Abstract
Purpose:
Computed tomography (CT) images enable capturing specific manifestations of tuberculosis (TB) that are undetectable using common diagnostic tests, which suffer from limited specificity. In this study, we aimed to automatically quantify the burden of Mycobacterium tuberculosis (Mtb) using biomarkers extracted from x-ray CT images.
Procedures:
Nine macaques were aerosol-infected with Mtb and treated with various antibiotic cocktails. Chest CT scans were acquired in all animals at specific times independently of disease progression. First, a fully automatic segmentation of the healthy lungs from the acquired chest CT volumes was performed and air-like structures were extracted. Next, unsegmented pulmonary regions corresponding to damaged parenchymal tissue and TB lesions were included. CT biomarkers were extracted by classification of the probability distribution of the intensity of the segmented images into three tissue types: (1) Healthy tissue, parenchyma free from infection; (2) soft diseased tissue, and (3) hard diseased tissue. The probability distribution of tissue intensities was assumed to follow a Gaussian mixture model. The thresholds identifying each region were automatically computed using an expectation-maximization algorithm.
Results:
The estimated longitudinal course of TB infection shows that subjects that have followed the same antibiotic treatment present a similar response (relative change in the diseased volume) with respect to baseline. More interestingly, the correlation between the diseased volume (soft tissue + hard tissue), which was manually delineated by an expert, and the automatically extracted volume with the proposed method was very strong (R2 ≈ 0.8).
Conclusions:
We present a methodology that is suitable for automatic extraction of a radiological biomarker from CT images for TB disease burden. The method could be used to describe the longitudinal evolution of Mtb infection in a clinical trial devoted to the design of new drugs.
Keywords: Tuberculosis, Imaging biomarker, Lung segmentation, Computer tomography, Macaque model
Introduction
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (Mtb), which mainly affects the lungs. According to the 2017 WHO [1] report, TB caused 10.3 million incident cases, one third of the world’s population is estimated to have latent TB and there is a risk of disease chronification due to the high incidence of multidrug resistance TB. A common root for these disquieting data lies in the poor characterization of TB. The tests to identify TB infection (i.e., tuberculin skin test and TB antigen interferon-gamma release assays) are unable to characterize the disease as a dynamic continuum (from latent to active TB), yet such an approach is essential for the development of effective drugs [2]. Consequently, the World Health Organization has implemented an urgent plan to drastically reduce the global burden of TB.
To achieve this ambitious goal, novel techniques for the characterization of the disease as a continuous spectrum (from latent to acute to chronic disease) must be implemented and the response to treatment must be assessed [2]. Needless to say, the identification of specific TB biomarkers would be a fundamental enabler [2, 3]. In this context, the use of x-ray studies becomes essential owing to the sensitivity of radiological findings [3], and consequently, also does the computer-assisted identification of imaging biomarkers [3, 4]. In particular, it would speed the radiological evaluation, and would facilitate the longitudinal quantitative assessment of TB in large studies.
Previous necessary steps for extraction of an automatic biomarker from radiological images are data acquisition and segmentation of the region of interest [4]. In the case of TB infection, including non-clinical models and natural human disease, the literature focuses on methods to solve these tasks or to manually extract a biomarker [5–10].
Here, we propose a complete methodology for automatic extraction of biomarkers from x-ray computed tomography (CT) studies that effectively quantifies changes in TB disease burden and thus enables an easy assessment of the response to treatment.
