Abstract
Salmonella enterica has been documented as one of the leading causes of salmonellosis throughout the world and is most commonly associated with the consumption of contaminated food products. Thus, this research was aimed at studying the antimicrobial susceptibility pattern and detection of quinolone resistance in Salmonella spp isolated from food of animal origin. Thirty-six Salmonella isolates comprising 8 from poultry and 28 from seafood (clams) were identified, serotyped and characterized for their antimicrobial susceptibility against 10 different antibiotics. Plasmid DNA was isolated from all the isolates by alkaline lysis, quinolone resistant non-typhoidal S. Weltevreden were examined for mutation in the DNA gyrase coding gene. Among the 36 Salmonella isolates, 20 were S. weltevreden (8 from poultry and 12 from seafood) and 16 were S. Typhimurium (from seafood). All the isolates showed multiple resistance to nalidixic acid, tetracycline, co-trimoxazole and nitrofurantoin, but, interestingly, the isolates were 100% susceptible to ampicillin, chloramphenicol and gentamicin. Resistant isolates from the study carried the genes responsible for resistance to respective antibiotics. The strain S130 isolated in the study showed single point mutation, Asp87Gly, at position 87 in quinolone resistance determining region. It revealed mutation in quinolone resistance determining region as a cause for quinolone resistance in non-typhoidal Salmonellae. The occurrence of genes accountable for plasmid mediated resistance to quinolones (viz., qnrA, qnrB and qnrS) in plasmid of non-typhoidal Salmonellae isolates provides evidence for plasmid mediated quinolone resistance.
Keywords: mutation, multi-drug resistant, non-typhoidal Salmonellae, plasmid mediated quinolone resistance, quinolone resistance determining region
Introduction
Non-typhoidal Salmonellae (NTS) has emerged as a major group of foodborne pathogens which causes gastroenteritis, septicaemia, endocarditis and subsequent infections like intra-abdominal infections, and pulmonary infections[1]. NTS have a wide range of hosts and are strongly associated with animals and agricultural products[2]. Hence salmonellosis can easily occur or spread by consumption of contaminated foods of animal origin, such as those from poultry, fish, eggs, beef and dairy products[3]. Both the developed and developing countries have recently reported continuous increase in the incidence of human salmonellosis in patients who are immunosuppressed, or at the extremes of age[4]. Traditionally, recommended regimens for the treatment for salmonellosis include the traditional first-line drugs like ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole[5]. But emerging drug resistance over the past 25 years has limited the usefulness of these antimicrobial agents. Currently, quinolone and third-generation cephalosporin antibiotics are preferred for the treatment against salmonellosis[5].
Quinolones and fluoroquinolones are an important class of broad spectrum antibiotics which possesses strong antimicrobial activity against many human pathogenic bacteria. They are usually used to treat urinary tract infections (UTI) and gastrointestinal infections[6]. This group of antibiotics mainly act on the bacterial DNA gyrase and topoisomerase IV, which are important in DNA replication[7].
In last recent years, resistance to quinolones and fluoroquinolones in NTS has been increasingly reported among human and veterinary isolates[1]. In the developing countries, this may be partly related to lack of legislation on the use of antibiotics. Bacteria may acquire resistance to quinolones via mutations in the gyrA, gyrB, parC and parE genes[11–12]. All mutations described reside within the quinolone resistance determining region of the A subunit of DNA gyrase, i.e. amino acids 67–122 and for some of the isolates the mechanism of resistance has been studied. In addition to mutation based resistance, Martinez et al.[13] found quinolone resistance genes (qnrA, qnrB and qnrS) in plasmid and confirmed that resistance can also occur via plasmid mediated quinolone resistance.
Globally, the impact of NTS on human health is very high and earlier reports have documented the emergence of multidrug resistant Salmonella in developing and developed countries[5,14–16]. The globalization of the food trade has increased the likelihood of international incidents of food contamination through travels, migration and distribution of food[14]. This increases the wide spread of resistance determinants in bacteria associated with food and even in clinical pathogens. The analysis of food samples from animal origin could provide useful information on resistance pattern of Salmonella against different antimicrobial agents. Hence, the present study attempted to determine antibiotic susceptibility and possible causes of resistance to quinolones in NTS strains isolated from food of animal origin.
