Abstract
H2B ubiquitylation (H2Bub)-dependent H3K4 methylation is mediated by the multisubunit Set1 complex (Set1C) in yeast, but precisely how Set1C subunits contribute to this histone modification remains unclear. Here, using reconstituted Set1Cs and recombinant H2Bub chromatin, we identified Set1C subunits and domains involved in the H2Bub-dependent H3K4 methylation process, showing that the Spp1 PHDL domain, in conjunction with the Set1 n-SET domain, interacts with Swd1/Swd3 and that this interaction is essential for H2Bub-dependent H3K4 methylation. Importantly, Set1C containing an Spp1-Swd1 fusion bypasses the requirement for H2Bub for H3K4 methylation, suggesting that the role of H2Bub is to induce allosteric rearrangements of the subunit-interaction network within the active site of Set1C that are necessary for methylation activity. Moreover, the interaction between the Set1 N-terminal region and Swd1 renders the Spp1-lacking Set1C competent for H2Bub-dependent H3K4 methylation. Collectively, our results suggest that H2Bub induces conformational changes in Set1C that support H3K4 methylation activity.
INTRODUCTION
Evolutionarily conserved histone H3 lysine 4 (H3K4) methylation plays important roles in many cellular processes, including transcription, DNA replication and repair, meiotic recombination, and class-switch recombination, among others (1–4). Defects in histone H3K4 methylation are closely correlated with human pathologies (5,6), emphasizing the importance of understanding underlying molecular mechanisms that govern H3K4 methylation.
In contrast to metazoans, which have multiple H3K4 methyltransferase enzymes, the budding yeast Saccharomyces cerevisiae contains a single H3K4 methyltransferase (Set1) that forms a multisubunit complex (Set1 complex) with seven other subunits (Swd1, Swd3, Bre2, Sdc1, Spp1, Swd2 and Shg1) (7), thus providing a good model system for studying the molecular basis of the regulation of H3K4 methylation. Genetic studies have revealed contributions of each subunit to cellular H3K4 methylation levels (8–11). However, these studies do not exclude the possibility that various cellular factors indirectly affect H3K4 methylation, emphasizing the necessity of biochemical analyses using defined factors to directly assess the precise roles of each subunit in the Set1 complex (Set1C) during the H3K4 methylation process.
The requirement for prior mono-ubiquitylation at histone H2B lysine 123 (H2Bub) is one of most interesting features of H3K4 methylation in yeast (12–14). Such histone modification crosstalk is also observed in at least a subset of human H3K4 methyltransferase complexes (15,16), emphasizing the importance of understanding the mechanistic details of this trans-tail histone modification for the development of H3K4 methylation-related therapies. Previous studies have proposed at least three mechanistic models for H2Bub-dependent H3K4 methylation that are not mutually exclusive (17): (i) H2Bub-mediated alteration of the nucleosome configuration to one favorable for H3K4 methylation (18), (ii) H2Bub-dependent recruitment of Set1C to chromatin (19) and (iii) H2Bub-induced conformational changes in the catalytic region, resulting in altered catalytic properties of Set1C (15). Despite extensive investigation, a definitive answer for how Set1C recognizes and methylates H2Bub-chromatin has yet to be provided.
Here, using a biochemically defined system reconstituted with recombinant H2Bub-containing chromatin (20) and recombinant Set1Cs (15), we provide mechanistic insights into the functions of Set1C subunits/domains during the H2Bub-dependent H3K4 methylation process. Our study reveals a previously unrecognized function of Spp1 and the N-terminal region of Set1 in mediating H2Bub-dependent H3K4 methylation that involves crosstalk among subunits that induces allosteric activation of the catalytic activity of Set1C.
MATERIALS AND METHODS
cDNA, plasmids, baculoviruses, recombinant proteins and Set1 complex purifications
The cDNAs for yeast genes were PCR-amplified from yeast genomic DNA. For protein expression in yeast, cDNAs were subcloned into pRS416 (ATCC). For GST-tagged proteins, cDNAs were subcloned into pGEX4T (Amersham), expressed in Escherichia coli, and purified on Glutathione-Sepharose 4B beads (GE Healthcare). For baculovirus-mediated expression, cDNAs were subcloned into pFASTBAC1 (Gibco-Invitrogen) with or without an epitope tag and baculoviruses were generated according to the manufacturer's instruction (Gibco-Invitrogen). For expression of FLAG-tagged proteins, and for reconstitution of Set1 complexes containing FLAG-Set1 or FLAG-Set1 fragments, Sf9 cells were infected with combinations of baculoviruses and proteins/complexes were affinity purified on M2 agarose (Sigma) as described (21). The expression and purification of recombinant Xenopus histones, semi-synthetic H2Bub, histone octamers, NAP1 and the ACF complex were as described (20,21).
In vitro chromatin assembly for histone methyltransferase assays
Procedures for chromatin assembly with the recombinant ACF/NAP1 system were as described (21). Briefly, the reaction containing core histone octamer (700 ng) and NAP1 (2.4 μg) in 55 μl HEG buffer (25 mM HEPES [pH 7.6], 0.1 mM EDTA and 10% glycerol) was incubated on ice for 30 min. After the further addition of the ACF complex (160 ng) and the p53ML plasmid (22, 700 ng), the reaction was adjusted to 25 mM HEPES [pH 7.6], 0.1 mM EDTA, 10% glycerol, 50 mM KCl, 3.2 mM ATP, and 5 mM MgCl2 in a final volume of 70 μl and incubated at 27°C for 4 h.
In vitro histone methyltransferase assays
For free histone H3 methyltransferase assays, reactions containing 100 ng recombinant histone H3 and purified Set1C (containing 30 ng Bre2 subunit) in 20 μl reaction buffer (25 mM HEPES [pH 7.6], 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA and 10% glycerol) supplemented with 100 μM SAM (S-adenosyl methionine, NEB) were incubated at 30°C for 1 h. For recombinant chromatin methyltransferase assays, reactions containing 350 ng (histone amount) recombinant chromatin (35 μl, assembled as above), purified Set1C (containing 30 ng Bre2 subunit) and 100 μM SAM were adjusted to 40 μl with HEG buffer and incubated at 30°C for 1 h. Proteins were resolved by SDS-PAGE and subjected to immunoblotting.
Protein interaction assays
For GST-pull down assays, 1 μg of GST or GST-tagged proteins immobilized on Glutathione-Sepharose 4B beads were incubated with 200 ng of purified proteins in the binding buffer (20 mM Tris–Cl [pH 7.9], 150 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.05% NP-40 and 0.2 mg/ml BSA) at 4°C for 3 h and then beads were extensively washed with the binding buffer. For peptide-pull down assays, 1 μg of biotinylated histone H3 peptides (1–20 amino acid residues) immobilized on Streptavidin Agarose beads (Pierce) were incubated with 500 ng of purified proteins in the binding buffer (50 mM Tris–Cl [pH 7.5], 150 mM NaCl and 0.05% NP-40) at 4°C for overnight and then beads were extensively washed with the binding buffer. Bound proteins were analyzed by immunoblotting.
Yeast strains
All yeast strains used in this study were derived from the BY4741 strain and summarized in Supplementary Table S1.
Whole cell extracts and immunoblot analyses
Yeast whole cell extracts were prepared from cells in exponential growth phase either in YPD or in synthetic media by TCA (trichloroacetic acid) extraction and then subjected to immunoblot analyses.
Antibodies
Polyclonal anti-Spp1 antibody was obtained from the Géli laboratory. The following antibodies were obtained commercially: anti-H3 (Abcam, ab1791); anti-H3K4me1 (Abcam, ab8895); anti-H3K4me2 (Abcam, ab7766); anti-H3K4me3 (Active Motif, AM39159); anti-FLAG (Sigma, A8592); anti-HA (Abcam, ab9110).