Materials and Methods
Computed Tomography Images
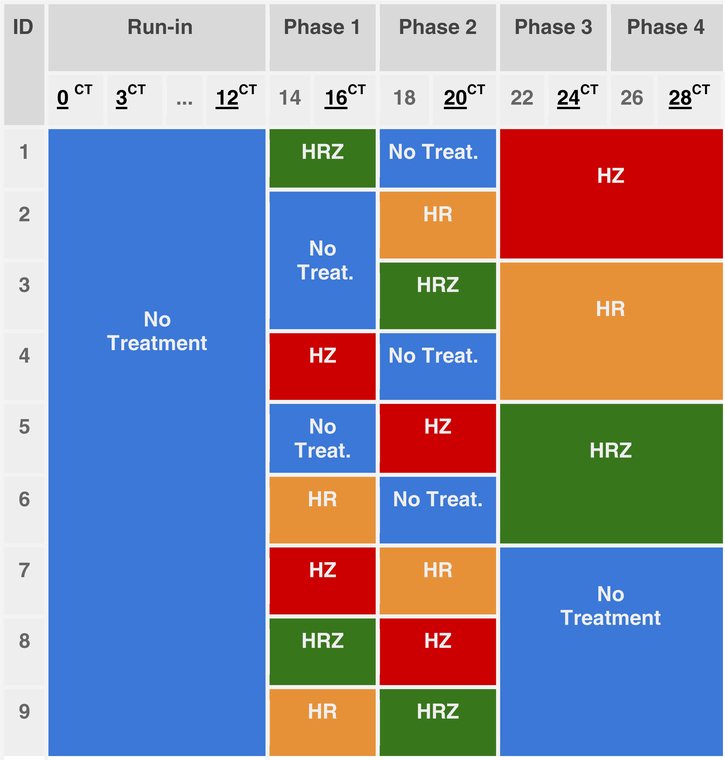
Nine male Indonesian cynomolgus macaques [11] were challenged by exposure to Mtb aerosols [12] and subsequently treated with a series of different antibiotic cocktails of isoniazid (H), rifampicin (R), and pyrazinamide (Z) in four phases (Table 1), using a Pigeon Control Balanced study design [13]. Our dataset comprises 63 chest CT scans, as each subject was imaged at seven time points (0, 3, 12, 16, 20, 24, and 28 weeks after aerosol exposure to Mtb), as shown in Table 1. Following aerosol challenge with M. tuberculosis, infection was confirmed using an ex vivo IFN-g ELISPOT assay which is a T cell-based assay that enumerates individual activated TBspecific effector T cells. All animals made responsesto antigens ESAT6 and CFP10 which are diagnostic for TB infection. Pulmonary changes consistent with TB-induced disease were identified from CT scans by an expert radiologist with 30 years’ experience of interpreting chest CT scans blinded to the animal’s treatment and clinical status. Pulmonary changes included the presence of nodules, cavitation, conglomeration, consolidation (an indicator of alveolar pneumonia), a “Tree-in-bud” pattern (an indicator of bronchocentric pneumonia) and lobular collapse. At the end of the study gross and microscopic changes in pathology were identified that were consist with TB-induced disease and Mycobacterium tuberculosis was cultured from extra pulmonary tissues that included the lung-associated lymph nodes, liver, spleen and kidney.
Table 1.
Antibiotic cocktail per week and subject. Each color represents the treatment (isoniazid (H), rifampicin (R), and pyrazinamide (Z)) taken by a subject during each treatment phase. Weeks with the CT superindex indicate the acquisition of a computed tomography volume at that week.
 |
The chest CT scans were acquired with a 16-slice Lightspeed CT scanner (General Electric Healthcare, Milwaukee, WI, USA), voxel spacing of 0.23 mm × 0.23 mm × 0.625 mm and in-plane size of 512 pixels × 512 pixels. All animal procedures and the study design were approved by the Public Health England Animal Welfare and Ethical Review Body, Porton Down, UK and authorized under an appropriate UK Home Office project license.
Lung Segmentation
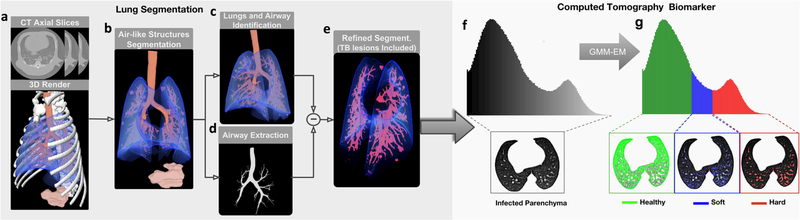
The procedure is illustrated in the left part of Fig. 1. Initially, air-like organs (e.g., healthy lungs, airway tree, stomach) present in the chest CT scans (Fig. 1a) are identified using an adaptive thresholding method based on Hu, et al. [14] (Fig. 1b). To subsequently isolate the object formed by the lungs and airways, the topology and connectivity of the organs are exploited (Fig. 1c). The intricate structure of the airways tree is computed using a region-growing algorithm, which propagates by simulating a spherical wavefront ruled by active contours [15] (Fig. 1d). Once computed, the airway tree structure is removed from the segmented lungs. After that, unsegmented inner lung tissue corresponding to TB lesions is added to the lung mask by means of a morphological hole-filling process. Finally, the lung boundaries are refined to include in the segmentation lesions attached to the pleura and to discard motion artifacts (Fig. 1e). For that, voxels at the segmented lung boundaries with larger gray levels are used as seeds to grow level sets and Geodesic Active Contours (GAC) [16]. The outputs are contours both for the lesions and for the fuzzy boundaries (produced by the respiration motion), and they can be identified using morphological information.