Materials and methods
Isolation of bacteria and antibiogram analysis
Mollusc (clam) samples (132) from the fish market and landing centers in Mangalore and also chicken samples (78) from the poultry and chicken market outlets in Mangalore (Table 1) were collected aseptically. Samples were brought to the laboratory and immediately processed for analysis. Salmonella strains were isolated from the samples by culture-based conventional method as recommended by the FDA Bacteriological Analytical Manual[18] and identified using standard biochemical tests as previously described[19]. Pre-enriched samples (10 g of chicken or clam meat) in lactose broth (90 mL) were enriched in selenite cysteine broth (SCB) and streaked in selective xylose lysine deoxy-cholate agar (XLD) plate. Typical colonies from each of the selective plates were sub-cultured on to Luria Bertani agar and subjected to a battery of biochemical tests for identification of Salmonella. The biochemically identified isolates were further confirmed by polymerase chain reaction (PCR) with invA and hns gene primers (Table 2). PCR confirmed isolates were then serotyped at the National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, India. Antibiotic susceptibility test was performed for all the isolates by disk diffusion method using standard procedure[26] on Mueller-Hinton agar as recommended by the Clinical and Laboratory Standards Institute. The antibiotics included in the test were nalidixic acid (30 mcg), tetracycline (30 mcg), co-trimoxazole (25 mcg), ciprofloxacin (5 mcg), chloramphenicol (30 mcg), ampicillin (10 mcg), gentamicin (10 mcg), nitrofurantoin (300 mcg), imipenem (10 mcg) and piperacillin-tazobactam (100/10 mcg). Minimum inhibitory concentration (MIC) was performed using Ezy MICTM strips (HiMedia Laboratories Pvt. Ltd., India) for nalidixic acid and ciprofloxacin. Multiple antibiotic resistance (MAR) index for each resistance pattern was calculated by using the formula by Singh et al.[27].
Tab.1.
Occurrence and distribution of Salmonella in seafood (clams) and poultry samples.
| Si No. | Source | No. of sampling | No. of positive samples | Percentage of positive samples | Serotypes (No.) |
|---|---|---|---|---|---|
| 1 | Seafood | 132 | 28 | 21.2 | S. Weltevreden (12) S. Typhimurium (16) |
| 2 | Poultry | 78 | 8 | 10.2 | S. Weltevreden (8) |
| Total | 210 | 36 | 17.1 |
Tab.2.
List of primers used for detection of antibiotic resistant determinants in the study.
| Target | Gene | Sequence 5′- 3′ | Size | Tm (°C) | Reference |
|---|---|---|---|---|---|
| Salmonella | invA | GTGAAATTATCGCCACGTTCGGGCAA | 284 bp | 64 | Rahn et al.[20] |
| TCATCGCACCGTCAAAGGAACC | |||||
| hns | TACCAAAGCTAAACGCGCAGCT | 156 bp | 60 | Jones et al.[21] | |
| TGATCAGGAAATCTTCCAGTTGC | |||||
| Gyrase | gyrA | CTGAAGCCGGTACACCGTCG | 290 bp | 55 | Casin et al.[22] |
| TCGGCCATCAGTTCGTGGGC | |||||
| Quinolone resistance protein | qnrA | ATTTCTCACGCCAGGATTTG | 516 bp | Robicsek et al.[23] | |
| GATCGGCAAAGGTTAGGTCA | |||||
| qnrB | GATCGTGAAAGCCAGAAAGG | 469 bp | |||
| ACGATGCCTGGTAGTTGTCC | |||||
| qnrS | ACGACATTCGTCAACTGCAA | 417 bp | |||
| TAAATTGGCACCCTGTAGGC | |||||
| Antibiotic resistant genes | tetA | TTGGCATTCTGCATTCACTC | 494 bp | Menggen et al.[24] | |
| GTATAGCTTGCCGGAAGTCG | |||||
| tetB | CAGTGCTGTTGTTGTCATTAA | 571 bp | |||
| GCTTGGAATACTGAGTGTTAA | |||||
| tetC | CTTGAGAGCCTTCAACCCAG | 418 bp | |||
| ATGGTCGTCATCTACCTGCC | |||||
| tetS | TGGAACGCCAGAGAGGTATT | 660 bp | Aarestrup et al.[25] | ||
| ACATAGACAAGCCGTTGACC | |||||
| Sul1 | TTTCCTGACCCTGCGCTCTAT | 425 bp | Menggen et al.[24] | ||
| GTGCGGACGTAGTCAGCGCCA | |||||
| Sul2 | CCTGTTTCGTCCGACACAGA | 435 bp | |||
| GAAGCGCAGCCGCAATTCAT | |||||
| Sul3 | ATGAGCAAGATTTTTGGAATCGTAA | 792 bp | |||
| CTAACCTAGGGCTTTGGATATTT |