RESULTS
The PHDL (PHD-Like) domain within Spp1 is essential for H2B ubiquitylation-dependent H3K4 methylation activity of the C762 complex
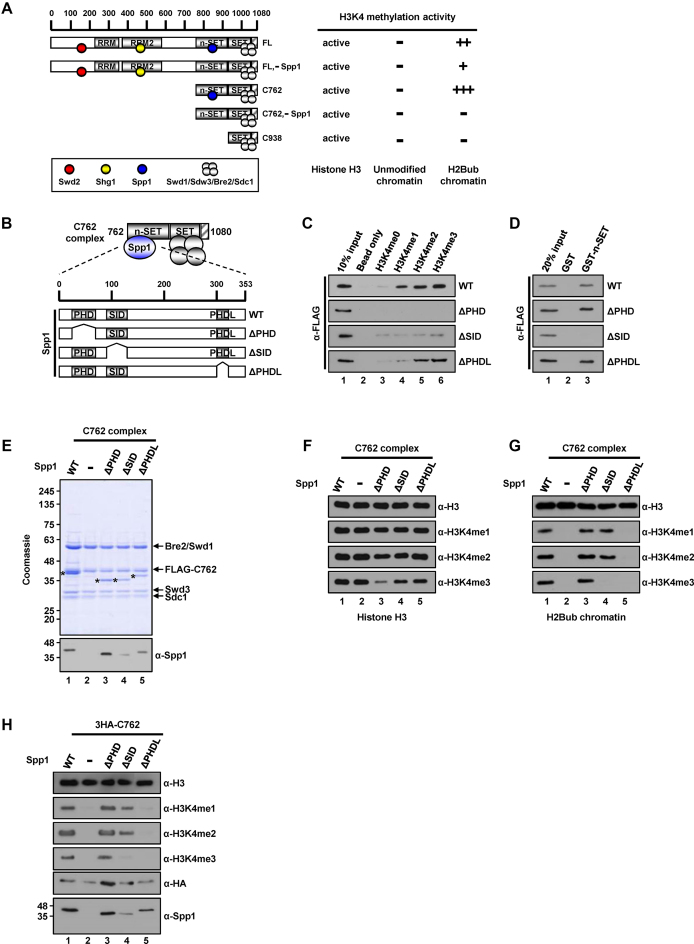
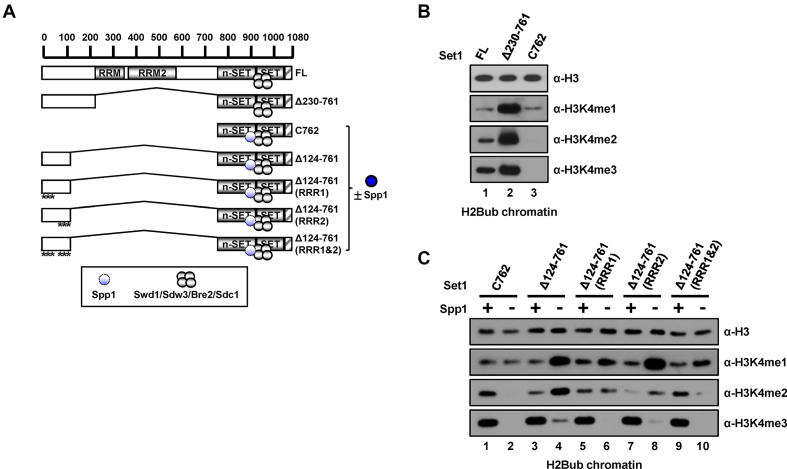
We previously demonstrated that the C762 complex, a partial Set1 complex that contains a Set1 fragment (residues 762–1080) encompassing the n-SET, SET and post-SET domains together with the five associated subunits, Spp1, Swd1, Swd3, Bre2 and Sdc1, exhibits strong H3K4 methylation activity toward H2Bub-containing nucleosomes (15). Furthermore, a C762-based complex lacking Spp1 was unable to methylate H2Bub chromatin, despite its intrinsic ability to methylate free histone H3 (H3) in vitro (summarized in Figure 1A, Supplementary Figure S1A and B) and in vivo (15,23). These results indicate that, at least in the absence of Set1 N-terminal residues 1–761 (see below), Spp1 acts in conjunction with the n-SET domain of Set1 and the Set1C catalytic core (i.e. C938 complex)—composed of SET and post-SET domains (Set1 residues 938–1080) plus Swd1, Swd3, Bre2 and Sdc1 (Supplementary Figure S2A)—to mediate H2Bub-dependent H3K4 methylation.
Figure 1.
Requirements of Spp1 domains for H3K4 methylation activity of the C762 Set1 complex. (A) A schematic diagram of full-length (FL) Set1 and C762 Set1 fragment along with associating subunits deduced from previous study (15). RRM (RNA recognition motif), RRM2 (RNA recognition motif 2), n-SET, SET, and post-SET (hatched box) domains within Set1 protein are depicted. H3K4 methylation activities of purified Set1Cs on free histone H3 and recombinant chromatin templates assembled with unmodified H2B or H2Bub-containing octamers (Supplementary Figure S1A and B) are summarized on the right. (B) A schematic diagram of the C762-based Set1 complexes containing wild-type (WT) or mutant Spp1. (C) Requirement of the PHD domain of Spp1 for binding to histone H3 peptides with mono- (H3K4me1), di- (H3K4me2), tri- (H3K4me3), or no (H3K4me0) methylation at lysine 4 residue. Biotin pull-down assays employed biotinylated H3 peptides and purified FLAG-Spp1 proteins (Supplementary Figure S1E) and peptide-bound proteins were scored by immunoblotting with anti-FLAG antibody. (D) Requirement of the SID domain of Spp1 for binding to the n-SET domain of Set1. GST pull-down assays employed GST-tagged n-SET fragment (Set1 residues 762–937) (Supplementary Figure S1F) and purified FLAG-Spp1 proteins (Supplementary Figure S1E) and bound proteins were scored by immunoblotting with anti-FLAG antibody. (E) SDS-PAGE/Coomassie blue staining and anti-Spp1 immunoblot analyses of purified C762-based Set1 complexes reconstituted with baculoviruses expressing FLAG-tagged C762 Set1, Swd1, Swd3, Bre2, Sdc1, and either WT or mutant Spp1. Spp1 polypeptides are marked by asterisks. (F and G) Requirement of the PHDL domain of Spp1 for in vitro H3K4 methylation activity of Set1C on H2Bub chromatin. Free histone H3 (F) and H2Bub chromatin (G) were subjected to in vitro histone methyltransferase (HMT) assays with indicated purified C762-based Set1 complexes (E). H3K4 methylation status was monitored by immunoblotting with indicated antibodies. (H) Requirement of the PHDL domain of Spp1 for H3K4 methylation in yeast cells. Plasmids expressing WT or mutant Spp1 were introduced into the set1Δspp1Δ yeast strain that contains a chromosomal gene expressing HA (three copies)-tagged C762 Set1. Yeast whole cell extracts were analyzed by immunoblotting with indicated antibodies. H2Bub, K120-ubiquitylated H2B; H3K4me1, K4-monomethylated H3; H3K4me2, K4-dimethylated H3; H3K4me3, K4-trimethylated H3.
Accordingly, we analyzed the C762 complex as a model to define the molecular function of Spp1 in H2Bub-dependent H3K4 methylation. A multispecies alignment predicted a PHD (plant homeodomain), an SID (Set1-interacting domain), and a PHDL (PHD-Like) domain within Spp1, all of which are well conserved among different species (Supplementary Figure S1C and D). To test whether each domain has a function similar to that predicted from studies on CFP1, a mammalian homolog of Spp1, we first purified FLAG-tagged Spp1 mutant proteins in which individual domains were deleted (Figure 1B, Supplementary Figure S1E). In a test for binding to synthetic peptides containing methylated H3K4, only the Spp1 mutant lacking PHD (ΔPHD-Spp1) exhibited a complete defect in interactions with H3K4-methylated peptides (Figure 1C), despite the fact that this mutant contains a PHDL domain, which has significant homology with the PHD2 of CFP1 (Supplementary Figure S1D). These results suggest that the ability of Spp1 to recognize methylated H3K4 (24,25) is attributable to the PHD rather than the PHDL domain. We also observed decreased interactions with H3K4-methylated peptides in the Spp1 mutant lacking SID (ΔSID-Spp1), which might be due to alteration of protein characteristics caused by deletion of this domain. Given previous observations that the SID within CFP1 binds to human Set1A and Set1B proteins (26) and that Spp1 selectively interacts with the n-SET domain of Set1 (15), we next examined direct interactions between purified Spp1 mutant proteins and the purified n-SET domain (Set1 residues 762–937) (Supplementary Figure S1F). These analyses revealed that ΔSID-Spp1 failed to bind to the n-SET domain (Figure 1D), confirming that the SID also plays a role in the association of Spp1 with the yeast Set1 complex.
To determine the domain within Spp1 involved in H3K4 methylation, we reconstituted C762-based Set1 complexes and affinity purified them from Sf9 cells infected with baculoviruses expressing FLAG-C762 Set1, four untagged catalytic core subunits (Swd1, Swd3, Bre2 and Sdc1), and either wild-type (WT) or mutant Spp1. All Spp1 mutant proteins, surprisingly including ΔSID-Spp1 which is unable to bind the n-SET domain, were stably integrated into the complex with all other subunits (Figure 1E). These results strongly suggest that Spp1 can be retained in the complex through secondary interactions with Set1C components other than the n-SET domain (see below).
