used the NlaIII restriction enzyme for digestion of 3C libraries, which were prepared as previously described41.
Briefly, aliquots of ~107 cells were fixed with 2% formaldehyde for 10 minutes, which has previously been shown to result in optimal cis/trans ratios and digestion efficiencies48. The reaction was quenched with 125 mM glycine, after which cells were washed in cold PBS, resuspended in cold lysis buffer (10 mM Tris pH 8, 10 mM NaCl, 0.2% Igepal CA-630, 1× cOmplete Protease Inhibitor Cocktail), incubated on ice for 20 minutes, and centrifuged. Pelleted cells were snap frozen, resuspended in restriction buffer and Dounce homogenized on ice. Cells were subsequently treated with 0.28% SDS for 1 hour and 1.67% Triton X-100 for 1 hour, before digestion with NlaIII. Three aliquots of 600 U of NlaIII restriction enzyme were added several hours apart over a total incubation time of 16–24 hours on a shaking thermomixer at 37 °C. The digestion reaction was heat-inactivated at 65 °C for 20 minutes and cooled on ice. Digested chromatin was ligated with 720 U of T4 DNA ligase overnight on a shaking thermomixer at 16 °C. Ligated DNA was de-crosslinked and Proteinase K (3 U) treated overnight at 65 °C. Samples were treated with RNase (7.5 mU) and DNA was purified using phenol-chloroform extraction and precipitation with ethanol and sodium acetate at −80 °C. After washing with 70% ethanol, the resulting 3C libraries were resuspended in water and stored at −20 °C.
To generate 3C library fragments containing multiple ligation junctions, we optimized the subsequent sonication step to generate DNA fragments of ~450 bp, using a Covaris S220 Focused Ultra-Sonicator (one cycle of 55 s; duty cycle: 10%; intensity: 4; cycles per burst: 200). DNA clean-up and size selection after sonication were performed with Ampure XP beads in a 0.7:1 bead-to-sample ratio. Illumina TruSeq adaptors were added using NEBNext DNA Library Prep reagents, according to the manufacturer’s protocol. DNA clean-up steps were performed with Ampure XP beads in a 1.8:1 bead-to-sample ratio to minimize loss of material. Adapter-ligated libraries were amplified and indexed using Agilent Herculase II reagents in a 7-cycle PCR reaction, according to the manufacturer’s protocol.
To enrich for 3C library fragments containing viewpoints of interest, we performed a double oligonucleotide capture step, using custom designed oligonucleotides targeting the middle of the viewpoint restriction fragments (Supplementary Table 5). The capture enrichment was performed in a single, multiplexed reaction, in which we pooled 1.5 μg of amplified, adapter-ligated material per library. The biotinylated capture oligonucleotides were pooled in equimolar ratios to a total concentration of 2.9 nM. The capture steps were performed using Nimblegen SeqCap EZ reagents, as previously described41.
We initially performed an experiment with 3 independent biological replicates of primary erythroid cells, which was sequenced using the Illumina MiSeq platform (V2 chemistry; 250 bp paired-end reads). In silico trimming of the 250 bp paired-end reads showed that there was little benefit of using 500 cycles of sequencing compared to 300 cycles for capturing multiple reporters in the reads. Therefore, to generate data at sufficient depth, we performed a second multiplexed experiment with 7 additional technical replicates (derived from 3 independent biological replicates) of erythroid and ES cells, which was sequenced using the Illumina NextSeq platform (V2 chemistry; 150 bp paired-end reads).