Analysis of antibiotic resistance determinants by PCR
Genomic DNA was extracted from the bacterial culture by CTAB (Cetyl trimethyl ammonium bromide) method as described by Ausubel et al.[28]. All the resistant isolates were analyzed for the presence of specific genes encoding for resistance to particular antibiotics using the primers listed in the Table 2. PCR was carried out in 30 µL reaction mixture containing 10X buffer (100 mmol/L Tris-HCl, pH 8.3, 20 mmol/L MgCl2, 500 mmol/L KCl, 0.1% gelatin), 200 mmol/L of dNTPs (dATP, dTTP, dGTP, dCTP), 10 pmol each of forward and reverse primers and 1.0 unit of Taq DNA polymerase enzyme with 2 µL of template DNA. Amplification was carried out in a MJ-Research Thermo Cycler (PTC-200, USA) with the optimized PCR program consisted of an initial denaturation at 94°C for 5 minutes, followed by 35 cycles with each cycle consisting of 94°C for 60 seconds, annealing temperature of X °C (based on primer used) for 60 seconds and extension at 72°C for 30 seconds. The final extension was set to 72°C for 10 minutes.
Detection of mutation in gyrA gene from quinolone resistant isolates
Bacterial strains resistant to quinolone (nalidixic acid/ ciprofloxacin) were selected and subjected to mutation study at the gyraseA gene. The gyrA gene was amplified by PCR using oligonucleotide primers (Table 1) with extracted genomic DNA (as described above), under specified cyclic conditions. Amplified PCR product was purified using a QIAquick PCR purification Kit (Qiagen, Hilden, Germany) and sequenced by Bio serve Biotechnologies (INDIA) Pvt. Ltd, Hyderabad, India. The obtained nucleotide sequences were analyzed using blast programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences were aligned and compared with the sequence of the gyrA gene of quinolone sensitive strain reported in the GenBank (Accession no. EU898544) by using multalign program (http://multalin.toulouse.inra.fr/multalin/multalin.htm). The amino acids were deduced from the DNA sequences obtained through a Molecular Tool Kit web program (http://www.vivo.colostate.edu/molkit). The nucleotide sequences were then submitted to GenBank.
Evaluation of Plasmid mediated quinolone resistance
Plasmid DNA was isolated from all the isolates by alkaline lysis method described by Sambrook et al.[29]. Isolated plasmid DNA was profiled using agarose gel electrophoresis in 1X Tris-acetate-EDTA buffer on 0.8% agarose gel. The separated bands were extracted by using gel extraction kit (Chromous Biotech, Bangalore, India). Extracted plasmid DNA was used as template to check quinolone resistance genes (qnrA, qnrB and qnrS) located in plasmid or chromosome, using the oligonucleotide primers (Table 2) by PCR.
Statistical analysis
Descriptive statistics was used to describe frequency and percentages of Salmonella positive isolates from food samples.
Results
Identification of bacterial strain and antimicrobial susceptibility tests
All the 36 strains isolated from different sources in the study were identified as Salmonella spp. by conventional methods and confirmed by PCR for invA and hns gene by yielding a product of 284 bp and 156 bp, respectively (Fig. 1). Of total 36 isolates, eight strains were isolated from 78 poultry samples and 28 were isolated from 132 seafood (clams) samples (Table 2). Confirmed isolates were serotyped and subjected to further studies. Among 36 Salmonella isolates, 20 were S. Weltevreden (8 from poultry and 12 from seafood) and 16 were S. Typhimurium (from seafood). All 36 isolates (100%) examined in the study were multi-drug resistant (MDR) i.e., resistant to more than two antibiotics and showed MAR index ranging from 0.40-0.50 (Table 3). Table 3 further shows the antibiogram of the results which includes the isolates/sources and their respective resistant gene to the tested antibiotics.