Fig. 1.
Lung Segmentation. From a the chest CT scan, b all aerated regions are identified. c The lungs, including the airways, are isolated successively by exploiting topological information. d Next, the airways tree is removed from the lungs. e Finally, to include the non-segmented damaged tissue, the lung segmentation is refined using a hole-filling method based on active contours. CT Biomarkers: The segmented lung volume is classified based on f the gray-level distribution in g three TB-associated tissue volumes: healthy, soft diseased, and hard diseased. Note: GMM-EM (Gaussian mixture model–Expectation maximization algorithm).
Computed Tomography Biomarker Extraction
In order to automatically retrieve quantitative information as a CT biomarker, our technique exploits the model proposed by Chen, et al. [5] and later used in [6–8]. In this study, based on the probability distribution of the voxels, measured in Hounsfield Units (HU), the authors divided lung tissue into three disease-associated volumes according to two thresholds selected by experts, as follows:
Healthy tissue: This volume corresponds to the region of the lung occupied by aerated parenchyma (without visible lesions)
Soft diseased tissue: This volume corresponds to lower density abnormal tissue, mainly correlated with small-tomedium nodular lesions, ground glass opacities and diffuse pulmonary infections. It may correspond to both healing lesions or to new forming lesions.
Hard diseased tissue: This volume corresponds to the higher-density abnormalities (large nodular lesions, fluidfilled cavities, consolidations, fibrosis, and bronchial thickening). It is likely that abnormal densities become less dense as they return to normal lung tissue (i.e., healing or response to treatment).
This approach of Chen et al. assigns a discrete tissue class (healthy, soft diseased, or hard diseased) to a range of values distributed around an expected intensity within certain variability, thus making it possible to capture subtle differences in the composition of each kind of tissue (Fig. 1, g). The expected intensity value and variability of each class are intrinsically determined by the threshold selected. This empiric approach can be modeled using the well-known Gaussian mixture model (GMM) and the most likely volume separation obtained through the expectation-maximization (EM) algorithm [17]. The GMM is formulated as:
| (1) |
where x is a vector of observed features (the intensity values of each voxel represented in the histogram), K is the number of expected Gaussians (K = 3 corresponding to healthy, soft diseased, and hard diseased tissues), and N represents each of the overlapped normal distributions (k) of the voxels’ gray level, with πk as the a priori probability, μk the mean, and Σk the covariance, respectively, for each distribution. These parameters are computed using the EM algorithm and selecting the set of three Gaussians, whose overlap is most similar to the known histogram (Eq. 1). This way, each voxel is assigned to a lung tissue class depending on which of the three fitted Gaussians yields the highest probability for its intensity.
Gold Standard Computer Tomography Biomarker
In order to measure the performance of the proposed automatic biomarker extraction method, we generated a ground truth: the aforementioned healthy, soft and hard diseased tissue volumes were manually extracted by an expert from the 63 original CT scans using the method proposed in [5]. Expert delineated a region-of-interest (ROI) around the main lesions for each subject and used the ROI to measure the volume of each lesion and its composition at different stages of treatment.
