Abstract
Cytochrome P450 3A5 (CYP3A5) and cytochrome P450 3A4 (CYP3A4) are the predominate enzymes responsible for tacrolimus metabolism. The presence of CYP3A4 and CYP3A5 genetic variants significantly affects tacrolimus clearance and dose requirements. CYP3A5*3 is a loss-of-function variant resulting in no CYP3A5 enzyme production. CYP3A4*22 is a variant that reduces production of functional CYP3A4 protein. Caucasians commonly carry these variant alleles but are very rarely homozygous for both CYP3A5*3 and CYP3A4*22. This report describes four kidney transplant recipients who carry a rare genotype combination (CYP3A5*3/*3 and CYP3A4*22/*22). These patients were identified from a larger cohort of Caucasian kidney transplant recipients (n=1366). To understand the significance of this genotype combination on tacrolimus troughs and doses, we compared these patients to recipients without this combination. Patients homozygous for both variants are at risk for profound reductions in metabolism of CYP3A substrates. A 342% and a 90.6% increase in the median dose-normalized trough was observed, when the CYP3A5*3/*3 and CYP3A4*22/*22 genotype combination was compared to the CYP3A5*1/*1 and CYP3A4*1/*1 genotype combination and the CYP3A5*3/*3 and CYP3A4*1/*1 genotype combination, respectively. These four individuals only required on average 2.5 mg/day of tacrolimus. Knowledge of these genotypes would be useful in selecting appropriate tacrolimus doses to avoid overexposure.
Keywords: CYP3A5, CYP3A4, tacrolimus, single nucleotide polymorphisms, Transplant, kidney, pharmacogenetics
Tacrolimus is a highly effective immunosuppressant used as maintenance therapy after solid organ transplantation. Tacrolimus pharmacokinetics are variable mainly due to unpredictable bioavailability and metabolism requiring therapeutic monitoring of trough concentrations.1 Low bioavailability is largely attributed to high first-pass metabolism and efflux from intestinal cells. It is extensively metabolized in the gut and liver with < 0.5% excreted unmetabolized in the feces.1 Biliary excretion is responsible for approximately 95% of tacrolimus metabolite excretion, and urinary excretion accounts for < 3%.1 Cytochrome P450 3A5 (CYP3A5) and cytochrome P450 3A4 (CYP3A4) are thought to be the predominate isozymes contributing to tacrolimus metabolism.2, 3 Four major metabolites (13-O-desmethyl tacrolimus, 15-O-desmethyl tacrolimus, 31-O-desmethyl tacrolimus, and 12-O-desmethyl tacrolimus) account for the majority of tacrolimus clearance by CYP3A.4 When CYP3A5 is expressed, it has twice the intrinsic clearance of CYP3A4.5 The relative contribution of CYP3A5 versus CYP3A4 within an individual is highly dependent on the presence of CYP3A4 or CYP3A5 activity.6
The presence of CYP3A4 or CYP3A5 variants significantly influences the metabolism of tacrolimus.7 A single-nucleotide polymorphism (SNP) in intron 3 of the CYP3A5 gene (CYP3A5*3, rs776746) significantly reduces tacrolimus clearance and lowers dosing requirements.8, 9 CYP3A5*3 is a loss-of-function variant that leads to the production of a nonfunctional protein caused by creation of a cryptic splice site resulting in incorporation of exon 3B.10 This leads to CYP3A5 nonsense–mediated mRNA decay.10, 11 The genetic variant CYP3A4*22 (rs35599367), in which a C-to-T substitution in intron 6 deletes a SF2/AF2 binding site, has also been associated with reduced tacrolimus metabolism.12, 13 It results in 255–base pair partial intron 6 retention and a splice variant that terminates before exon 7, leading to reduced production of functional mRNA and enzyme activity when compared to the wild-type. When the CYP3A4*22 variant was transfected into HepG2 cells, higher alternative splice variant formation was observed. No difference was observed when this variant is expressed in intestine-derived LS-174T cells, suggesting a tissue-specific effect.12, 13
A previous case report described tacrolimus troughs in a kidney transplant recipient with Alport syndrome who was homozygous for the CYP3A5*3 allele and homozygous for a variant in exon 5 of the CYP3A4 gene (802 C>T, CYP3A4*26) that results in a premature stop codon R268*.14 This patient had extremely high tacrolimus trough concentrations and was eventually stabilized on delayed-release tacrolimus 0.5 mg 3 times weekly, leading to trough concentrations between 5 and 10 ng/ml.