We then assessed the contribution of each domain within Spp1 to H3K4 methylation activity using an in vitro (cell-free) histone methyltransferase (HMT) assay, first employing free H3 as the substrate. Consistent with the dispensability of Spp1 in a C762-based complex for methylation of free H3 (Figure 1A), all complexes exhibited comparable levels of H3K4 mono-, di-, and trimethylation (H3K4me1, H3K4me2 and H3K4me3, respectively) (Figure 1F), although H3K4me3 levels were somewhat decreased for the ΔPHD-Spp1–containing complex (Figure 1F, lane 3). We next tested the H3K4 methylation activity of complexes towards a more physiologically relevant H2Bub-containing chromatin template (Figure 1G). The complex containing ΔPHD-Spp1 generated levels of all three H3K4 methylation states similar to those observed with the complete C762 complex (Figure 1G, lane 3), and the ΔSID-Spp1–containing complex exhibited H3K4me1/me2 activity, but no H3K4me3 activity (Figure 1G, lane 4). Importantly, we further found that the complex reconstituted with ΔPHDL-Spp1 was totally inactive with respect to H3K4 methylation (Figure 1G, lane 5), similar to the case for the C762-based complex reconstituted without Spp1 (Figure 1G, lane 2).
To confirm the critical role of the PHDL domain for H3K4 methylation in intact cells, we generated a yeast strain carrying a chromosomal copy of HA-C762 Set1 derived from an spp1-deletion background. After reintroduction of plasmids expressing either WT or mutant Spp1, we monitored H3K4 methylation levels (Figure 1H). Consistent with results obtained using a reconstituted recombinant protein system, the ΔSID-Spp1 strain exhibited a selective loss of H3K4me3 (Figure 1H, lane 4), whereas the ΔPHDL-Spp1 strain showed almost undetectable levels of all three H3K4 methylation states (Figure 1H, lane 5). Hence, these data strongly suggest that the PHDL domain within Spp1 plays a critical role in C762 complex-mediated, H2Bub-dependent H3K4 methylation of the chromatin template.
The PHDL domain mediates interactions between Spp1/n-SET and Swd1/Swd3 within the catalytic core of the Set1 complex
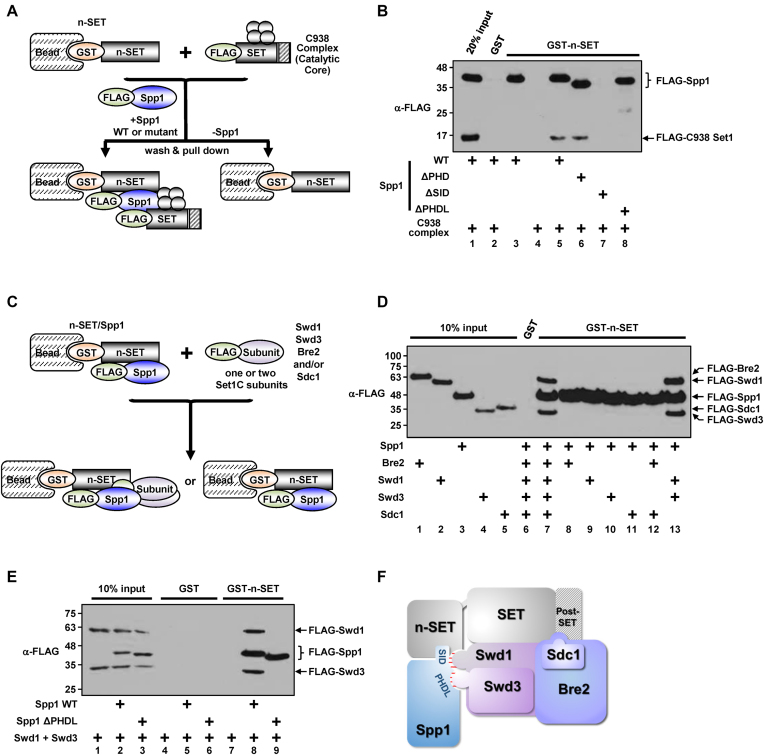
We previously showed that Spp1 mediates physical interactions between the n-SET domain and the catalytic core (15). With Spp1 mutant proteins lacking individual domains in hand, we next sought to detail the role of Spp1 as a bridging molecule within the C762 complex and determine its relationship with the H2Bub-dependent H3K4 methylation activity of the complex. To determine the Spp1 domain required for bridging the n-SET domain and the catalytic core, we performed in vitro protein-interaction assays employing a purified n-SET fragment, the C938 complex and either WT or mutant Spp1, as shown schematically in Figure 2A. Consistent with our previous observation (15), a GST-immobilized n-SET fragment was able to bind the C938 complex only in the presence of Spp1 (Figure 2B, lane 5 versus lane 4). The addition of ΔPHD-Spp1 also resulted in association of the C938 complex with n-SET (Figure 2B, lane 6), whereas addition of ΔSID-Spp1 did not (Figure 2B, lane 7), reflecting its inability to bind to n-SET (Figure 1D). Importantly, we found that ΔPHDL-Spp1 is unable to mediate interactions between the n-SET domain and the C938 complex (Figure 2B, lane 8), strongly suggesting that the defective H3K4 methylation activity of the ΔPHDL-Spp1–containing complex towards an H2Bub chromatin template (Figure 1G) might involve failure of ΔPHDL-Spp1 to mediate trans-interactions between the n-SET domain and the catalytic core.
Figure 2.
Direct domain-subunit interactions within the C762 Set1 complex. (A) A Schematic representation of the in vitro protein interaction assays in (B). Four gray-colored circles indicate Swd1, Swd3, Bre2 and Sdc1. (B) Requirement of the SID and PHDL domains of Spp1 for trans-interaction between the n-SET domain and the catalytic core of Set1C. The purified C938 complex (Supplementary Figure S2A) was tested for binding to GST-n-SET in the presence of purified WT or mutant Spp1. (C) A Schematic representation of the in vitro protein interaction assays in (D) and (E). (D) Physical interaction of Swd1 and Swd3 with Spp1 bound to the n-SET domain. Combinations of purified Set1C subunits (Supplementary Figure S2B) as indicated were tested for binding to GST-n-SET in the presence of purified Spp1. Note that FLAG-Sdc1 ran slower than FLAG-Swd3 in this assay where a gradient SDS-PAGE gel was used for better resolution of protein migrations. (E) Requirement of the PHDL domain for Spp1-mediated interaction of the n-SET domain with Swd1/Swd3 heterodimer. Purified Swd1 and Swd3 proteins were tested for binding to GST-n-SET in the presence of either purified WT or ΔPHDL-Spp1. (F) A schematic model of subunit/domain interactions in the C762 Set1 complex. A surface created by the n-SET domain and Spp1 interacts with heterodimeric Swd1/Swd3 (highlighted by red lines). Direct association of heterodimeric Swd1/Swd3 and Bre2/Sdc1 with SET and post-SET domains has been established by previous studies (15,33,34). The SID domain may also contribute to the proper subunit-interaction network in the catalytic region of Set1C, as inferred from the selective loss of H3K4me3 in the ΔSID-Spp1–containing complex (Figure 1G and H).
Taken together with the above results, the presence of ΔSID-Spp1 in the purified C762-based complex (Figure 1E) led us to postulate that, in addition to the n-SET domain, Spp1 can also interact with one or more components in the catalytic core. To test this idea, we examined interactions of n-SET domain-bound Spp1 (Spp1/n-SET) with purified Set1C subunits (Supplementary Figure S2B), as shown schematically in Figure 2C. We found that Spp1/n-SET selectively interacted with Swd1 and Swd3 in a reaction containing all four subunits (Swd1, Swd3, Bre2 and Sdc1) (Figure 2D, lane 7), but not with any individual subunit alone (Figure 2D, lanes 8–11). Moreover, Swd1 and Swd3 were found to be sufficient for Spp1/n-SET binding (Figure 2D, lane 13), confirming the minimal subunit requirement. Finally, and importantly, we further confirmed that the PHDL domain plays a key role in the interaction between Spp1/n-SET and Swd1/Swd3 (Figure 2E, lane 9 versus lane 8). Collectively, these results suggest that the n-SET domain of Set1 and the PHDL domain of Spp1 together create a binding platform for interaction with heterodimeric Swd1/Swd3 in the catalytic core (Figure 2F), an interaction that is correlated with H2Bub-dependent H3K4 methylation (Figure 1G and H).