Evaluation
To create a more general biomarker of the TB disease burden, besides the volumes defined in the previous section, we also included the total volume of diseased tissue, defined as:
| (2) |
Additionally, to avoid the effects of the temporal changes in the whole lung volume due to the subjects’ intervariability (growth during the 28 weeks, weight change, variability in lung inflation in each serial CT, etc.), instead of the absolute diseased tissue volume, we applied the relative value defined as:
| (3) |
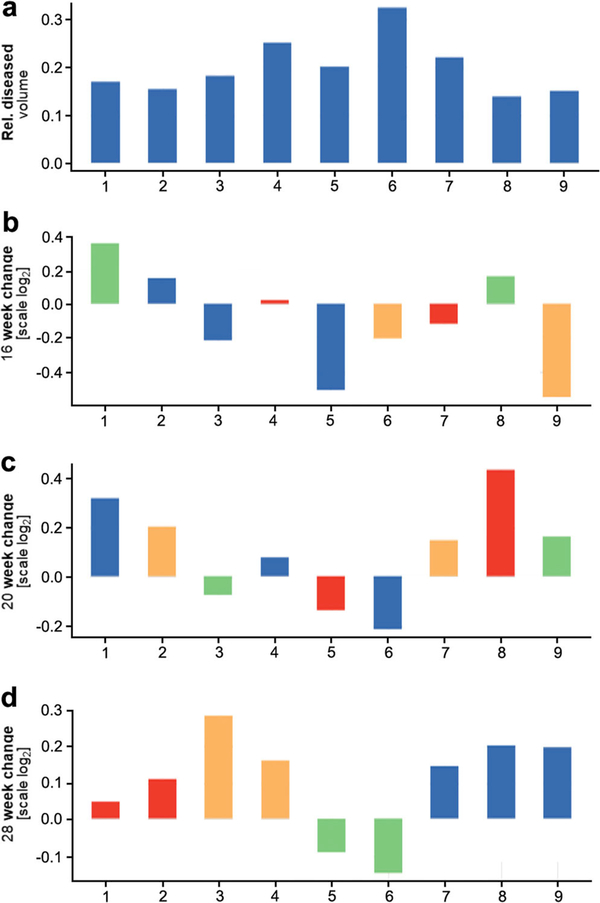
To evaluate the longitudinal change, we used a multirow bar plot known as a waterfall (Fig. 2). The first row shows the relative diseased volume of each subject at baseline. The remainder presents the change in the volume at a concrete time point on a log2 scale. Namely, the change in the relative diseased volume is computed as follows:
| (4) |
Fig. 2.
Longitudinal course of TB infection. The waterfall plot depicts the longitudinal change in the relative diseased tissue volume between a baseline and the representative weeks (b 16, c 20, and d 28 weeks after infection) as the log2 fold change (log2 (week change/baseline)) and the correlation with treatment outcomes:  no-D (no drugs),
no-D (no drugs),  HR (isoniazid + rifampicin),
HR (isoniazid + rifampicin),  HRZ (isoniazid + rifampicin + pyrazinamide),
HRZ (isoniazid + rifampicin + pyrazinamide),  HZ (isoniazid + pyrazinamide). Subjects taking the same antibiotic cocktail present a similar response (improvement or worsening) with respect to the baseline.
HZ (isoniazid + pyrazinamide). Subjects taking the same antibiotic cocktail present a similar response (improvement or worsening) with respect to the baseline.
Results
Figure 2 shows a waterfall plot for the nine subjects (horizontal axis) at only four of the seven time points for clarity; the first row contains the relative diseased volume at week 3 after infection, while the remainder represents the log2 change at weeks 16, 20, and 28 with respect to the first row (baseline). Beyond quantitative meaningful differences between equal treatments, it can be observed how subjects taking the same drug cocktail (each treatment is shown with a different color) at the end of the study (week 28) present a similar response to treatment (relative change in the diseased volume with respect to baseline). Thus, we see how subjects 7, 8, and 9, who did not receive treatment during the last phase of the study, present an increase of almost 0.2 units in the log2 scale, whereas subjects 5 and 6, who were treated with a mix of isoniazid + rifampicin + pyrazinamide, present a decrease of around 0.1 units.
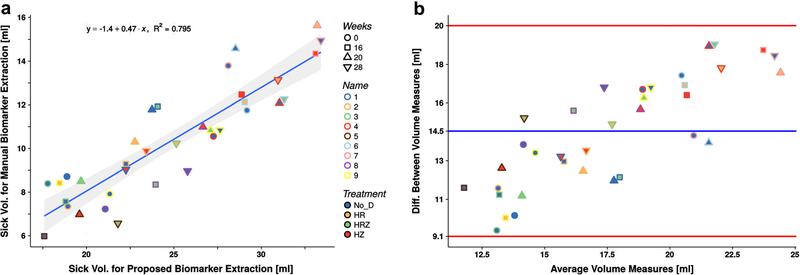
Figure 3 presents the relative diseased volume obtained using the manual delimitation of regions compared with the volume estimated by the proposed automatic extraction method for each of the 63 segmented lungs in the dataset (BComputed Tomography Images^) together with the corresponding Bland-Altman plot in order to show the agreement between methods. The similarity between measures is mostly independent of the subject, treatment, and study time point (none of these factors show remarkable bias). The correlation coefficient was good (R ≅ 0.8, p < 1 × 10−4), with a tendency to obtain higher values for the automatically obtained volumes, pointed by a bias factor of 0.47. The Bland-Altman plot depicts all the values within the 95 % limits of agreement.