This case report describes four kidney transplant recipients who carry a rare genotype combination (CYP3A5*3/*3 and CYP3A4*22/ *22). To understand the significance of this genotype combination on tacrolimus troughs and doses, we compared these four patients to kidney transplant recipients without this combination.
Materials and Methods
Genotypes from 1366 Caucasian kidney transplant recipients enrolled in the multicenter Deterioration of Kidney Allograft Function (DeKAF) Genomics study (www.ClinicalTrials.gov, NCT01714440) were evaluated. Deterioration of Kidney Allograft Function Genomics is a prospective, observational study to identify genetic variants associated with transplant outcomes through a genome-wide association study (GWAS). The 1366 Caucasian kidney transplant recipients were divided into 6 groups based on their CYP3A5*3 and CYP3A4*22 genotype combination. From this cohort, four transplant recipients who were homozygous for both CYP3A5*3 and CYP3A4*22 alleles were identified. These patients would be expected to have profoundly reduced tacrolimus clearance since tacrolimus is both a CYP3A4 and CYP3A5 substrate. To understand the influence of this genotype, tacrolimus troughs, doses, and dose-normalized troughs in the first 6 months post transplant of these four recipients were compared to patients not carrying this genotype combination. All 1366 patients received oral immediate-release tacrolimus therapy with mycophenolate for maintenance immunosuppression post transplant. Induction therapy varied among the patients. Most patients received a short course of induction corticosteroids at time of transplant and no steroid maintenance. Tacrolimus trough concentrations were obtained as part of routine clinical care in all patients and were measured from whole blood. Tacrolimus concentrations were measured with Clinical Laboratory Improvement Amendments (CLIA)-approved assays and 95% were measured by liquid chromatography–mass spectrometry. Two trough concentrations were obtained in each of weeks 1–8, and in each of months 3–6, for a maximum of 24 trough concentrations per subject. Tacrolimus doses were adjusted based on institution-specific trough goals; generally, the goals were 8–12 ng/ml in months 0–3 and 6–10 ng/ml in months 4–6. Tacrolimus troughs and doses were obtained from the medical record in all patients. Dose-normalized trough values were calculated using the trough concentration (ng/ml) divided by the total daily dose (mg). All patients were also evaluated for acute tacrolimus-related nephrotoxicity, which has been previously described.15 Tacrolimus-related nephrotoxicity was defined as any rise in serum creatinine that resulted in lowering of the tacrolimus dose. The study was carried out in accordance with the Declaration of Helsinki and was approved by the institutional review boards of the enrolling centers. Statistical analyses were not conducted given the small number of patients in groups 1 and 6.
Concomitant use of angiotensin-converting enzyme inhibitors (ACEi), anti-cytomegalovirus (CMV) prophylaxis, dihydropyridine calcium channel blockers (CCBs, mainly amlodipine), and corticosteroids was recorded for all patients. In the four patients of interest, time on one of these concurrent therapies was calculated and defined as the percentage of time the patient reported taking a concurrent medication around the time of tacrolimus trough measurements.
The patients’ genotypes were determined from DNA isolated from pretransplant peripheral blood lymphocytes. Genotypes were determined with the Affymetrix Transplant GWAS chip and have been previously described.16, 17 The CYP3A5*3 (rs776746) and CYP3A4*22 (rs35599367) genotypes were taken from this chip.