Physical linkage between Spp1 and Swd1 renders the complex competent for nucleosomal H3K4 methylation in the absence of H2B ubiquitylation
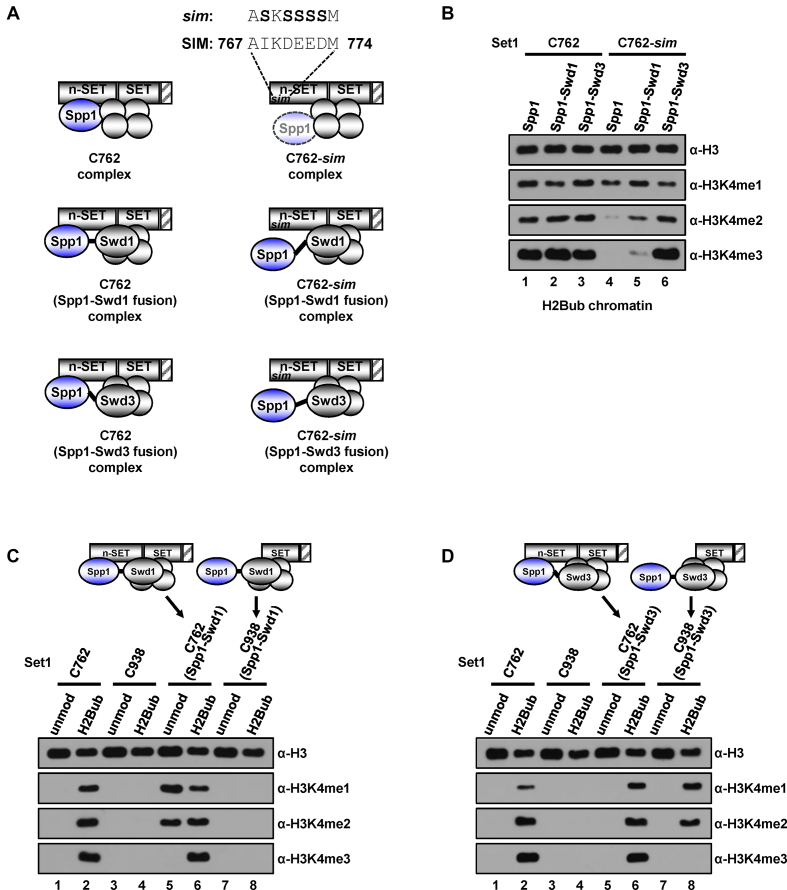
Deletion of the n-SET domain results in the concomitant disappearance of Spp1 from Set1C; the resulting Set1C is incompetent for H2Bub-dependent H3K4 methylation (15). Similar to the case above, where Spp1 plays a critical role in the catalytic activity of the C762 complex, this situation complicates the task of defining the role of the n-SET domain because the n-SET domain may participate in the H3K4 methylation process (i) as a catalytic player in conjunction with Spp1 or (ii) by merely presenting Spp1 to the catalytic core. To differentiate these mechanisms, we performed the following experiments using purified Set1Cs (Supplementary Figure S3A and B). First, we reconstituted a C762-based complex containing either an Spp1-Swd1 or an Spp1-Swd3 fusion protein (Figure 3A, middle and bottom left, respectively). We found that these complexes exhibited H3K4 methylation activities comparable to that of the intact C762 complex (Figure 3B, lanes 2 and 3 versus lane 1), indicating that a physical linkage between Spp1 and Swd1 or Swd3 does not disrupt the intrinsic enzymatic activity of the complex. Second, we introduced serine substitutions at several amino acid residues in the Spp1-interacting motif (SIM), located within n-SET, to uncouple Spp1 from the n-SET domain (Figure 3A, upper right). Consistent with a previous report (25), these mutations led to a defect in interactions of Spp1 with the n-SET domain (Supplementary Figure S3D and E) and caused almost a complete disappearance of Spp1 in the reconstituted complex (Supplementary Figure S3A, anti-Spp1 immunoblot, lane 4). The resulting C762-sim complex generated H3K4me1 and significantly decreased levels of H3K4me2, but failed to generate H3K4me3 (Figure 3B, lane 4). The interpretation of these results is that the residual amount of Spp1 within the complex is responsible for generating H3K4me1, but is insufficient for generating H3K4me2 and H3K4me3. Third, combining the above two strategies, we generated a complex in which Spp1 is physically linked to Swd1, but is unable to interact with the n-SET domain (Figure 3A, middle right). Tests of H2Bub-dependent H3K4 methylation activity showed that the resulting complex generated significant, albeit somewhat decreased, levels of all three H3K4 methylation states (Figure 3B, lane 5). Similarly, we found that a C762-sim–based complex containing an Spp1-Swd3 fusion (Figure 3A, lower right) also generated significant levels of all three H3K4 methylation states (Figure 3B, lane 6). Importantly, these results indicate that physical association of Spp1 with the catalytic core is sufficient to support the H3K4 methylation activity of Set1C, even if the interaction of Spp1 with the n-SET domain is disrupted.
Figure 3.
H3K4 methylation activities of the subunit fusion-containing Set1 complexes. (A) Schematic representations of anticipated subunit associations in the purified complexes used in an in vitro HMT assay in (B). Four gray-colored circles indicate Swd1, Swd3, Bre2 and Sdc1. Amino acid substitutions to serine (bold) in the SIM (Spp1-interacting motif) are depicted in the C762-sim Set1 complex (upper right). In Spp1 and either Swd1 or Swd3 fusion proteins, two subunits are connected by the linker amino acids (GGGGGGAAA, depicted by short black lines). Depiction of faint Spp1 in the C762-sim Set1 complex implies its inefficient retention in the complex (Supplementary Figure S3A). (B) H3K4 methylation activities of Set1Cs containing physically-associated subunits. H2Bub chromatin templates were subjected to in vitro HMT assays with purified C762-based Set1 complexes containing subunit fusions (Supplementary Figure S3A). (C and D) Spp1-Swd1 fusion bypasses requirement of H2Bub for H3K4 methylation activity of the C762-based Set1 complex (C). Spp1-Swd3 fusion bypasses requirement of the n-SET domain for H2Bub-dependent H3K4 methylation activity of the C938-based Set1 complex (D). Chromatin templates assembled with unmodified H2B- or H2Bub-containing octamers were subjected to in vitro HMT assays with purified Set1Cs (Supplementary Figure S3B) that all have intact intrinsic H3K4 methyltransferase activities on free H3 (Supplementary Figure S3C). Subunit compositions and subunit fusions within the complexes are depicted at the top. unmod, unmodified H2B.
In a test for H2Bub dependence, we surprisingly found that the Spp1-Swd1 fusion-containing, C762-based complex is able to generate H3K4me1 and H3K4me2 in the absence of H2Bub (Figure 3C, lane 5). This strongly suggests that the forced proximity between Spp1 and Swd1 bypasses the requirement for H2Bub for the H3K4 methylation activity of Set1C. Importantly, this further suggests the role of H2Bub on the nucleosome is to induce rearrangements of subunit interactions in the catalytic region of Set1C that render the complex competent for H3K4 methylation (see Discussion). Moreover, we found that a C938-based complex containing an Spp1-Swd3 fusion also generated H3K4me1 and H3K4me2 in an H2Bub-dependent manner (Figure 3D, lanes 7 and 8), indicating that the forced proximity between Spp1 and Swd3 bypasses the requirement for an n-SET domain for the H2Bub-dependent H3K4 methylation activity of Set1C.
Collectively, these results suggest that a role of the n-SET domain is to locate Spp1 at the proper position for interacting with the catalytic core, an interaction that is required for the H3K4 methylation activity of Set1C. Failure of the C762-based complex containing an Spp1-Swd1 fusion (Figure 3C, lane 5) and the C938-based complex containing an Spp1-Swd3 fusion (Figure 3D, lane 8) to generate H3K4me3 is probably attributable to the unnatural linkage between Spp1 and Swd1 or Swd3, which prevents enzymatic activity from proceeding beyond the H3K4 di-methylation state.