Fig.1. Gel-electrophoresis of PCR amplified products of Salmonella isolates and their gyrA gene.
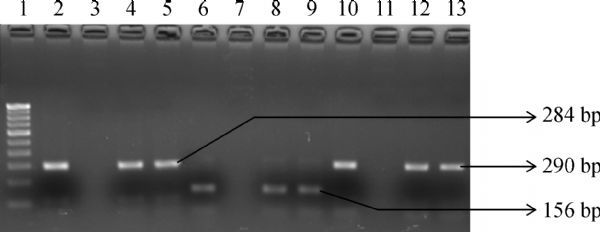
Lane1: 100 bp DNA ladder; Lane 2: invA gene positive control (284 bp); Lane 3: invA gene negative control; Lane 4 and 5: Positive Salmonella isolates (positive invA gene); Lane 6: Positive control for hns gene (156 bp); Lane 7: Negative control for hns gene; Lane 8 and 9: Positive Salmonella isolates (positive hns gene); Lane 10: Positive control for gyrA gene (290 bp); Lane 11: Negative control for gyrA gene; Lane 12 to 13: gyrA gene of isolates
They showed resistance to nalidixic acid, tetracycline and co-trimoxazole. Twenty two (61%) isolates showed resistance to nitrofurantoin, 16 (44%) were resistant to piperacillin-tazobactam and four (11%) were resistant to imipenem. But surprisingly all the isolates were susceptible to ampicillin, chloramphenicol, ciprofloxacin and gentamicin. Isolates resistant to nalidixic acid showed MIC of more than 256 µg/mL and the MIC of isolates for ciprofloxacin was ranged between 0.032 to 1.00 µg/mL.
Fig.2. Multialigned sequences of gyrA gene of S130 (present study) and TW14359 (from GenBank).
Highlight (oval shape) indicates the change in nucleotide sequence at 259 from adenine (A) to guanine (G) which leads to change in amino acid at position 87 (in QRDR region).
PCR analysis for antibiotic resistant genes
The isolates were tested for presence of antibiotic resistance genes viz. tetA, tetB, tetC, and tetS for tetracycline and sul1, sul2, sul3 for sulphonamides. All (100%) tetracycline resistant isolates amplified 494 bp of PCR product with tetA gene, 32 (89%) of them amplified 660 bp of PCR product with tetS gene, but none of them tested positive for tetB and tetC. Among the isolates resistant to co-trimoxazole, eight (23%) yielded 425 bp PCR product when amplified with sul1 gene primer, nevertheless none of the resistant isolates showed positive for sul2 and sul3 gene.
Tab.3.
Antibiotic sensitivity/resistance pattern of Salmonella isolates.
| Strain | Source | Antibiotic resistant pattern-Phenotype | Salmonella Serovar | Antibiotic resistant pattern-Genotype | MAR index |
|---|---|---|---|---|---|
| S130 | Poultry | NA,TE,COT,NIT,IPM, | S. Weltevreden | TetA,Sul1, TetS, qnrA, qnrS | 0.50 |
| S131 | Poultry | NA,TE,COT,NIT, | S. Weltevreden | TetA,Sul1, qnrA, qnrS | 0.40 |
| S132 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrB, qnrS | 0.50 |
| S133 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA, qnrS | 0.50 |
| S188 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S189 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, | 0.50 |
| S190 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, | 0.50 |
| S191 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA, qnrS | 0.50 |
| S192 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, | 0.40 |
| S193 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S194 | Seafood | NA,TE,COT,NIT,PIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.50 |
| S195 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA. qnrS | 0.50 |
| S210 | Poultry | NA,TE,COT,NIT,IPM, | S. Weltevreden | TetA,Sul1, TetS, qnrA, qnrS | 0.50 |
| S211 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, | 0.50 |
| S222 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, | 0.40 |
| S223 | Seafood | NA,TE,COT,NIT,PIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.50 |
| S224 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA, qnrS | 0.50 |
| S225 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrB, qnrS | 0.50 |
| S226 | Poultry | NA,TE,COT,NIT, | S. Weltevreden | TetA,Sul1, qnrA, qnrS | 0.40 |