Results
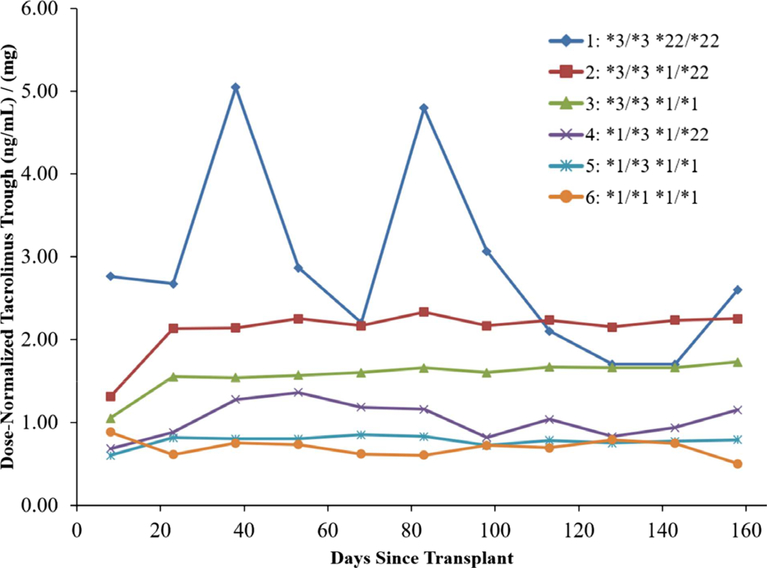
The 1366 transplant recipients were divided into six groups based on their CYP3A5*3 and CYP3A4*22 allele status. Group 1 consisted of the four subjects with the CPY3A5*3/*3 and CYP3A4*22/*22 genotype combination. Groups 2–6 contained recipients with the CYP3A5*3/*3 and CYP3A4*1/*22, CYP3A5*3/*3 and CYP3A4*1/*1, CYP3A5*1/*3 and CYP3A4*1/*22, CYP3A5*1/*3 and CYP3A4*1/*1, and CYP3A5*1/ *1 and CYP3A4*1/*1 genotype combinations, respectively. Table 1 describes the demographics, genotype distribution, dose-normalized tacrolimus trough concentrations, trough concentrations, and total daily dose for each group. Group 3 (CYP3A5*3/*3 and CYP3A4*1/*1) and group 5 (CYP3A5*1/*3 and CYP3A4*1/*1) had the most common genotypes, representing 76.7% and 11.8% of the study population, respectively. Group 1 accounted for only 0.29% of the population. Figure 1 displays the median dose-normalized trough concentrations for groups 1–6.
Table 1.
Comparison of Caucasian Patients’ Demographics, Dose-Normalized Tacrolimus Trough Concentrations, Tacrolimus Trough Concentrations, and Total Daily Tacrolimus Dose by CYP3A5 and CYP3A4 Genotype Status in the First 6 Months Post transplant
| Group 1 (n=4, 0.29%) | Group 2 (n=133, 9.7%) | Group 3 (n=1048, 76.7%) | Group 4 (n=13, 0.95%) | Group 5 (n=161, 11.8%) | Group 6 (n=7, 0.51%) | |
|---|---|---|---|---|---|---|
| Genotype | CYP3A5*3/*3 and CYP3A4 *221*22 | CYP3A5*3/*3 and CYP3A4*1/*22 | CYP3A5*3/*3 and CYP3A4*1/*1 | CYP3A5*1/*3 and CYP3A4*1/*22 | CYP3A5*1/*3 and CYP3A4*1/*1 | CYP3A5*1/*1 and CYP3A4*1/*1 |
| Age, mean, SD (yrs) | 49.9, 13.6 | 51.9, 13.1 | 51.2, 12.8 | 47.4, 13.4 | 51.3, 14.0 | 58.7, 13.4 |
| BMI, mean, SD (kg/m2) | 27.8, 3.7 | 27.9, 5.1 | 28.3, 5.6 | 28.4, 4.8 | 27.8, 5.2 | 26.7, 5.6 |
| Weight, mean, SD (kg) | 84.2, 13.1 | 82.5, 17.9 | 83.9, 20.0 | 88.1, 19.0 | 83.8, 18.3 | 73.5, 20.8 |
| Living donor transplant (%) | 75 | 63.2 | 66.5 | 53.9 | 68.3 | 71.4 |
| Female (%) | 25 | 37.6 | 37.2 | 23.1 | 32.9 | 57.1 |
| Diabetes at time of transplant (%) | 25 | 33.1 | 39.7 | 46.2 | 36.6 | 42.9 |
| Simultaneous pancreas kidney transplant (%) | 0 | 6.0 | 9.0 | 7.7 | 8.1 | 0 |
| Dose-normalized tacrolimus trough, median, IQR (ng/ml/mg of total daily dose) | 3.05, 2.12–1.32 | 2.18, 1.50–3.33 | 1.60, 1.10–2.40 | 1.06, 0.65–1.50 | 0.80, 0.56–1.20 | 0.69, 0.52–1.15 |
| Tacrolimus trough, median, IQR (ng/ml) | 8.25, 6.45–10.50 | 8.70, 7.00–10.60 | 8.50, 6.80–10.5 | 8.00, 6.20–9.50 | 7.80, 5.90–9.70 | 5.70, 4.10–8.40 |
| Daily tacrolimus dose, median, IQR (mg) | 2.50, 2.00–3.00 | 4.00, 2.50–6.00 | 5.00, 4.00–8.00 | 7.00, 5.00–12.00 | 9.00, 6.00–13.00 | 8.00, 6.00–13.00 |
BMI = body mass index; IQR = interquartile range; SD = standard deviation.