Set1 N-terminal residues 1–229 play a role in H2B ubiquitylation-dependent H3K4 methylation activity in the absence of Spp1
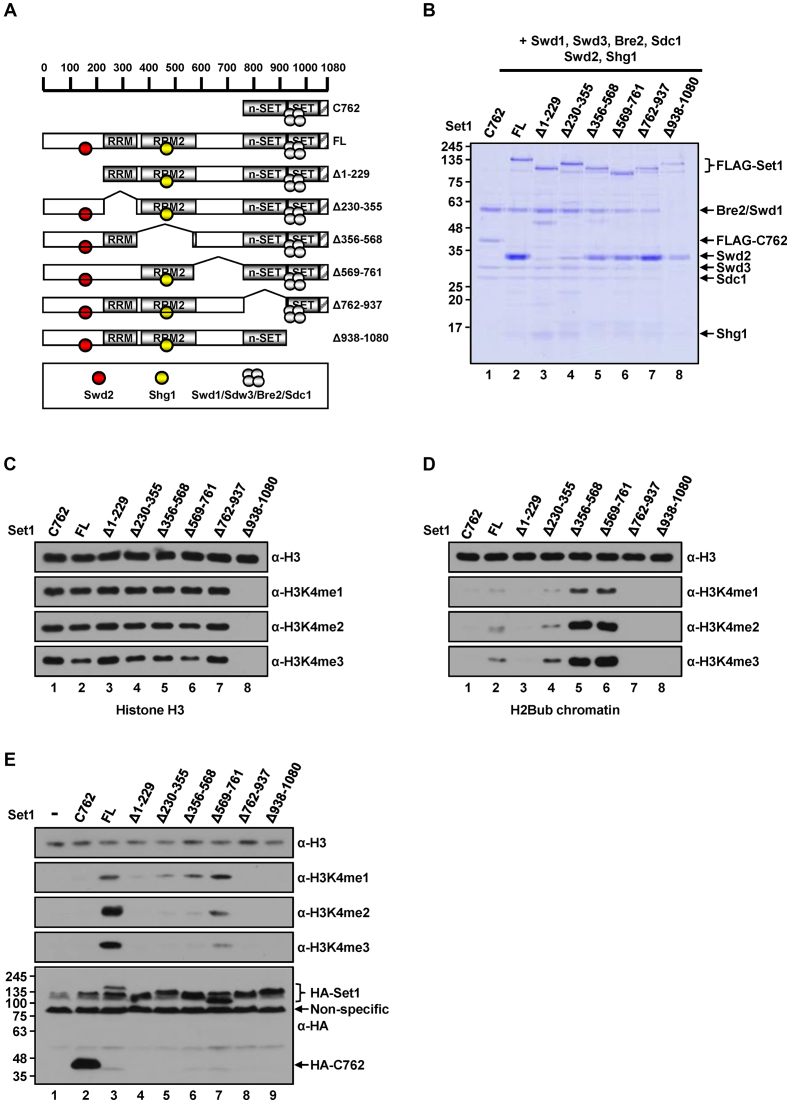
In contrast to the strict requirement for Spp1 for the H2Bub-dependent H3K4 methylation activity of the C762 complex, Spp1-deficient Set1C containing full-length Set1 exhibited significant H3K4 methylation activity in vitro (15) (summarized in Figure 1A, Supplementary Figure S1A and B) and in vivo (10,11,15), although this activity was moderately decreased compared with that of complete Set1C. This conditional requirement for Spp1 raises the possibility that some region(s) within the first 761 amino acid residues of Set1 play a redundant role in H2Bub-dependent H3K4 methylation, through a mechanism similar to (or possibly distinct from) Spp1. To determine region(s) required for H3K4 methylation activity of Set1C in the absence of Spp1, we generated a series of deletion mutants of FLAG-tagged Set1 and then reconstituted them into complexes with six other (untagged) subunits (Swd1, Swd3, Bre2, Sdc1, Swd2 and Shg1), but without Spp1 (Figure 4A). Compositional analyses revealed that all purified complexes contained the expected subunits, as depicted in Figure 4A (Figure 4B).
Figure 4.
Requirement of the Set1 N-terminal region for H3K4 methylation activity of the Set1 complex in the absence of Spp1. (A) A schematic diagram of Set1Cs containing FL Set1 and derived Set1 fragments along with associating subunits. Note that Spp1 is absent in all complexes. (B) SDS-PAGE/Coomassie blue staining of purified Set1Cs reconstituted with baculoviruses expressing FLAG-Set1 or FLAG-Set1 fragments and six (untagged) Set1C subunits. (C and D) Requirement of the Set1 N-terminal 1-229 residues for H2Bub-dependent H3K4 methylation activity of the Spp1-deficient Set1Cs. Free histone H3 (C) and H2Bub chromatin (D) were subjected to in vitro HMT assays with indicated purified Set1Cs (B). (E) Requirement of the Set1 N-terminal 1-229 resides for H3K4 methylation in yeast cells that lack Spp1. Plasmids expressing indicated HA (three copies)-tagged FL Set1 or Set1 fragments were introduced into the set1Δspp1Δ yeast strain. Yeast whole cell extracts were analyzed by immunoblotting with indicated antibodies.
In functional in vitro assays, all complexes, except the Δ938–1080 Set1 complex, generated comparable levels of all three H3K4 methylation states on free H3 (Figure 4C), confirming that the catalytic core has intrinsic H3K4 methylation activity. Importantly, tests of H3K4 methylation activity towards an H2Bub-containing chromatin substrate (Figure 4D) revealed that the Δ1–229 Set1 complex showed no H3K4 methylation activity (Figure 4D, lane 3). However, complexes with Set1 internal deletions exhibited markedly enhanced (Δ356–568 and Δ569–761, Figure 4D, lanes 5 and 6) or comparable (Δ230–355, Figure 4D, lane 4) H3K4 methylation activity relative to the full-length Set1-containing complex (Figure 4D, lane 2). Notably, a common feature of these H3K4 methylation-competent complexes is inclusion of the first N-terminal 229 amino acid residues of Set1 and the presence of associating Swd2. However, the fact that these complexes gained H3K4 methylation activity is not attributable to Swd2, because we previously showed that a partial Set1C composed of five subunits (full-length Set1, Swd1, Swd2, Bre2 and Sdc1), but lacking Swd2, Shg1 and Spp1, possesses H2Bub-dependent H3K4 methylation activity (15, see also Figure 6B, lane 1). Finally, as expected, and consistent with our previous report (15), deletion of n-SET (Δ762–937) and SET plus post-SET (Δ938–1080) domains resulted in complete loss of H3K4 methylation activity (Figure 4D, lanes 7 and 8).
Figure 6.
Requirement of the basic motifs within the Set1 N-terminal region for H3K4 methylation activity of the Spp1-deficient Set1 complex. (A) A schematic diagram of Set1Cs containing Set1 and its derived fragments along with associating subunits. Asterisks indicate arginine to alanine substitutions. Dotted outline and faint depiction for Spp1 indicate its conditional presence in the complexes depending on the inclusion of Spp1 during complex reconstitution. Note that Swd2 and Shg1 are not included for all complex preparations. (B and C) In vitro HMT assays with H2Bub chromatin and indicated purified Set1Cs (Supplementary Figure S6A). Note that all purified complexes used in (B) do not contain Spp1.
To confirm these results in intact cells, we expressed HA-tagged full-length Set1 and Set1 fragments in the set1Δspp1Δ yeast strain and examined global levels of H3K4 methylation (Figure 4E). Consistent with results from our cell-free reconstitution system, the three strains that expressed the N-terminal 1-229 and C-terminal 762–1080 (n-SET, SET and post-SET domains) residues of Set1 in common showed all three H3K4 methylation states (Figure 4E, lanes 5–7), whereas those with a deletion of any of these regions showed no detectable activity (Figure 4E, lanes 4, 8 and 9). Similar results were obtained using purified complexes and yeast strains containing Spp1 in the same series of experiment as deployed in Figure 4, although the Spp1-independent role of the Set1 N-terminal region in H3K4 methylation activity could not be addressed in this experimental context owing to the presence of Spp1 (Supplementary Figure S4). Collectively, these results strongly suggest that the N-terminal region of Set1 contains a previously unrecognized motif required for H2Bub-dependent H3K4 methylation that was unveiled in the absence of Spp1.
Basic motifs within the Set1 N-terminal region physically interact with Swd1
Given that Set1 N-terminal residues 1-229 compensate for the absence of Spp1 in the C762-based complex in supporting H2Bub-dependent H3K4 methylation activity, as indicated above, we considered the possibility that communication between the Set1 N-terminal region and the catalytic core might also be involved in the H2Bub-dependent H3K4 methylation activity of Set1C. To test this possibility, we examined direct interactions between purified Set1 fragments (Supplementary Figure S5A and B) and purified Set1C subunits constituting the catalytic core. These analyses revealed a direct interaction of Swd1 with the Set1 1–229 fragment (Figure 5A, Swd1 immunoblot, lane 3). None of the tested subunits alone showed direct binding to SET or post-SET domains (Figure 5A, lane 8) or the n-SET domain (Figure 5A, lane 7), possibly reflecting the necessity of prior interactions between these subunits for their incorporation into Set1C.
Figure 5.
Direct interaction between the Set1 N-terminal region and Swd1. (A) Direct binding of purified FLAG-tagged Set1C catalytic core subunits (Swd1, Swd3, Bre2 and Sdc1) (Supplementary Figure S2B) to GST-tagged Set1 fragments (Supplementary Figure S5B) relative to GST. Note that the binding condition in this assays (150 mM salt) was less stringent compared to that of similar experiments in our previous report (300 mM salt) (15). (B) Schematic representations of the Set1 1–229 RRR1, RRR2, RRR1&2 fragments containing amino acids substitution from arginine (underlined) to alanine (bold). (C) Direct binding of purified FLAG-Swd1 to GST-tagged Set1 1–229 WT and mutant fragments (Supplementary Figure S5C). (D) Direct binding of purified FLAG-Swd1 and FLAG-Swd2 to GST-tagged Set1 fragments (Supplementary Figure S5D).