| S227 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S228 | Poultry | NA,TE,COT,NIT,IPM, | S. Weltevreden | TetA,Sul1, TetS, qnrA, qnrS | 0.50 |
| S229 | Seafood | NA,TE,COT,NIT,PIT, | S. Typhimurium | TetA, TetS, | 0.50 |
| S230 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA, qnrS | 0.50 |
| S278 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, | 0.40 |
| S279 | Poultry | NA,TE,COT,NIT, | S. Weltevreden | TetA,Sul1, qnrA, qnrS | 0.40 |
| S280 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S281 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, | 0.40 |
| S282 | Seafood | NA,TE,COT,NIT,PIT, | S. Typhimurium | TetA, TetS, | 0.50 |
| S283 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA, qnrS | 0.50 |
| S284 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S296 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S297 | Poultry | NA,TE,COT,NIT,IPM, | S. Weltevreden | TetA,Sul1, TetS, qnrA, qnrS | 0.50 |
| S298 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, | 0.40 |
| S299 | Poultry | NA,TE,COT,NIT, | S. Weltevreden | TetA,Sul1, qnrA, qnrS | 0.40 |
| S300 | Seafood | NA,TE,COT,NIT, | S. Typhimurium | TetA, TetS, qnrB, qnrS | 0.40 |
| S301 | Seafood | NA,TE,COT,NIT,PIT, | S. Weltevreden | TetA, TetS, qnrA, qnrS | 0.50 |
NA: nalidixic acid, TE: tetracycline, COT: co-trimoxazole, NIT: nitrofurantoin, PIT: piperacillin-tazobactam, IPM: imipenem. MAR index= Number of resistance antibiotics/total number of antibiotics tested.
Analysis of gyrA gene sequence of quinolone resistant isolates
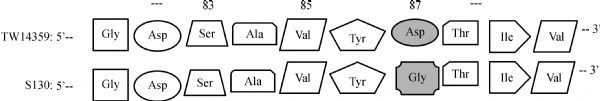
Nalidixic acid resistant isolates were selected and studied for detection of mutation by using gyrA primers, since it has been reported that a point mutation in the gyrA gene coding for the A subunit of gyrase could cause resistance for quinolone. PCR amplification with gyrA primer yielded 290 bp product (Fig. 1) and the product was purified and sequenced. The obtained nucleotide sequences were compared with other gyrA gene sequence of quinolone sensitive strain. In the gyrA sequence of isolate S130, transition of nucleotide from guanine to adenine (G→A) at 259th position in the quinolone resistance determining region was observed (Fig. 2). This point mutation in gyrA sequence resulted in the replacement of aspartic acid by glycine (asp-87-gly) at position 87 (Fig. 3). But the gyrA sequence of other isolates was similar with the sensitive strain sequence. Sequence of S130 was deposited at GenBank under the accession number KC570463. Of interest, the gyrA sequence of S130 also showed change in nucleotide (point mutation) at other different points but that mutation did not result change in amino acid.
Fig.3. Partial amino acid sequence of gyraseA gene of strain TW14359 and isolate S130.

Amino acid in the sequences at the position 87 (highlighted) attesting QRDR region indicates point mutation Asp87Gly.
Fig.4. Detection of PCR amplified products of quinolone resistance protein coding genes.
Lane 1:100 bp DNA ladder, Lane 3 to 5: qnrA (516 bp), Lane 7 to 9: qnrB (469 bp), Lane 11 to 13: qnrS (417 bp) and Lanes 2, 6, 10 negative controls.
Plasmid mediated quinolone resistance
Apart from the mutations in the chromosomal gene of quinolone resistant isolates, the plasmid mediated quinolone resistance was tested for the presence of qnr genes (qnrA, qnrB and qnrS). Total 26 (72%) isolates were tested positive for any of the three tested qnr genes. Out of the 36 isolates, 15 (42%) tested positive for qnrA gene by yielding 516 bp of PCR product, 11 (30%) amplified 469 bp PCR product of qnrB gene and 26 (72%) isolates amplified 417 bp PCR product of qnrS (Fig. 4).