Figure 1.
Median dose-normalized trough concentrations for group 1 (CYP3A5*3/*3 and CYP3A4*22/*22, diamond, n=4), group 2 (CYP3A5*3/*3 and CYP3A4*1/*22, square, n=133), group 3 (CYP3A5*3/*3 and CYP3A4*1/*1, triangle, n=1048), group 4 (CYP3A5*1/*3 and CYP3A4*1/*22, “x,” n=13), group 5 (CYP3A5*1/*3 and CYP3A4*1/*1, cross, n=161), and group 6 (CYP3A5*1/*1 and CYP3A4*1/*1, circle, n=7) calculated by 15-day intervals spanning the first 6 months post transplant. [Color figure can be viewed at wileyonlinelibrary.com]
Group 1 (CYP3A5*3/*3 and CYP3A4*22/*22) had the highest dose-normalized tacrolimus troughs (3.05 ng/ml/mg) and received the lowest dose (2.5 mg/day) of the six groups. Dose-normalized tacrolimus troughs incrementally increased with each addition of a variant allele (CYP3A5*3 and CYP3A4*22). The median tacrolimus dose in group 1 (2.5 mg/day) was > 3-fold lower relative to groups 5 and 6 (8–9 mg/day). For the patients in group 1, the median tacrolimus trough was similar (8.25 ng/ml) to groups 2–5 and higher compared to group 6 (5.70 ng/ ml) with no variants.
The number of CYP3A5*22 alleles present was important. The addition of a second CYP3A4*22 allele increased the median dose-normalized tacrolimus trough from 2.18 to 3.05 ng/ml/mg (40.2% increase) when comparing group 2 (CYP3A5*3/*3 and CYP3A4*1/*22) to group 1 (CYP3A5*3/*3 and CYP3A4*22/*22), respectively. A 90.6% increase (1.60 ng/ml/mg compared to 3.05 ng/ml/mg) in the median dose-normalized trough was observed when comparing group 3 (CYP3A5*3/*3 and CYP3A4*1/*1, the most frequent allele combination) to group 1 (CYP3A5*3/ *3 and CYP3A4*22/*22). Furthermore, a 342% higher (3.05 ng/ml/mg vs 0.69 ng/ml/mg) median dose-normalized trough was observed in group 1 (CYP3A5*3/*3 and CYP3A4*22/*22) relative to group 6 (CYP3A5*1/*1 and CYP3A4*1/*1).
One of the four patients in group 1 experienced tacrolimus-induced nephrotoxicity (defined as any rise in serum creatinine that resulted in lowering of the tacrolimus dose) by day 25. In the overall cohort, acute tacrolimus-related nephrotoxicity occurred in 16% of the population. In group 1, one of the four patients received corticosteroids for maintenance therapy, whereas the three others reported no corticosteroids use except for a short course with induction. ACEi, anti-CMV prophylaxis, and CCB use varied among the four patients in group 1 (Table 2).
Table 2.
Time on Concurrent Drug Therapies in Group 1 Patients in the First 6 Months Post transplant
| Group 1 – Time on Concurrent Drug Therapies (%) |
||||
|---|---|---|---|---|
| Therapy | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
| ACEi | 60 | 0 | 0 | 0 |
| Anti-CMV | 0 | 66.7 | 64.3 | 27.8 |
| Maintenance corticosteroids | 0 | 0 | 0 | 100 |
| CCB | 70 | 33.3 | 35.7 | 61.1 |
Concurrent drug therapies of ACEi, anti-CMV therapy, maintenance corticosteroids, and dihydropyridine CCBs were recorded at time of tacrolimus trough measurement. Time on a concurrent drug therapy was defined as the percentage of time the patient reported taking a medication in each drug category. ACEi = angiotensin-converting-enzyme inhibitors; CCB = calcium channel blocker; CMV = cytomegalovirus.