It has been previously reported that Swd1 contains acidic patches at its C-terminus, which are important for Set1 protein stability and proper levels of H3K4 methylation (27). Accordingly, we considered the possibility of charge-based interactions between the Set1 N-terminal region and Swd1, and thus sought to identify Set1 region(s) enriched with basic amino acid residues. Interestingly, we found three consecutive arginine resides at two locations (RRR1 and RRR2) in Set1 residues 1–229 (Figure 5B) that could potentially create basic binding platform(s) for the acidic region of Swd1. In support of this idea, we found that a purified Set1 1–229 fragment containing arginine-to-alanine substitutions in RRR1 and, to a lesser extent, a fragment containing substitutions in RRR2, exhibited significantly decreased binding to Swd1 (Figure 5C, lanes 4 and 5 versus lane 3, Supplementary Figure S5C); moreover, combining mutations in RRR1 and RRR2 resulted in an almost complete loss of interaction with Swd1 (Figure 5C, lane 6). These results suggest that the Set1 N-terminal region physically interacts with the catalytic core of Set1C through charge-based binding to Swd1.
We previously showed that Swd2 also binds to Set1 residues 1-229 (15). To examine the binding of Swd1 and Swd2 to Set1 in greater detail, we further subdivided the Set1 1–229 fragment (Supplementary Figure S5D) and performed protein interaction analyses using purified Swd1 and Swd2. We found that Swd1 and Swd2 directly bound to regions of Set1 encompassing residues 1–123 and 124–229, respectively (Figure 5D), and furthermore that Swd1 and Swd2 could bind simultaneously to the Set1 1-229 fragment (Supplementary Figure S5E). These data imply that Swd2 does not interfere with physical interactions of the Set1 N-terminal region with Swd1.
Set1 N-terminal basic motifs are involved in H2B ubiquitylation-dependent H3K4 methylation
The above-described results raise the question of whether the Set1 N-terminal region alone is sufficient to render the Spp1-deficient C762-based complex competent to methylate H3K4. To address this, we generated baculovirus plasmids that express polypeptides composed of Set1 N-terminal regions and a C762 fragment incorporating combinatorial arginine-to-alanine substitutions in the RRR1 and RRR2 motifs, and then reconstituted Set1C with four catalytic core subunits (Swd1, Swd3, Bre2 and Sdc1) in the presence and absence of Spp1 (Figure 6A). All purified complexes (Supplementary Figure S6A) exhibited intrinsic H3K4 methylation activity towards free H3 (Supplementary Figure S6B), as expected based on their common possession of all components of the catalytic core.
In an HMT assay using H2Bub chromatin as a substrate, the full-length Set1-containing complex lacking Spp1 generated all three H3K4 methylation states (Figure 6B, lane 1), confirming that five subunits—Set1, Swd1, Swd3, Bre2 and Sdc1—constitute the minimal set of subunits sufficient for H2Bub-dependent H3K4 methylation activity (15). Importantly, in the absence of Spp1, the complex containing Δ230–761 Set1 (N-terminal 1–229 residues fused to C762) exhibited significant levels of H3K4 methylation, whereas the Spp1-deficient C762-based complex was almost unable to methylate H3K4 (Figure 6B, lane 2 versus lane 3). In addition, the complex containing Δ124–761 Set1 (N-terminal 1–123 residues fused to C762) was also able to generate H3K4 methylation, exhibiting somewhat increased levels of H3K4me1 and H3K4me2 and reduced levels of H3K4me3 relative to the Spp1-containing complex (Figure 6C, lane 4 versus lane 3). These results indicate that the Swd1-interacting Set1 N-terminal residues 1–123 are able to compensate for Spp1 in supporting the H3K4 methylation activity of the Spp1-deficient C762-based complex. We also found that mutations in individual RRR1 and RRR2 motifs resulted in a marked reduction in H3K4 methylation activity (Figure 6C, lanes 6 and 8 versus lane 4), and that combining these mutations led to a greater decrease in H3K4 methylation activity (Figure 6C, lane 10 versus lane 4). In addition, we confirmed that the H3K4 methylation activities of all these complexes required H2Bub on chromatin (Supplementary Figure S6C). Collectively, these observations lend strong support to the idea that the Set1 N-terminal region plays a previously unrecognized role in mediating H2Bub-dependent H3K4 methylation through direct interaction with Swd1.
DISCUSSION
H3K4 methylation is regulated by various players, including the H2B ubiquitylation machinery, Set1C itself, and many other cellular factors. To directly assess the roles of subunits and domains of Set1C in H2Bub-dependent H3K4 methylation without interference from other cellular factors, we used biochemically defined in vitro HMT assays employing reconstituted Set1Cs and H2Bub chromatin templates in conjunction with yeast genetic analyses. Our findings provide a number of valuable insights into the mechanism of action of domains and subunits within Set1C in H2Bub-dependent H3K4 methylation: (i) the PHDL domain of Spp1, in conjunction with the n-SET domain of Set1, interacts with the catalytic core of Set1C, an interaction that is critical for the H2Bub-dependent H3K4 methylation activity of Set1C; (ii) the proximity between Spp1 and the Swd1/Swd3 heterodimer bypasses the requirement for an n-SET domain or H2B ubiquitylation for the H3K4 methylation activity of Set1C and (iii) a charge-based interaction between the N-terminal region of Set1 and Swd1 regulates the H2Bub-dependent H3K4 methylation activity of Set1C, at least in the absence of Spp1. Taken together, these results provide a mechanistic basis for the functions of Set1C subunits in the H2Bub-dependent H3K4 methylation process.
Roles of Spp1 domains in the structure and H3K4 methylation activity of the Set1 complex
Our ability to reconstitute Set1Cs with Spp1 mutants allowed us to address the functions of individual domains of Spp1. The SID of Spp1 is responsible for direct interaction with the n-SET domain of Set1, but is dispensable for retention of Spp1 in Set1C (Figure 1). Instead, we found that the PHDL domain located at the C-terminus of Spp1 is critical for the association of Spp1 with Set1C in an n-SET domain-dependent manner (Figure 2, see below). These results are consistent with the previously reported requirement for the C-terminus of CFP1, which contains the PHD2 domain, for interaction with human Set1A and Set1B complexes (26). However, the SID likely still contributes to the full H3K4 methylation activity of Set1C, because its deletion resulted in selective loss of H3K4me3 (Figure 1G and H). One plausible interpretation is that the SID contributes to the proper binding of Spp1 to the n-SET domain, which in turn creates a binding surface suitable for interaction with Swd1/Swd3 as well as full H3K4 methylation activity. We found that the PHD of Spp1 was not directly involved in the enzymatic activity of Set1C (Figure 1), excluding the possibility that Spp1 acts through binding of methylated H3K4 to bring Set1C to chromatin to methylate H3K4 in the other tail in the same nucleosome or a neighboring nucleosome during the methylation process. Alternatively, it is possible that, by binding methylated H3K4, the PHD contributes to the maintenance of cellular H3K4 methylation levels by protecting against demethylation (25) (see below). Also, distinct from its function as a Set1C component, the PHD of Spp1 appears to play a more critical role in meiotic recombination by binding H3K4me3 enriched in the vicinity of meiotic double-strand breaks (24,25). Mammalian Set1 complexes can target the promoter region of actively transcribing genes through recognition of unmethylated CpG dinucleotides by the CXXC domain of the CFP1 subunit (28–30), a function that is important for the proper distribution of H3K4 methylation on chromatin (31). In a related observation, a multispecies alignment also revealed a putative DNA-binding domain in Spp1 (32); however, we did not detect DNA binding activity of Spp1 in vitro under conditions in which human CFP1 exhibited a strong affinity for DNA (Supplementary Figure S1G and H). Thus, in light of the lack of DNA methylation in yeast, targeting of the yeast Set1 complex to chromatin may be regulated differently than that of the mammalian Set1 complexes, which is affected by DNA methylation status.
Conformational change in the catalytic region of the Set1 complex
Although several plausible models have been proposed (17), no clear evidence has been presented to provide a definitive answer for how Set1C recognizes and methylates H2Bub chromatin. The current study presents data that bear on this tantalizing question, providing several lines of evidence supporting the conclusion that an H2Bub-induced conformational change in the catalytic region of Set1C (encompassing the n-SET domain, Spp1, Swd1, Swd3, and SET plus post-SET domains, which also connect Bre2 and Sdc1) renders the complex competent for nucleosomal H3K4 methylation. Our in vitro protein-interaction studies demonstrated a complex subunit-interaction network that correlates with the H2Bub-dependent H3K4 methylation activity of Set1C. The n-SET domain and Spp1 together create a binding platform that is required for interaction with heterodimeric Swd1/Swd3; importantly, this implies that all essential components in the catalytic region required for H2Bub-dependent H3K4 methylation activity are interconnected (Figure 2F). In support of our demonstration of the subunit interaction network, very recent structural analyses of the Set1C catalytic region showed that Swd1/Swd3 heterodimer, in conjunction with the SET domain of Set1, creates an inter-subunit pocket next to the active site and that proper positioning of Swd1 and Swd3 is crucial for formation of the catalytic pocket and substrate selectivity (33,34). Hence, consistent with our results, these studies raise a possibility that rearrangement of subunit interactions in the catalytic site of Set1C accompanies by recognition of correct substrate. In line with this, it is possible that recognition or sensing of ubiquitin on the surface of the nucleosome core triggers allosteric changes in the catalytic region, which in turn results in alteration of the catalytic properties of Set1C towards nucleosome substrates. The most compelling observation in this context is our demonstration that an Spp1-Swd1 fusion bypassed the requirement for H2Bub for the H3K4 methylation activity of Set1C (Figure 3); thus, we postulate that the role of H2Bub is to cause rearrangements in the subunit-interaction network that ultimately generate H3K4 methylation-competent crosstalk among subunits. In support of our model, recognition of histone modifications has been proposed to induce a similar allosteric activation of the HMT activity of a different multisubunit complex. Structural and biochemical analyses of the PRC2 complex have revealed that recognition of H3K27me3 by the EED subunit causes a conformational change in the PRC2 complex that leads to activation of the H3K27 methyltransferase activity of the SET domain catalytic subunit, EZH2 (35–37).