Discussion
Previously, non-typhoidal Salmonella (NTS) infections were commonly treated with ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole. With the emergence of resistance to those first line antibiotics quinolones, fluoroquinolones and third-generation cephalosporins are used for treatment[30]. But their widespread use has developed a selective pressure on NTS and triggered resistance against them[2]. Earlier reports from different developing countries revealed high multi-drug resistance rate in NTS isolated from humans and poultry[2]. Correspondingly, data from the present study harbingers the acquirement of multi-drug resistance (MDR) in NTS isolated from the poultry and seafood products. All 36 isolates (both from poultry and seafood) showed resistance to at least more than three antibiotics tested in the study. The isolates showed 100% resistance to nalidixic acid, tetracycline, co-trimoxazole and nitrofurantoin. Of 36 isolates, 16 (44%) showed resistance to piperacillin-tazobactam and 4 (11%) were resistant to imipenem.
Of late many researchers observed that due to low level use of first line antibiotics, they have re-emerged and are used to treat multi-drug bacterial isolates[31]. Snehali and Mohammed[32] reported that the older antibiotics such as polymyxins, co-trimoxazole, aminoglycosides and chloramphenicol are more active and can be used to treat many of the MDR, XDR and PDR strains. However, interestingly in our study all the isolates (100%) were susceptible to ampicillin, chloramphenicol and gentamicin. It prominently demonstrates that these antibiotic compounds can be effectively used to treat against multi-drug NTS infections. But currently reason remains unclear and therefore further studies need to be carried out with NTS isolated from the study against these compounds.
Quinolones were highly recommended and was a first choice for treatment of serious intestinal Salmonella infections in adults but development of resistance reduced the efficacy of provisional treatment[33]. Acquisition of this desperate resistance to quinolones has been developed due to point mutations in chromosome at quinolone resistance-determining region (QRDR) of DNA gyrase and topoisomerase IV gene and along with plasmid mediated resistance mechanisms[5]. Change in a single nucleotide (mutation) in gyraseA gene sequence, which resulted in change in confirmation of protein (DNA gyrase) will prevent binding of antimicrobial agents and carrying out antimicrobial activity, and it is the most common resistance mechanism described till now in various Gram negative bacteria[34]. Mutations at codon Ser83 or/and Asp87 in quinolone resistance determining region of gyrA are the two most common sites leading to resistance in quinolone[35]. Previous studies have indicated that in Salmonella the most common residues associated with mutation are at serine 83 and aspartate 87, either alone or together.[35] This study described the mutation in gyrase gene which has been clustered in a QRDR due to observed resistance for quinolone (nalidixic acid) group of antibiotic. Giraud et al.[35] reported that a single point mutation in gyrA gene of Salmonella is adequate to cause high level of quinolone resistance, the same was observed in the present study. A single point mutation of strain S130 at position 87 in the quinolone resistance determining region, caused substitution in amino acid from aspartic acid to glycine. This showed reduced susceptibility to ciprofloxacin (MIC 0.032 µg/mL). In the study, some silent mutations were observed at different points which make no changes in the protein or enzyme (gyrase A) conformation.
Apart from development of quinolone resistance by mutation Martinez et al.[13] recognized the plasmid mediated quinolone resistance determinant, qnr protein (qnrA1) on a conjugative plasmid isolated from ciprofloxacin resistant Klebsiella pneumoniae. Similarly, the isolates from the present study harbored qnr genes in their plasmid provided evidence that the plasmid mediated resistance transfer and it is in agreement with Hernandez et al.[35]. The results from the study were corroborated with earlier reports and 26 (72%) of 36 isolates carried at least one or more qnr genes in their plasmid which is an accountable for resistance to nalidixic acid and reduction in susceptibility against ciprofloxacin. Among 36 isolates, 15 (42%) carried qnrA, 11 (30%) carried qnrB and 26 (72%) carried qnrS genes. This demonstrated that the plasmid mediated quinolone resistance can be transferred between Salmonella populations from different sources.
In conclusion, the occurrence of high rate of antimicrobial resistance among NTS strains isolated from food of animal origin has emerged as a serious problem in India. Our findings in the study clearly demonstrated the acquirement of multi-drug resistance in NTS isolated from poultry and seafood. Besides, interestingly, the first line antibiotics showed their antimicrobial activity against multi-drug resistant NTS. Since the antibiotic resistant determinants were found on the plasmid (mobile genetic element), there is a strong chance for transmission of resistance among bacterial population. This study clearly demonstrates the need for regular monitoring of antibiotic use and an effective antibiotic policy to prevent the indiscriminate use of antibiotics in different fields.