Discussion
The median dose-normalized tacrolimus troughs increased with the accumulation of inactive or reduced activity CYP3A5 and CYP3A4 alleles (Figure 1). The individuals homozygous for both CYP3A5*3 and CYP3A4*22 had the lowest total daily tacrolimus dose requirements (2.5 mg/day) compared to all the other groups. This is a rare genotype combination and occurred in only 0.29% of our study population. The CYP3A4*22 allele frequency in African and European populations is approximately 0% and 5%, respectively (phase3browser.1000genomes.org). The CYP3A5*3 allele frequency in African and European populations is approximately 18% and 94%, respectively (phase3browser.1000genomes.org). We believe, after an extensive literature review, that this is the first in-depth description of patients who are homozygous for both the CYP3A5*3 and CYP3A4*22 alleles receiving tacrolimus. Therefore, little is known about this genotype combination. CYP3A5*3 and CYP3A4*22 allele status can explain up to 50% of dose-normalized tacrolimus trough concentration variability in kidney transplant recipients.18 In a mixed-model analysis, dose-normalized tacrolimus trough concentrations were reported to be 43% higher in CYP3A4*22 carriers than in non carriers and 43.3% higher in CYP3A5*3 homozygotes than in CYP3A5*1 carriers after adjusting for age, sex, and ethnicity.19 In another study, similar tacrolimus trough concentrations were achieved with a 33% lower daily tacrolimus dose in individuals with CYP3A4*1/*22 genotype compared to the homozygote wildtype (CYP3A4*1/*1).19 Individuals homozygous for the CYP3A5*3 allele, who also carried one CYP3A4*22 allele, were reported to have a 26% higher dose-adjusted tacrolimus trough concentrations compared to those who did not carry a CYP3A4*22 allele.20 Other investigators also found that European white kidney transplant recipients who carried one CYP3A4*22 allele required lower tacrolimus doses.18, 19 A study in Dutch and German kidney transplant patients found that individuals who were CYP3A5*3 homozygotes and also carried CYP3A4*22 alleles required lower mean tacrolimus doses than those who were CYP3A5*1 carriers who also carried one or two CYP3A4*22 alleles (4.8 and 12.5 mg/ day, respectively).21 Researchers observed higher dose-normalized tacrolimus trough concentrations at 3 months post transplant in individuals carrying a CYP3A4*22 allele when compared to CYP3A4*1 homozygotes.22 These studies highlight the important effect of CYP3A5*3 and CYP3A4*22 variants on reducing tacrolimus clearance.
An in vitro study evaluating the effect of CYP3A5 polymorphisms on tacrolimus metabolic clearance showed that the formation of a major inactive tacrolimus metabolite, 13-O-desmethyl tacrolimus, was 13.5 times greater in human liver microsomes from CYP3A5*1 carriers compared to CYP3A5*3 homozygotes.5 This suggests that metabolite formation may be altered in transplant recipients with variants. The effect of CYP3A4*22 on tacrolimus metabolite formation is not known. The clinical implication of differing metabolite formation has not been studied.
We predicted tacrolimus elimination would be nearly absent in the four individuals who carried the rare genotype combination (CYP3A5*3/*3 and CYP3A4*22/*22); however, tacrolimus was eliminated in these patients, albeit substantially less than in the other groups. This may be explained by several possibilities: (i) despite two CYP3A4*22 alleles, partially active enzyme is produced and facilitates clearance; (ii) compensatory tacrolimus metabolism via other CYP enzymes or tacrolimus glucuronidation occurs; (iii) increased elimination of non metabolized tacrolimus occurs in the presence of reduced CYP3A4 and CYP3A5 enzyme activity; and (iv) the CYP3A4*22 variant reduces only hepatic enzyme, leaving intact CYP3A4 intestinal metabolism.