Fig. 3.
Biomarker evaluation. a Correlation between the biomarker (relative diseased volume) extracted manually and using the proposed method (R2 = 0.8, p < 10−4); the 95 % confidence interval is drawn as the shadow of the regression line. b The BlandAltman plot presents a good agreement between measures with the regression bias of 0.47.
Discussion
Our results confirm that automatic biomarker extraction provides a satisfactory estimation of the volumes of interest by statistically modeling of the decision making process. In particular, our approach automatically assigns thresholds to each image at the three modeled Gaussian cross-points (“Computed Tomography Biomarker Extraction”). The method is able to assess the longitudinal course of TB disease burden (Fig. 2), as seen in the similarities in the treatment response. Given the experimental design, in which the subjects are not treated with the same antibiotic throughout the study (Table 1), these similarities are observed, as expected, at the end (week 28), and there is a correspondence between the automatically and manually obtained results, as indicated by the strong correlation coefficient (R2 = 0.8). This relationship is biased by a factor of 0.47 in favor of the volumes obtained with the proposed method, even when the agreement between methods is suitable (Fig. 3). This difference has two explanations: (a) The difficulty arising from the manual delimitation of complex three dimensional structures, which are often intricate in healthy tissue, prevents the whole region of interest from being segmented, thus resulting insmaller volumes; (b) The automatic extraction of the biomarker tends to include small vessels (that were not excluded from the lung mask), increasing the estimated diseased volume. The inclusion of vessels as damaged lung tissue is undesirable and can be mitigated using the relative diseased volume. Moreover, it is reasonable to assume that this extra volume remains constant over time and does not affect the changes with respect to baseline observed in Fig. 2.
It is important to note that the proposed method is mainly intended to capture differences in the infection burden of subjects (i.e., establish a continuous spectrum between latent and active TB). Therefore, extra-large air-filled cavities (i.e., the manifestationsoftissue destruction)arenotincludedasdamaged tissue. This can cause small drifts in the data correlation (i.e., break the linear relationship between the manual and automatic estimations) for highly infected subjects.
The limitations mentioned will be addressed in future studies by using improved variations of the statistical model, for example, including semiquantitative annotations performed by radiologist. Moreover, the automatic estimation could also take advantage of well-known radiomics techniques [18].
Conclusion
We propose a fully automatic method for computer-assisted extraction of a radiological imaging biomarker for the longitudinal characterization of gradual changes in TB disease burden. The proposed technique yields similar estimations to those obtained manually by a trained specialist. The method has the potential to be used as a tool for quantifying disease burden in clinical trials aimed at establishing effective antibiotic regimes for TB.
Acknowledgments
Funding Information The research leading to these results received funding from the Innovative Medicines Initiative (www.imi.europa.eu) Joint Undertaking under grant agreement no. 115337, whose resources comprise funding from EU FP7/2007–2013 and EFPIA companies in-kind contribution. This study was partially funded by projects TEC2013–48552C2–1-R, RTC-2015–3772-1, TEC2015–73064-EXP and TEC2016–78052R from the Spanish Ministry of Economy, Industry and Competitiveness (MEIC), TOPUS S2013/MIT-3024 project from the regional government of Madrid and by the Department of Health, UK. LEV is funded by the Intramural Research Program of NIAID, NIH. The CNIC is supported by the MEIC and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015–0505).
Footnotes
Conflicts of Interest. All animal procedures and the study design were approved by the Public Health England Animal Welfare and Ethical Review Body, Porton Down, UK and authorized under an appropriate UK Home Office project license.
The authors declare they have no conflicts of interest.