Tri-C – Data analysis
Tri-C data were analyzed using a combination of publicly available tools and customized scripts. Trim_galore (Babraham Institute, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to remove adapter sequences in the reads. Where possible, paired-end reads were reconstructed into single reads using FLASH with interleaved output settings49. A custom script was used to perform an in silico restriction enzyme digestion, after which the reads were aligned to the reference genome using Bowtie1 (-p 1 -m 2 -v 3 --best --strata). The aligned reads were analyzed with custom scripts to identify captured reads containing the targeted viewpoints of interest. PCR duplicates were removed by excluding reads that had the same start and end coordinates of each individual restriction fragment. Restriction fragments in unique captured reads were defined as interacting ‘reporter’ fragments if they were located outside a ~1 kb exclusion zone around the viewpoint restriction fragment. An overview of the numbers of detected reporters is shown in Supplementary Table 6. Reads with two or more reporters were used to calculate multi-way interaction counts between reporter fragments for each viewpoint. These interaction counts were normalized for a total of 1,000,000 interactions on the chromosome analyzed. To allow the data to be represented in symmetrical contact matrices, interaction counts were binned in 500 bp bins and corrected for the number of restriction fragments present in each bin. The contact matrices were scaled to 400–1,000 normalized interactions per bin, depending on the viewpoint analyzed. To allow for direct comparisons between erythroid and ES cells, equivalent scales were used to display the multi-way interactions with each viewpoint in both cell types.
Note that both globin genes are duplicated and that interactions with these genes and their promoters cannot be distinguished bioinformatically. Bowtie maps these duplicated sequences to either of the genes randomly, which explains the high interaction signal between the globin genes in both erythroid and ES cells. As these signals do not represent reliable interactions, they were not considered in the statistical analyses described below.
Statistical analysis of differential multi-way interactions in 10 erythroid and 7 ES replicates was performed using the Bioconductor package DESeq2. For these analyses, we considered interactions between all bins on the chromosome containing the viewpoint fragment of interest that contained interaction counts in at least 2 replicates. However, we would not expect to find chromosome-wide changes in multi-way interactions with cis-regulatory elements that normally only interact within a ~100 kb domain. The displayed p-values therefore represent p-values prior to multiple testing adjustment. However, even after adjusting for multiple testing, which is very stringent considering all possible triplets that can be formed between 500 bp bins on an entire chromosome, most reported significant interactions with R2 in the α-globin locus were significant at a level of p < 0.05, with the exception of the interactions between R3 and the promoters, and R4 and the promoters, which had an adjusted p-value < 0.1 (Fig. 4c). All reported significant differential interactions with HS2 in the β-globin locus were also significant at a level of p < 0.1 after multiple testing adjustment (Supplementary Fig. 9c). In contrast, all enriched interactions in Tri-C matrices from the viewpoint of the CTCF-binding site in both the α- and β-globin locus had adjusted p-values > 0.5 (Fig. 5c and Supplementary Fig. 10c). This indicates that CTCF-binding sites do not form specific higher-order structures in which they interact simultaneously. As the data depth for the CTCF viewpoints is similar to the enhancer viewpoints (Supplementary Table 6), it is unlikely that the absence of CTCF hubs in our data reflect insufficient power or coverage to detect such structures.
To calculate the enrichment of triplets containing two major globin enhancers and one promoter relative to the pair-wise interactions between these elements in both erythroid and ES cells, we counted the number of unique triplets containing all three elements in each replicate. We normalized these counts for a total of 1,000,000 reads containing triplets with the viewpoint of interest and calculated their frequencies relative to the normalized number of triplets containing pair-wise interactions with either of these elements. For example, in the α-globin locus we calculated the normalized R2-R1-Hba triplet counts and expressed these as a proportion of the normalized counts of triplets containing R2 and either R1 or Hba (Fig. 3d and Supplementary Fig. 9d). Using an unpaired, two-tailed t-test, we show that the numbers of R2-R1-Hba triplets in the α-globin locus and HS2-HS1-Hbb triplets in the β-globin locus are significantly enriched in erythroid cells (p = 0.0002 and p < 0.0001), even after accounting for increased pair-wise interactions between these elements (p = 0.0005 and p < 0.0001) (Fig. 3e and Supplementary Fig. 9e).