Because there is currently no evidence for direct, stable interactions of H2Bub with any components of Set1C, one unanswered question in our scenario relates to how Set1C recognizes H2Bub on chromatin. In this context, we postulate that interaction of n-SET/Spp1 with the Swd1/Swd3 heterodimer, together with other components in the catalytic region, generates a new surface that potentially senses H2Bub on chromatin. Our failure to detect a direct interaction between H2Bub and Set1C is possibly attributable to the dynamic nature of the catalytic region within Set1C, reflecting its modulation by H2Bub.
Involvement of the N-terminal region of Set1 in mediating H2B ubiquitylation-dependent H3K4 methylation in the absence of Spp1
Several studies have reported that the Set1 N-terminal region affects cellular H3K4 methylation levels through a number of mechanisms, including regulation of Set1 protein stability (38), recruitment and/or localization of Set1C to chromatin (15,23,39), and binding to nascent mRNA (40,41). However, none of these studies implicated direct function of the Set1 N-terminal region in the catalytic activity of Set1C. In this regard, our previous study demonstrated that the inhibitory central region of Set1 (residues 230–761) is counteracted by the Set1 N-terminus (residues 1–229) (15). The current study provides evidence that the Set1 N-terminus participates directly in the catalytic activity of Set1C by communicating with the catalytic region of Set1C (Figure 5 and 6). This interaction is also supported by previous reports by other groups showing the interaction of Swd1 with a Set1 polypeptide as it emerges from the ribosome (e.g. the Set1 N-terminal region) during translation (42), as well as by evidence for coimmunoprecipitation of Swd1/Swd3 with a Set1 fragment incorporating residues 1-900 (10). In addition, our demonstration that Set1 N-terminal residues 1-123 converts the Spp1-deficient C762 complex into an H3K4 methylation-competent form (Figure 6) provides a plausible explanation for why the full-length Set1-containing complex is able to methylate H3K4 in the absence of Spp1. These observations raise the interesting question of whether the interaction of the Set1 N-terminus with the catalytic region of Set1C is conditional only in the absence of Spp1. In this case, we postulate that the Set1 N-terminal region acts as a backup for Spp1 that can be dispensed with in Set1C during meiosis (25).
We note that a parallel experiment using an spp1Δ strain that expresses the Δ124–761 Set1 fragment showed only weak H3K4me1 signals, and no H3K4me2 or H3K4m3 signals (Supplementary Figure S6D). Chromatin immunoprecipitation analyses used to assess localization of Set1 revealed that the Δ124–761 Set1 fragment showed markedly decreased recruitment to chromatin relative to full-length Set1 (Supplementary Figure S6E and F). Thus, it is likely that defective physical association of Δ124–761 Set1 with chromatin resulted in more severely reduced enzymatic activity in cells compared with that observed in the defined in vitro system, where substrates are much more readily accessible to the enzyme. In addition, a recent genetic study suggested that Spp1 protects methylated H3K4 from demethylation activity in cells (25), an observation that may account for the decreased H3K4 methylation levels in the Δ124–761 Set1 strain lacking Spp1 (Supplementary Figure S6D).
Models of Spp1- and Set1 N-terminal region-mediated H2B ubiquitylation-dependent H3K4 methylation
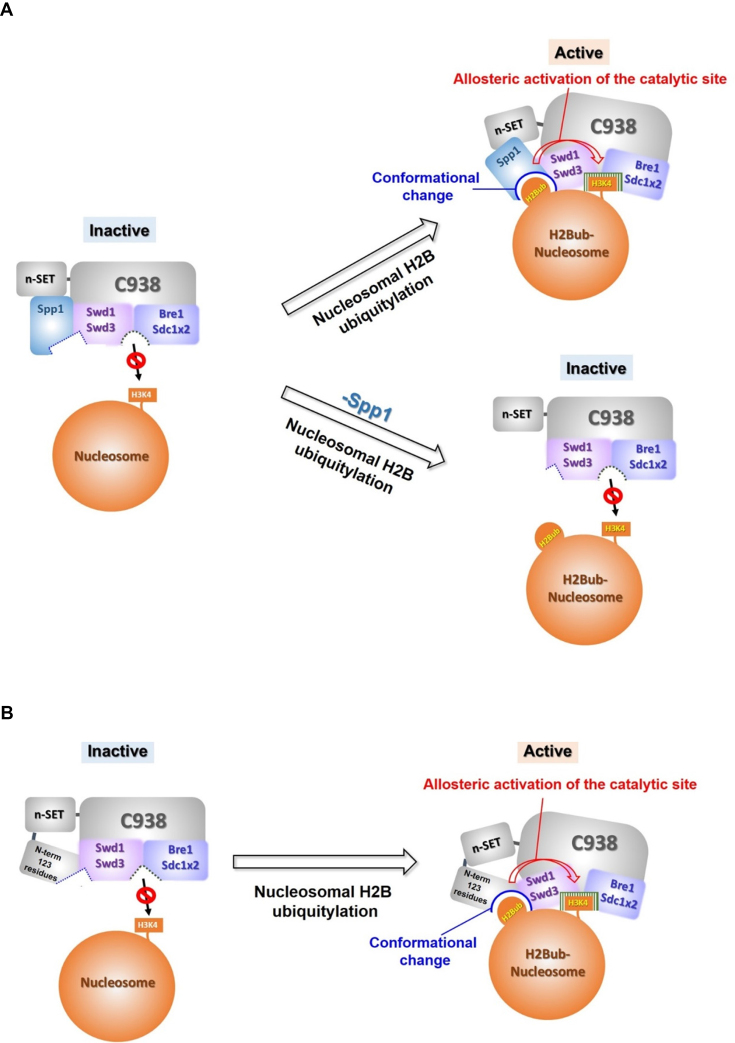
It is unlikely that H2Bub-dependent recruitment of Set1C to nucleosomes fully accounts for the requirement for H2Bub for H3K4 methylation activity. This is because, despite intensive efforts, no convincing evidence for a direct, stable interaction of ubiquitin or H2Bub chromatin with any components of Set1C has been reported. On the contrary, it has been reported that accumulation of Set1 on chromatin is little affected by impaired H2B ubiquitylation (10,15). Instead, our study favors a model in which H2Bub induces a conformational change in the catalytic region of Set1C. According to our proposed model, in the absence the N-terminal region of Set1, the active site of the catalytic region (e.g., C762 complex) is predicted to be present in a form that is inactive toward nucleosomal substrates owing to occlusion by the chromatin structure. Upon sensing ubiquitin on nucleosomal H2B, the catalytic region undergoes allosteric changes that could involve shifting of Spp1 and/or Swd1 positions within the active site, changes that, in turn, render Set1C enzymatically active for nucleosomal H3K4 methylation. This conformational change does not occur in the absence of Spp1 (Figure 7A). We also envision that, in the absence of Spp1, H2Bub can induce a rearrangement of the subunit-interaction network within the active site of Set1C, which is also physically connected to the Set1 N-terminal region (Figure 7B). One intriguing question in this context is whether the Set1 N-terminal region participates in the H2Bub-dependent H3K4 methylation process through the same mechanism as Spp1. Previous reports of H2Bub-mediated conformational changes that reinforce the repositioning of human Dot1L for efficient H3K79 methylation (43,44) lend support to our models. Structural analyses, ideally involving Set1C and an H2Bub nucleosome, should provide detailed information relevant to key aspects of these models. The mechanistic insights into Set1C subunits provided here set the stage for detailed structural analyses of Set1C.
Figure 7.