References
- 1.World Health Organization (2017) Global tuberculosis report [Technical Report]
- 2.Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, Ginsberg A, Swaminathan S, Spigelman M, Getahun H, Menzies D, Raviglione M (2016) Tuberculosis. Nat Rev Dis Primers 2:16076. [DOI] [PubMed] [Google Scholar]
- 3.Nachiappan AC, Rahbar K, Shi X, Guy ES, Mortani Barbosa EJ Jr, Shroff GS, Ocazionez D, Schlesinger AE, Katz SI, Hammer MM (2017) Pulmonary tuberculosis: role of radiology in diagnosis and management. Radiographics 37:52–72 [DOI] [PubMed] [Google Scholar]
- 4.Galbán CJ, Han MK, Boes JL, Chughtai KA, Meyer CR, Johnson TD, Galbán S, Rehemtulla A, Kazerooni EA, Martinez FJ, Ross BD (2012) Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med 18:1711–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen RY, Dodd LE, Lee ME et al. (2014) PET/CT imaging correlates with treatment outcome in patients with multidrug-resistant tuberculosis. Sci Transl Med 6:166–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Via LE, Schimel D, Weiner DM, Dartois V, Dayao E, Cai Y, Yoon YS, Dreher MR, Kastenmayer RJ, Laymon CM, Carny JE, Flynn JAL, Herscovitch P, Barry CE III (2012) Infection dynamics and response to chemotherapy in a rabbit model of tuberculosis using [18F] 2-fluoro-deoxy-D-glucose positron emission tomography and computed tomography. Antimicrob Agents Chemother 56:4391–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Via LE, Weiner DM, Schimel D, Lin PL, Dayao E, Tankersley SL, Cai Y, Coleman MT, Tomko J, Paripati P, Orandle M, Kastenmayer RJ, Tartakovsky M, Rosenthal A, Portevin D, Eum SY, Lahouar S, Gagneux S, Young DB, Flynn JAL, Barry CE III (2013) Differential virulence and disease progression following mycobacterium tuberculosis complex infection of the common marmoset (callithrix jacchus). Infect Immun 81:2909–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Via LE, England K, Weiner DM, Schimel D, Zimmerman MD, Dayao E, Chen RY, Dodd LE, Richardson M, Robbins KK, Cai Y, Hammoud D, Herscovitch P, Dartois V, Flynn JAL, Barry CE III (2015) A sterilizing tuberculosis treatment regimen is associated with faster clearance of bacteria in cavitary lesions in marmosets. Antimicrob Agents Chemother 59:4181–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallis RS, Kim P, Cole S, Hanna D, Andrade BB, Maeurer M, Schito M, Zumla A (2013) Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis 13:362–372 [DOI] [PubMed] [Google Scholar]
- 10.Mansoor A, Bagci U, Foster B (2015) Segmentation and image analysis of abnormal lungs at CT: current approaches, challenges, and future trends. Radiographics 35:1056–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell JL, Mee ET, Almond NM, Cutler K, Rose NJ (2012) Characterization of MHC haplotypes in a breeding colony of Indonesian cynomolgus macaques reveals a high level of diversity. Immunogenetics 64:123–129 [DOI] [PubMed] [Google Scholar]
- 12.Sharpe S, White A, Gleeson F, McIntyre A, Smyth D, Clark S, Sarfas C, Laddy D, Rayner E, Hall G, Williams A, Dennis M (2016) Ultra low dose aerosol challenge with Mycobacterium tuberculosis leads to divergent outcomes in rhesus and cynomolgus macaques. Tuberculosis 96:1–12 [DOI] [PubMed] [Google Scholar]
- 13.Pigeon JG, Raghavarao D (1987) Crossover designs for comparing treatments with a control. Biometrika 74:321–328 [Google Scholar]
- 14.Hu S, Hoffman EA, Reinhardt JM (2001) Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans Med Imaging 20:490–498 [DOI] [PubMed] [Google Scholar]
- 15.Ceresa M, Artaechevarria X, Muñoz-Barrutia A, Ortiz-de-Solorzano C (2010) Automatic leakage detection and recovery for airway tree extraction in chest CT images. Proc IEEE Int Symp Biomed Imaging 568–571 [Google Scholar]
- 16.Caselles V, Kimmel R, Sapiro G (1997) Geodesic active contours. Int J Comput Vis 22:61–79 [Google Scholar]
- 17.Bishop C (2006) Pattern Recognition and Machine Learning [Book]
- 18.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749–762 [DOI] [PubMed] [Google Scholar]