References
- 1. Hohmann EL. Nontyphoidal salmonellosis[J]. Clin Infect Dis, 2001, 32(2): 263–269. 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 2. Al-Dawodi R, Farraj MA, Essawi T. Antimicrobial resistance in non-typhi Salmonella enterica isolated from humans and poultry in Palestine[J]. J Infect Dev Ctries, 2012, 6(2): 132–136 . [DOI] [PubMed] [Google Scholar]
- 3. Rice DH, Hancock DD, Roozen PM, et al. Household contamination with Salmonella enterica [J]. Emerg Infect Dis, 2003, 9(1): 120–122. 10.3201/eid0901.020214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han J, Gokulan K, Barnette D, et al. Evaluation of virulence and antimicrobial resistance in Salmonella enterica serovar Enteritidis isolates from humans and chicken- and egg-associated sources[J]. Foodborne Pathog Dis, 2013, 10(12): 1008–1015. 10.1089/fpd.2013.1518 [DOI] [PubMed] [Google Scholar]
- 5. Crump JA, Sjölund-Karlsson M, Gordon MA, et al. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella Infections[J]. Clin Microbiol Rev, 2015, 28(4): 901–937. 10.1128/CMR.00002-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oliveira CM, Ribeiro AR, Santos LR, et al. Antibiotic resistance in Salmonella enteritidis isolated from broiler carcasses[J]. Braz J Microbiol, 2006, 37(3): 368–371 10.1590/S1517-83822006000300030. [DOI] [Google Scholar]
- 7. Guan X, Xue X, Liu Y, et al. Plasmid-mediated quinolone resistance--current knowledge and future perspectives[J]. J Int Med Res, 2013, 41(1): 20–30. 10.1177/0300060513475965 [DOI] [PubMed] [Google Scholar]
- 8. Randall LP, Coldham NG, Woodward MJ. Detection of mutations in Salmonella enterica gyrA, gyrB, parC and parE genes by denaturing high performance liquid chromatography (DHPLC) using standard HPLC instrumentation[J]. J Antimicrob Chemother, 2005, 56(4): 619–623. 10.1093/jac/dki293 [DOI] [PubMed] [Google Scholar]
- 9. Haugum K, Aas L, Lindstedt BA. Effect of quinolone antibiotics and chemicals on mutation types in Salmonella enterica serovars Enteritidis, Hadar and Virchow[J]. Division for Infectious Diseases Control, Norwegian Institute of Public Health, N-0403 Oslo, Norway 2006; 1–5.
- 10. Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid[J]. Lancet, 1998, 351(9105): 797–799. 10.1016/S0140-6736(97)07322-4 [DOI] [PubMed] [Google Scholar]
- 11. Saha SK, Talukder SY, Islam M, et al. A highly ceftriaxone-resistant Salmonella typhi in Bangladesh[J]. Pediatr Infect Dis J, 1999, 18(4): 387. 10.1097/00006454-199904000-00018 [DOI] [PubMed] [Google Scholar]
- 12. Akinyemi KO, Bamiro BS, Coker AO. Salmonellosis in Lagos, Nigeria: incidence of Plasmodium falciparum-associated co-infection, patterns of antimicrobial resistance, and emergence of reduced susceptibility to fluoroquinolones[J]. J Health Popul Nutr, 2007, 25(3): 351–358 . [PMC free article] [PubMed] [Google Scholar]
- 13. Adesiji YO, Alli O, Adekanle M, et al. Prevalence of Arcobacter, Escherichia coli, Staphylococcus aureus and Salmonella species in Retail Raw Chicken, Pork, Beef and Goat meat in Osogbo, Nigeria[J]. Sierra Leone J Biomed Res, 2011, 3(1): 8–12 10.4314/sljbr.v3i1.66644. [DOI] [Google Scholar]
- 14. Mensah P, Mwamakamba L, Kariuki S, et al. Strengthening foodborne diseases surveillance in the WHO African region: an essential need for disease control and food safety assurance[J]. Afr J Food Agric Nutr Dev, 2012, 12(4): 6336–6353. [Google Scholar]
- 15. Andrews WH, Hammack T. Salmonella.In Bacteriological Analytical Manual US Food and Drug Administration[J]. 2011. Available from: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/default.htm.