To investigate if the multi-way interactions with enhancers detected by Tri-C in erythroid cells are enriched over the underlying pair-wise contact distribution, we compared these Tri-C interactions to the pair-wise interactions detected by multiplexed Capture-C experiments (Fig. 4f and Supplementary Fig. 9f). As the resolution of the Capture-C contact matrices is 4 kb, we generated Tri-C matrices at comparable resolution. We normalized these matrices by extracting the regions of interest (α-globin: chr11:32,120,000–32,240,000; β-globin: chr7:110,930,000–111,070,000) and expressing the interactions counts relative to a total of 1,000 interactions in these regions. We calculated the average enrichment of multi-way contacts detected by Tri-C over the pair-wise distribution derived from Capture-C data and plotted these values in contact matrices. To examine the statistical significance of these enrichments, we compared 3 erythroid replicates and performed multiple unpaired, two-tailed t-tests, with the Holm-Sidak method to correct for multiple comparisons. As the Tri-C data are derived from a specific viewpoint of interest (one vs. all) and the multiplexed Capture-C data represent general interaction frequencies (all vs. all), we excluded the bins containing the R2 or HS2 viewpoint from analysis. Due to strong proximity signals in one vs. all 3C experiments, interactions between elements close to the viewpoint are expected to be enriched in Tri-C. For the bins neighboring the viewpoint bins, we therefore only considered bins with enrichments above the average enrichment of the entire row/column neighboring the viewpoint. We also excluded proximity contacts (< 4 kb, i.e. 1 bin form the matrix diagonal) from statistical analysis. We displayed statistically significant enriched interactions with a p-value < 0.01 after adjustment for multiple testing.
To examine whether multi-way interactions in triplets containing two major globin enhancers are more likely to occur with other cis-regulatory elements compared to all interactions with a single globin enhancer, we generated double-anchored interaction profiles, which display all fragments interacting in triplets containing two fixed viewpoints. We compared double-anchored R2-R1 interaction profiles in the α-globin locus and HS2-HS1 profiles in the β-globin locus to conventional interaction profiles containing fragments interacting with the R2 or HS2 viewpoint in all triplets. DESeq2 analysis based on raw counts of interacting restriction fragments in each of the 10 erythroid samples was used to analyze statistical significance of enriched multi-way interactions. Displayed p-values represent adjusted p-values after adjustment for multiple testing.
C-Trap – Experimental procedure
3C libraries were prepared as previously described41, using the DpnII restriction enzyme for digestion. Primers targeting the restriction fragments of interest were designed > 100 bp away from the restriction site (Supplementary Table 7), to allow for the selection of reads that were specifically amplified and of sufficient quality. PCR amplification was performed using the Takara Prime Star GXL 2-step amplification program for 10–30 kb amplicons, according to the manufacturer’s protocol. The Agencourt AMPure XP system was used to purify the PCR product and select fragments > 700 bp. The purified amplicons were adapter-ligated and sequenced using the Oxford Nanopore MinION platform (MAP-005 chemistry).
C-Trap – Data analysis
The MinION reads were basecalled using Metrichor and converted to fasta format using Poretools50. To select reads that were specifically amplified and of sufficient quality, a BLAST search against a database containing the sequences between the primers and the restriction sites of the targeted fragments was performed. Reads that matched > 60% of both primer sequences were selected. To map the trapped restriction fragments in the reads, a BLAST search against a database containing all the DpnII fragments in the genome was used. Custom scripts were used to iterate through the BLAST results and select the non-overlapping BLAST matches in the reads with a match > 60% of fragment length or > 100 bp (to avoid a skew towards smaller fragments) in order of significance of the BLAST scores. A summary of the read statistics is shown in Supplementary Table 8.
Statistical analyses
Statistical significance of differential pair-wise (Capture-C) and multi-way (Tri-C) interactions between erythroid and ES cells was analyzed using the Bioconductor package DESeq251. This method tests for differential signals by use of negative binomial generalized linear models, which is a common approach for the analysis of count-based data generated by next-generation sequencing. As DESeq2 normalizes the data internally, we ran the model using raw interaction counts per restriction fragment (Capture-C) or bin (Tri-C). The remaining statistical analyses were performed with Student’s two-tailed t tests using GraphPad Prism software.
URLs
Analyzed Capture-C data from all the viewpoints in the α- and β-globin loci are available in UCSC Genome Browser Track Hubs and can be accessed with the following links:
http://sara.molbiol.ox.ac.uk/public/hugheslab/CaptureC_alpha_globin/hub.txt
http://sara.molbiol.ox.ac.uk/public/hugheslab/CaptureC_beta_globin/hub.txt
Custom scripts used for the analysis of Capture-C and Tri-C data are available at:
Reporting Summary
Further information on experimental design and statistical analysis is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (Wellcome Trust Genomic Medicine and Statistics PhD Programme, reference 105281/Z/14/Z, A.M.O.; Wellcome Trust Strategic Award, reference 106130/Z/14/Z, D.R.H. and J.R.H.), the Medical Research Council (MRC Core Funding and Project Grant, reference MR/N00969X/1, V.J.B. and J.R.H.) and the National Institutes of Health (reference R01 HG003143, J.D.). J.D. is an investigator of the Howard Hughes Medical Institute.