Mechanistic models of H2B ubiquitylation-dependent H3K4 methylation involving Spp1 and the Set1 N-terminal region. (A) In the absence of the Set1 N-terminal region, we envision that H2B ubiquitylation causes conformational changes in the catalytic region of Set1C that involve rearrangements of the subunit-interaction network; this, in turn, induces allosteric activation of the catalytic site, rendering it accessible to nucleosomal H3K4. This conformational change does not occur in the absence of Spp1, as evidenced by the fact that Spp1 is essential for the interaction between n-SET/Spp1 and Swd1/Swd3, which is involved in sensing H2Bub and/or conformational changes. (B) In the absence of Spp1, we envision that basic motifs within the Set1 N-terminal region physically interact with Swd1, creating a surface for sensing H2Bub in conjunction with other subunits in the catalytic region of Set1C. This also induces allosteric activation of the catalytic site of Set1C, rendering it favorable for nucleosomal H3K4 methylation. Sdc1 × 2 indicates the dimeric association of Sdc1 in the catalytic region, which was established in recent reports (33,34).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Vincent Géli for anti-Spp1 antibody and Dr. Daeyoup Lee for valuable discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Samsung Science and Technology Foundation [SSTF-BA1702-13 to J.K.]; National Research Foundation of Korea [2013R1A1A1006638 to J.A.K.]; KRIBB Research Initiative Program (to J.A.K.); National Institutes of Health [R37 GM086868 and RO1 GM107047 to T.W.M.]. Funding for open access charge: Samsung Science and Technology Foundation [SSTF-BA1702-13].
Conflict of interest statement. None declared.
REFERENCES
- 1. Howe F.S., Fischl H., Murray S.C., Mellor J.. Is H3K4me3 instructive for transcription activation. Bioessays. 2017; 39:1–12. [DOI] [PubMed] [Google Scholar]
- 2. Rivera C., Gurard-Levin Z.A., Almouzni G., Loyola A.. Histone lysine methylation and chromatin replication. Biochim. Biophys. Acta. 2014; 1839:1433–1439. [DOI] [PubMed] [Google Scholar]
- 3. Acquaviva L., Drogat J., Dehé P.M., de la Roche Saint-André C., Géli V.. Spp1 at the crossroads of H3K4me3 regulation and meiotic recombination. Epigenetics. 2013; 8:355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daniel J.A., Nussenzweig A.. Roles for histone H3K4 methyltransferase activities during immunoglobulin class-switch recombination. Biochim. Biophys. Acta. 2012; 1819:733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greer E.L., Shi Y.. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012; 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyun K., Jeon J., Park K., Kim J.. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017; 49:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012; 81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krogan N.J., Dover J., Khorrami S., Greenblatt J.F., Schneider J., Johnston M., Shilatifard A.. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002; 277:10753–10755. [DOI] [PubMed] [Google Scholar]
- 9. Nagy P.L., Griesenbeck J., Kornberg R.D., Cleary M.L.. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dehé P.M., Dichtl B., Schaft D., Roguev A., Pamblanco M., Lebrun R., Rodríguez-Gil A., Mkandawire M., Landsberg K., Shevchenko A. et al. . Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J. Biol. Chem. 2006; 281:35404–35412. [DOI] [PubMed] [Google Scholar]
- 11. Nedea E., Nalbant D., Xia D., Theoharis N.T., Suter B., Richardson C.J., Tatchell K., Kislinger T., Greenblatt J.F., Nagy P.L.. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell. 2008; 29:577–587. [DOI] [PubMed] [Google Scholar]
- 12. Briggs S.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D.. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002; 418:498. [DOI] [PubMed] [Google Scholar]
- 13. Sun Z.W., Allis C.D.. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002; 418:104–108. [DOI] [PubMed] [Google Scholar]
- 14. Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A.. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002; 277:28368–28371. [DOI] [PubMed] [Google Scholar]
- 15. Kim J., Kim J.A., McGinty R.K., Nguyen U.T., Muir T.W., Allis C.D., Roeder R.G.. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell. 2013; 49:1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J., Guermah M., McGinty R.K., Lee J.S., Tang Z., Milne T.A., Shilatifard A., Muir T.M., Roeder R.G.. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009; 137:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soares L.M., Buratowski S.. Histone crosstalk: H2Bub and H3K4 methylation. Mol. Cell. 2013; 49:1019–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fierz B., Chatterjee C., McGinty R.K., Bar-Dagan M., Raleigh D.P., Muir T.W.. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011; 7:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J.S., Shukla A., Schneider J., Swanson S.K., Washburn M.P., Florens L., Bhaumik S.R., Shilatifard A.. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007; 131:1084–1096. [DOI] [PubMed] [Google Scholar]
- 20. McGinty R.K., Köhn M., Chatterjee C., Chiang K.P., Pratt M.R., Muir T.W.. Structure–activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem. Biol. 2009; 4:958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim J., Roeder R.G.. Nucleosomal H2B ubiquitylation with purified factors. Methods. 2011; 54:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J., Guermah M., Roeder R.G.. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010; 140:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thornton J.L., Westfield G.H., Takahashi Y.H., Cook M., Gao X., Woodfin A.R., Lee J.S., Morgan M.A., Jackson J., Smith E.R. et al. . Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 2014; 28:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Acquaviva L., Székvölgyi L., Dichtl B., Dichtl B.S., de La Roche Saint André C., Nicolas A., Géli V.. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013; 339:215–218. [DOI] [PubMed] [Google Scholar]
- 25. Adam C., Guérois R., Citarella A., Verardi L., Adolphe F., Béneut C., Sommermeyer V., Ramus C., Govin J., Couté Y. et al. . The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet. 2018; 14:e1007223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Butler J.S., Lee J.H., Skalnik D.G.. CFP1 interacts with DNMT1 independently of association with the Setd1 histone H3K4 methyltransferase complexes. DNA Cell Biol. 2008; 27:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mersman D.P., Du H.N., Fingerman I.M., South P.F., Briggs S.D.. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J. Biol. Chem. 2012; 287:2652–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voo K.S., Carlone D.L., Jacobsen B.M., Flodin A., Skalnik D.G.. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 2000; 20:2108–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tate C.M., Lee J.H., Skalnik D.G.. CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J. 2010; 277:210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown D.A., Di Cerbo V., Feldmann A., Ahn J., Ito S., Blackledge N.P., Nakayama M., McClellan M., Dimitrova E., Turberfield A.H. et al. . The SET1 complex selects actively transcribed target genes via multivalent interaction with CpG island chromatin. Cell Rep. 2017; 20:2313–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clouaire T., Webb S., Skene P., Illingworth R., Kerr A., Andrews R., Lee J.H., Skalnik D., Bird A.. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012; 26:1714–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murton B.L., Chin W.L., Ponting C.P., Itzhaki L.S.. Characterising the binding specificities of the subunits associated with the KMT2/Set1 histone lysine methyltransferase. J. Mol. Biol. 2010; 398:481–488. [DOI] [PubMed] [Google Scholar]
- 33. Hsu P.L., Li H., Lau H.T., Leonen C., Dhall A., Ong S.E., Chatterjee C., Zheng N.. Crystal structure of the COMPASS H3K4 methyltransferase catalytic module. Cell. 2018; 174:1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qu Q., Takahashi Y.H., Yang Y., Hu H., Zhang Y., Brunzelle J.S., Couture J.F., Shilatifard A., Skiniotis G.. Structure and conformational dynamics of a COMPASS histone H3K4 methyltransferase complex. Cell. 2018; 174:1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margueron R., Justin N., Ohno K., Sharpe M.L., Son J., Drury W.J. 3rd, Voigt P., Martin S.R., Taylor W.R., De Marco V. et al. . Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009; 461:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiao L., Liu X.. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015; 350:aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brooun A., Gajiwala K.S., Deng Y.L., Liu W., Bolaños B., Bingham P., He Y.A., Diehl W., Grable N., Kung P.P. et al. . Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 2016; 7:11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soares L.M., Radman-Livaja M., Lin S.G., Rando O.J., Buratowski S.. Feedback control of Set1 protein levels is important for proper H3K4 methylation patterns. Cell Rep. 2014; 6:961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soares L.M., He P.C., Chun Y., Suh H., Kim T., Buratowski S.. Determinants of histone H3K4 methylation patterns. Mol. Cell. 2017; 68:773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luciano P., Jeon J., El-Kaoutari A., Challal D., Bonnet A., Barucco M., Candelli T., Jourquin F., Lesage P., Kim J. et al. . Binding to RNA regulates Set1 function. Cell Discov. 2017; 3:17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sayou C., Millán-Zambrano G., Santos-Rosa H., Petfalski E., Robson S., Houseley J., Kouzarides T., Tollervey D.. RNA binding by histone methyltransferases Set1 and Set2. Mol. Cell. Biol. 2017; 37:e00165-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Halbach A., Zhang H., Wengi A., Jablonska Z., Gruber I.M., Halbeisen R.E., Dehé P.M., Kemmeren P., Holstege F., Géli V. et al. . Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 2009; 28:2959–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McGinty R.K., Kim J., Chatterjee C., Roeder R.G., Muir T.W.. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008; 453:812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou L., Holt M.T., Ohashi N., Zhao A., Müller M.M., Wang B., Muir T.W.. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat. Commun. 2016; 7:10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.