Investigators have found that CYP3A4*22 variant splicing may be liver specific13; therefore, tacrolimus metabolism in our patients may have occurred through intestinal CYP3A4 activity. Our patients did not receive intravenous tacrolimus, so we were unable to distinguish intestinal epithelial cell metabolism versus hepatic metabolism in these four patients. Other enzymes also may contribute to tacrolimus metabolism, including UGT1A4-mediated glucuronidation and/or other CYP enzymes. Recently, the existence of a tacrolimus-glucuronide metabolite was confirmed in vivo in human bile, indicating that tacrolimus may undergo enterohepatic circulation.23 In addition, tacrolimus is a known inhibitor of CYP2C8, and CYP2C8 genotypes have been linked to nephrotoxicity and delayed graft function among patients receiving tacrolimus, suggesting it may be a substrate for CYP2C8.24, 25 The impact of CYP2J2 on tacrolimus pharmacokinetics has also been studied, but was not linked to any clinical outcomes.24, 25
The seven patients identified in group 6 also present a unique subset as the CYP3A5*1/*1 and CYP3A4*1/*1 genotype combination was also rare (0.51%) in our patients of Caucasian descent. Individuals of African ancestry commonly carry the CYP3A5*1/*1 genotype, and this explains why they have significantly lower tacrolimus trough concentrations than Caucasians. The CYP3A5*1/*1 genotype is well known to lead to higher rates of tacrolimus clearance in humans17 and in cell culture studies.17, 26 These individuals are also of concern with a median tacrolimus trough of only 5.70 ng/ml (target generally > 8 ng/ml) that places them at higher risk for transplant rejection.
Drug–gene or drug–drug interactions may play a role in our reported observation. Exogenous glucocorticoid steroids are believed to induce CYP3A via the pregnane X receptor (PXR).27 All patients received corticosteroids beginning at the time of transplant, and only one of the four patients in group 1 received corticosteroids throughout the first 6 months. Initially, corticosteroids may have induced CYP3A, resulting in a phenoconversion to a higher metabolism phenotype in the first 1–2 weeks post transplant. CYP3A induction may have allowed patients in group 1 to tolerate the higher tacrolimus doses they initially received while doses were titrated down.
Tacrolimus is therapeutically monitored to identify out-of-range blood concentrations and allow for dose adjustments accordingly. However, most CYP3A substrate drugs are not monitored. The clinical implication of no or dramatically reduced CYP3A5 and CYP3A4 activity may be serious as these enzymes are widely involved in drug metabolism. The frequency of individuals who do not express CYP3A5 and are homozygous for the CYP3A4*22 allele is rare (0.29% in our population), but the implications for this subset of patients could be serious.
Definitive conclusions about the impact of two CYP3A4*22 alleles on drug metabolism will need to come from larger populations and additional patients, but this will likely be difficult due to the infrequency of this genotype combination. In addition, DeKAF Genomics did not collect information regarding hepatic function. Therefore, the influence of liver function on tacrolimus metabolism could not be accounted for in this case report. Our data suggest that individuals homozygous for both CYP3A5*3 and CYP3A4*22 display a reduced ability to eliminate tacrolimus compared to those heterozygous and homozygous for the wild-types. This case report contributes to the pharmacogenomic considerations in patients receiving tacrolimus and to understanding the metabolism phenotype produced by the combination of CYP3A5*3/*3 and CYP3A4*22/*22 genotypes.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
- 1.Möller A, Iwasaki K, Kawamura A, et al. The disposition of 14C-labeled tacrolimus after intravenous and oral administration in healthy human subjects. Drug Metab Dispos 1999;27 (6):633–6. [PubMed] [Google Scholar]
- 2.de Jonge H, de Loor H, Verbeke K, Vanrenterghem Y, Kuypers DR. In vivo CYP3A4 activity, CYP3A5 genotype, and hematocrit predict tacrolimus dose requirements and clearance in renal transplant patients. Clin Pharmacol Ther 2012;92 (3):366–75. [DOI] [PubMed] [Google Scholar]
- 3.Staatz CE, Tett SE. Clinical pharmacokinetics and pharmaco-dynamics of tacrolimus in solid organ transplantation. Clin Pharmacokinet 2004;43(10):623–53. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki K, Shiraga T, Nagase K, et al. Isolation, identification, and biological activities of oxidative metabolites of FK506, a potent immunosuppressive macrolide lactone. Drug Metab Dispos 1993;21(6):971–7. [PubMed] [Google Scholar]