- 16. MacFaddin JF. Biochemical tests for identification of medical bacteria[J], 3rd ed. Lippincott Williams & Wilkins, Philadelphia 2000.
- 17. Rahn K, De Grandis SA, Clarke RC, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella [J]. Mol Cell Probes, 1992, 6(4): 271–279. 10.1016/0890-8508(92)90002-F [DOI] [PubMed] [Google Scholar]
- 18. Jones DD, Law R, Bej AK. Detection of Salmonella spp. in oysters using polymerase chain reaction (PCR) and gene probes.[J]. J Food Sci, 1993, 58(6): 1191–1197 10.1111/j.1365-2621.1993.tb06146.x. [DOI] [Google Scholar]
- 19. Casin I, Breuil J, Darchis JP, et al. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans[J]. Emerg Infect Dis, 2003, 9(11): 1455–1457. 10.3201/eid0911.030317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robicsek A, Strahilevitz J, Sahm DF, et al. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States[J]. Antimicrob Agents Chemother, 2006, 50(8): 2872–2874. 10.1128/AAC.01647-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma M, Wang H, Yu Y, et al. Detection of antimicrobial resistance genes of pathogenic Salmonella from swine with DNA microarray[J]. J Vet Diagn Invest, 2007, 19(2): 161–167. 10.1177/104063870701900204 [DOI] [PubMed] [Google Scholar]
- 22. Aarestrup FM, Agerso Y, Gerner-Smidt P, et al. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark[J]. Diagn Microbiol Infect Dis, 2000, 37(2): 127–137. 10.1016/S0732-8893(00)00130-9 [DOI] [PubMed] [Google Scholar]
- 23. Bauer AW, Kirby WMM, Sherris JC, et al. Antibiotic susceptibility testing by a standardized single disk method[J]. Am J Clin Pathol, 1966, 45(4): 493–496 . [PubMed] [Google Scholar]
- 24. Singh S, Yadav AS, Singh SM, et al. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance[J]. Food Res Int, 2010, 43(8): 2027–2030 10.1016/j.foodres.2010.06.001. [DOI] [Google Scholar]
- 25. Ausubel F, Brent R, Kingston RE, et al. Short protocols in molecular biology(3rd Ed.)[J]. John Wiley & Sons, New York; 1995. [Google Scholar]
- 26. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual[J]. Small Scale Preparation of Plasmid DNA: Alkaline Lysis Method. Cold Spring Harbor Laboratory Press, New York; 1989; 1.25–1.27. [Google Scholar]
- 27. Hohmann EL. Nontyphoidal salmonellosis[J]. Clin Infect Dis, 2001, 32(2): 263–269. 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 28. Falagas ME, Michalopoulos A. Polymyxins: old antibiotics are back[J]. Lancet, 2006, 367(9511): 633–634. 10.1016/S0140-6736(06)68241-X [DOI] [PubMed] [Google Scholar]
- 29. Majumder S, Rahmatullah M. Efficacy of old antibiotics against commonly isolated bacterial isolates in a tertiary care hospital[J]. J Chem Pharm Res, 2015, 7(4): 1392–1396. [Google Scholar]
- 30. Helms M, Vastrup P, Gerner-Smidt P, et al. Excess mortality associated with antimicrobial drug-resistant Salmonella typhimurium[J]. Emerg Infect Dis, 2002, 8(5): 490–495. 10.3201/eid0805.010267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weigel LM, Steward CD, Tenover FC. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae[J]. Antimicrob Agents Chemother, 1998, 42(10): 2661–2667 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hakanen A, Kotilainen P, Huovinen P, et al. Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia[J]. Emerg Infect Dis, 2001, 7(6): 996–1003. 10.3201/eid0706.010613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reyna F, Huesca M, González V, et al. Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance[J]. Antimicrob Agents Chemother, 1995, 39(7): 1621–1623. 10.1128/AAC.39.7.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giraud E, Baucheron S, Cloeckaert A. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies[J]. Microbes Infect, 2006, 8(7): 1937–1944. 10.1016/j.micinf.2005.12.025 [DOI] [PubMed] [Google Scholar]
- 35. Hernández A, Sánchez MB, Martínez JL. Quinolone resistance: much more than predicted[J]. Front Microbiol, 2011, 2(22): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]