We thank D. Hay and M. Kassouf for advice and R. Gibbons and H. Ayyub for help with Oxford Nanopore MinION sequencing.
Footnotes
Accession Codes
All sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107940.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Sanyal A, Lajoie BR, Jain G & Dekker J The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanssen LLP et al. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nature Cell Biology 19, 952–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JM A tissue-specific self-interacting chromatin domain forms independently of enhancer-promoter interactions. 1–1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oudelaar AM et al. Between form and function: the complexity of genome folding. Hum. Mol. Genet 26, R208–R215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay D et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat Genet 48, 895–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender MA et al. The hypersensitive sites of the murine β-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood 119, 3820–3827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osterwalder M et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel N et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry MW, Boettiger AN, Bothma JP & Levine M Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol 20, 1562–1567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giorgetti L et al. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tolhuis B, Palstra R-J, Splinter E, Grosveld F & de Laat W Looping and Interaction between Hypersensitive Sites in the Active β-globin Locus. Molecular Cell 10, 1453–1465 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Nagano T et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flyamer IM et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens TJ et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beagrie RA et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinodoz SA et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell (2018). doi:10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang T et al. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic Acids Research 44, 8714–8725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darrow EM et al. Deletion of DXZ4on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci USA 113, E4504–E4512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ay F et al. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics 16, 121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavrilov AA, Chetverina HV, Chermnykh ES, Razin SV & Chetverin AB Quantitative analysis of genomic element interactions by molecular colony technique. Nucleic Acids Research 42, e36–e36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivares-Chauvet P et al. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 540, 296–300 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Rao SSP et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasmyth K Disseminating the Genome: Joining, Resolving, and Separating Sister Chromatids During Mitosis and Meiosis. Annual Review of Genetics 35, 673–745 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Fudenberg G et al. Formation of Chromosomal Domains by Loop Extrusion. CellReports 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanborn AL et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A 112, E6456–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao SSP et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wutz G et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen AS, Pustova I, Cattoglio C, Tjian R & Darzacq X CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6, e25776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagey MH et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes J, Mazza D, Nasmyth K & Uphoff S Scc2/Nipbl hops between chromosomal cohesin rings after loading. Elife 6, 11202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allahyar A et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet 295, 1306 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Giorgetti L et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157, 950–963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiariello AM, Annunziatella C, Bianco S, Esposito A & Nicodemi M Polymer physics of chromosome large-scale 3D organisation. Scientific Reports 6, 29775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbieri M et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci USA 109, 16173–16178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cattoni DI et al. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nature Communications 8, 1753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vian L et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell 173, 1165–1178.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dekker J & Mirny L The 3D Genome as Moderator of Chromosomal Communication. Cell 164, 1110–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbieri M et al. Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol 109, 1427 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 41.Davies JOJ et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Meth 86, 1202–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman-Aiden E et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Servant N et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kruse K, Hug CB, Hernández-Rodríguez B & Vaquerizas JM TADtool: visual parameter identification for TAD-calling algorithms. Bioinformatics 32, 3190–3192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes JR et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet 46, 205–212 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Davies JOJ, Oudelaar AM, Higgs DR & Hughes JR How best to identify chromosomal interactions: a comparison of approaches. Nat Meth 14, 125–134 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Schwartzman O et al. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat Meth 13, 685–691 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Oudelaar AM, Davies JOJ, Downes DJ, Higgs DR & Hughes JR Robust detection of chromosomal interactions from small numbers of cells using low-input Capture-C. Nucleic Acids Research (2017). doi:10.1093/nar/gkx1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magoč T & Salzberg SL FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loman NJ & Quinlan AR Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30, 3399–3401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.