- 5.Dai Y, Hebert MF, Isoherranen N, et al. Effect of CYP3A5 polymorphism on tacrolimus metabolic clearance in vitro. Drug Metab Dispos 2006;34(5):836–47. [DOI] [PubMed] [Google Scholar]
- 6.Kamdem LK, Streit F, Zanger UM, et al. Contribution of CYP3A5 to the in vitro hepatic clearance of tacrolimus. Clin Chem 2005;51(8):1374–81. [DOI] [PubMed] [Google Scholar]
- 7.Elens L, Bouamar R, Shuker N, Hesselink DA, van Gelder T, van Schaik RH. Clinical implementation of pharmacogenetics in kidney transplantation: calcineurin inhibitors in the starting blocks. Br J Clin Pharmacol 2014;77(4):715–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haufroid V, Mourad M, Van Kerckhove V, et al. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics 2004;14 (3):147–54. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson PA, Oetting WS, Brearley AM, et al. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation 2011;91(3):300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 2001;27(4):383–91. [DOI] [PubMed] [Google Scholar]
- 11.Busi F, Cresteil T. CYP3A5 mRNA degradation by nonsense-mediated mRNA decay. Mol Pharmacol 2005;68(3): 808–15. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 2011;11 (4):274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Sadee W. CYP3A4 intronic SNP rs35599367 (CYP3A4*22) alters RNA splicing. Pharmacogenet Genomics 2016;26(1):40–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werk AN, Lefeldt S, Bruckmueller H, et al. Identification and characterization of a defective CYP3A4 genotype in a kidney transplant patient with severely diminished tacrolimus clearance. Clin Pharmacol Ther 2014;95(4):416–22. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson PA, Schladt D, Israni A, et al. Genetic and clinical determinants of early, acute calcineurin inhibitor-related nephrotoxicity: results from a kidney transplant consortium. Transplantation 2012;93(6):624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li YR, van Setten J, Verma SS, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med 2015;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oetting WS, Schladt DP, Guan W, et al. Genomewide Association study of tacrolimus concentrations in African American kidney transplant recipients identifies multiple CYP3A5 alleles. Am J Transplant 2016;16(2):574–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elens L, van Schaik RH, Panin N, et al. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics 2011;12(10):1383–96. [DOI] [PubMed] [Google Scholar]
- 19.Elens L, Bouamar R, Hesselink DA, et al. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem 2011;57(11):1574–83. [DOI] [PubMed] [Google Scholar]
- 20.Lloberas N, Elens L, Llaudo I, et al. The combination of CYP3A4*22 and CYP3A5*3 single-nucleotide polymorphisms determines tacrolimus dose requirement after kidney transplantation. Pharmacogenet Genomics 2017;27(9):313–22. [DOI] [PubMed] [Google Scholar]
- 21.Bruckmueller H, Werk AN, Renders L, et al. Which genetic determinants should be considered for tacrolimus dose optimization in kidney transplantation? A combined analysis of genes affecting the CYP3A locus. Ther Drug Monit 2015;37 (3):288–95. [DOI] [PubMed] [Google Scholar]
- 22.Kurzawski M, Dąbrowska J, Dziewanowski K, Domański L, Perużyńska M, Droździk M. CYP3A5 and CYP3A4, but not ABCB1 polymorphisms affect tacrolimus dose-adjusted trough concentrations in kidney transplant recipients. Pharmacogenomics 2014;15(2):179–88. [DOI] [PubMed] [Google Scholar]
- 23.Tron C, Petitcollin A, Verdier MC, et al. Tacrolimus: does direct glucuronidation matter? An analytical and pharmacological perspective. Pharmacol Res 2017;124:164–6. [DOI] [PubMed] [Google Scholar]
- 24.Smith HE, Jones JP, Kalhorn TF, et al. Role of cytochrome P450 2C8 and 2J2 genotypes in calcineurin inhibitor-induced chronic kidney disease. Pharmacogenet Genomics 2008;18 (11):943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gervasini G, Garcia M, Macias RM, Cubero JJ, Caravaca F, Benitez J. Impact of genetic polymorphisms on tacrolimus pharmacokinetics and the clinical outcome of renal transplantation. Transpl Int 2012;25(4):471–80. [DOI] [PubMed] [Google Scholar]
- 26.Dorr CR, Remmel RP, Muthusamy A, et al. CRISPR/Cas9 genetic modification of CYP3A5*3 in HuH-7 human hepatocyte cell line leads to cell lines with increased midazolam and tacrolimus metabolism. Drug Metab Dispos 2017;45(8):957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodwin B, Redinbo MR, Kliewer SA. Regulation of CYP3A gene transcription by the pregnane X receptor. Annu Rev Pharmacol Toxicol 2002;42:1–23. [DOI] [PubMed] [Google Scholar]