Abstract
Runt-related transcription factor 1 (RUNX1) is a well-known master regulator of hematopoietic lineages but its mechanisms of action are still not fully understood. Here, we found that RUNX1 localizes on active chromatin together with Far Upstream Binding Protein 1 (FUBP1) in human B-cell precursor lymphoblasts, and that both factors interact in the same transcriptional regulatory complex. RUNX1 and FUBP1 chromatin localization identified c-KIT as a common target gene. We characterized two regulatory regions, at +700 bp and +30 kb within the first intron of c-KIT, bound by both RUNX1 and FUBP1, and that present active histone marks. Based on these regions, we proposed a novel FUBP1 FUSE-like DNA-binding sequence on the +30 kb enhancer. We demonstrated that FUBP1 and RUNX1 cooperate for the regulation of the expression of the oncogene c-KIT. Notably, upregulation of c-KIT expression by FUBP1 and RUNX1 promotes cell proliferation and renders cells more resistant to the c-KIT inhibitor imatinib mesylate, a common therapeutic drug. These results reveal a new mechanism of action of RUNX1 that implicates FUBP1, as a facilitator, to trigger transcriptional regulation of c-KIT and to regulate cell proliferation. Deregulation of this regulatory mechanism may explain some oncogenic function of RUNX1 and FUBP1.
INTRODUCTION
Hematopoiesis is an intricate process leading to the differentiation of stem cells into all hematopoietic lineages. It requires an elaborate network of transcription factors articulated with transcriptional regulators, and deregulation of those networks is implicated in hematologic malignancies. Runt-related transcription factor 1 (RUNX1) is one of these critical transcription factors playing a prominent role in hematopoiesis (1,2). RUNX1 is essential for definitive hematopoiesis in early development and adulthood, for megakaryocyte maturation, T- and B-cell lineages and neuronal development (3–10). In line with its founding role, loss- or gain-of-function variants of RUNX1 protein promote hematologic malignancies, notably in the most frequent pediatric subtype of leukemia, the pre-B cell acute lymphoblastic leukemia (ALL) (1).
RUNX1 functioning is complex and remains a matter of debate. RUNX1 acts as a transcriptional platform recruiting co-factors that modulate its transcriptional activity. RUNX1 thereby endorses activator or repressor functions (11,12). Transcriptional activation by RUNX1 requires its heterodimerization with Core Binding Factor Beta (CBFB) (13,14) and the recruitment of co-factors (CBP, P300, etc.), which display a lineage-specific or ubiquitous expression pattern, to direct specific biological programs (12,15). Pre-B ALL emerge from pro-B or pre-B lymphocytes where RUNX1 is essential for survival and development of B cell–specified progenitors and for the transition through the pre-B-cell stage (16). To increase our understanding about the molecular mechanisms that modulate the transcriptional activity of RUNX1 in pre-B cells and its physiological consequences, we aimed at identifying its specific partners. Using an unsupervised approach termed RIME (rapid immunoprecipitation mass spectrometry of endogenous proteins), we identified Far Upstream Element Binding Protein 1 (FUBP1), an ATP-dependent DNA helicase and a transcriptional regulator able to bind single-stranded DNA (ssDNA) and RNA (17–19), as a potential RUNX1-regulator. FUBP1 promotes cell proliferation, inhibits apoptosis, and enhances cell migration by modulating expression of transcripts such as c-MYC (MYC), P21 (CDKN1A) and Noxa (Pmaip1) (18,20–23). FUBP1 is also implicated in different processes in RNA regulation such as post-transcriptional regulation or splicing (19,24–27). Accordingly, FUBP1 endorses both oncogenic and tumor suppressor roles (21,22,28–31). Importantly, FUBP1 has been recently described as an important regulator of the expansion and self-renewal of hematopoietic stem cells (20,32,33). Potential cancer driving mutations of FUBP1 have also been reported in chronic and acute pre-B lymphocytic leukemias (34–36).
In this context, we hypothesized that FUBP1 could functionally cooperate with RUNX1. We demonstrate that the oncogene c-KIT (KIT) is one of FUBP1 and RUNX1 common target genes. We characterized two regulatory regions at +700 pb and +30 kb from Transcription Start Site (TSS) within the first intron of c-KIT which are bound by RUNX1 and FUBP1, together with active histone marks. c-KIT is a crucial player in hematopoietic stem cell maintenance and differentiation (37,38) and presents oncogenic functions (39,40). We uncovered here that upregulation of c-KIT by FUBP1 and RUNX1 impacts cell proliferation, which can be counteracted by the pharmacological c-KIT inhibitor, imatinib mesylate. Altogether, our data underscore a novel mechanism of regulation of the oncogene c-KIT that could play a role in pathophysiological context of RUNX1 and FUBP1 deregulation.
MATERIALS AND METHODS
Detailed experimental procedures for RNA extraction, RT-qPCR, generation of stable cell lines, co-immunoprecipitation, cells sorting, immunoblotting, chromatin immunoprecipitation (ChIP-Seq), luciferase assay, molecular simulation, immunophenotyping, and cell proliferation assays are included in supplemental materials and methods. Lists of primary antibodies and primers are outlined in Supplementary Tables S1 and S2.
Cell lines and patients’ cells
Pre-B leukemia cell line Nalm6 (ATCC® CRL-3273™) was maintained in RPMI-1640 medium containing 10% heat-inactivated fetal calf serum (FCS) and 1% antibiotics (penicillin/streptomycin). HEK293 cells (ATCC® CRL-1573™) were maintained in DMEM/10% FCS/1% antibiotics. Bone marrow cells from pre-B acute lymphoblastic leukemia patients were collected at diagnosis, after informed consent had been obtained, in accordance with the declaration of Helsinki. The protocol was approved by the ethics committee of Rennes Hospital (France).
Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME)
RIME was conducted with Nalm6 cells as previously described (41). The lysates were incubated with two anti-RUNX1 and normal rabbit IgG antibodies (Supplementary Table S1). Peptides were visualized using Scaffold 4 software (http://www.proteomesoftware.com/products/scaffold/). We selected proteins where the sum of peptide count for both RUNX1 antibodies is at least three and higher than 2-fold IgG background. Proteins identified by only one RUNX1 antibody were excluded.
Proximity ligation assay (PLA)
PLA was carried out with Duolink® In Situ Detection Reagents and Probes (DUO92007, DUO92002 and DUO92004, Sigma-Aldrich). The protocol was described in Debaize et al. (42). Quantification of PLA dots per nucleus was performed by automatic counting with ImageJ and values below the assay cut-off (set to two standard deviations over background signal) were considered negative for the interaction of interest (42).
Generation of stable cell lines
We used the constructs pLKO1-shFUBP1 (TRCN0000230197, Sigma Aldrich), pLenti-CMV-Puro-DEST-Flag-FUBP1, pLenti-CMV-Puro-DEST-Halotag-RUNX1 (from pFN21A #FHC01784, Kazusa collection, Promega), pSPAX2 (a gift from Didier Trono, Addgene plasmid #12260) and pCMV-VSV-G (a gift from Bob Weinberg, Addgene plasmid #8454) (43) to produce lentivirus to generate HEK293shFUBP1, Nalm6+FUBP1 and Nalm6+RUNX1 Nalm6+RUNX1+FUBP1 stable cell lines. Transfected cells were selected with 1.1 or 0.25 μg/ml puromycin (44). pLenti-CMV-Puro-DEST was a gift from Eric Campeau (Addgene plasmid #17452) (45). For details, see the supplemental Methods.
Chromatin immunoprecipitation (ChIP) and binding site analysis
The procedure was adapted from a previous study (46). Detailed procedure is described in the supplemental methods. All sequencing data are available at NCBI GEO ( GSE109377). Genomic regions were analyzed with CEAS (47) and GREAT (48) software to uncover enriched genomic regions.
Luciferase assay
Genomic DNA fragments derived from the human c-KIT gene and control DNA were cloned into pGL4.10-luc with a minimal promoter and transfected in HEK293shFUBP1 cells, in presence of pFN-Halotag-RUNX1 (#FHC01784, Kazusa collection, Promega) and/or pCDNA-Flag-FUBP1 and supplemented with empty pCDNA vector. For luciferase assays in Nalm6 cells, the +30 kb enhancer region was cloned with the minimal promoter into a MSCV Luciferase PGK-hygro vector (a gift from Scott Lowe, Addgene plasmid #18782) by the Gibson Assembly® Cloning Kit (NEB). To produce the +30 kb enhancer-MSCV-luciferase retrovirus, HEK293 cells were co-transfected with +30 kb enhancer-MSCV-luciferase together with pCMV-VSV-G and pGag-pol (49). Nalm6 transduced cells were selected with 300 μg/mL hygromycin. FUBP1 truncated expression vector (FUBP1del-CEN) was kindly provided by W. Keith Miskimins (50). Mutated RUNX1 (RUNX1R174Q, RUNX1R174Q-T161A) expression vectors were generated by site-directed mutagenesis.
Molecular simulation
Models of the KH3 FUBP1 subdomain complexed to single-stranded DNA were based on the nuclear magnetic resonance (NMR) solution structure 1J4W from the Protein Data Bank. YASARA software (http://www.yasara.org), Amber03 force field and standard protocols (51) were used to produce binding energies.
Immunophenotyping
Intracellular and surface stainings of c-KIT protein were performed by flow cytometry using APC-Mouse-Anti-Human CD117 antibody (Clone YB5.B8, BD Biosciences).
Cell proliferation and cell analysis
Proliferation assays were done by automatic counting with a Cellometer (Nexcelom). Apoptosis/necrosis analyses and inhibition assays were performed with automatic counting following Hoechst, propidium iodine and Yo-Pro stainings.
Xenograft transplantation and survival analysis
NOD/scid IL2 Rgnull mice (Charles River Laboratories, France) were maintained in the ARCHE Animal Housing Center (Rennes, France). Animal experiments were performed after authorization by the French Research Ministry, and according to European regulation. Four-week-old mice received two intraperitoneal injections of 20 μg/g busulfan (60 mg/10 ml, Pierre Fabre) on 2 days. They were then allowed to rest for 2 days before intravenous retro-orbital injection of 100 000 cells as previously described (44).
RESULTS
RUNX1 ChIP-Seq signal is enriched within transcriptionally active chromatin regions in pre-B cells
To investigate the transcriptional role of RUNX1 in human pre-B cells, we mapped its genome-wide distribution by chromatin immunoprecipitation followed by sequencing (ChIP-Seq) in pre-B Nalm6 cells and bone marrow cells isolated from pre-B leukemic patients. Antibody against RUNX1 was validated in our models (Supplementary Figure S1A and B). To correlate RUNX1 engagement with chromatin activity in Nalm6 cells, we ran ChIP-Seq of histones H3K4me1, H3K4me3 and H3K27ac, respectively markers of active/poised enhancers, active/poised promoters and active promoters/enhancers.
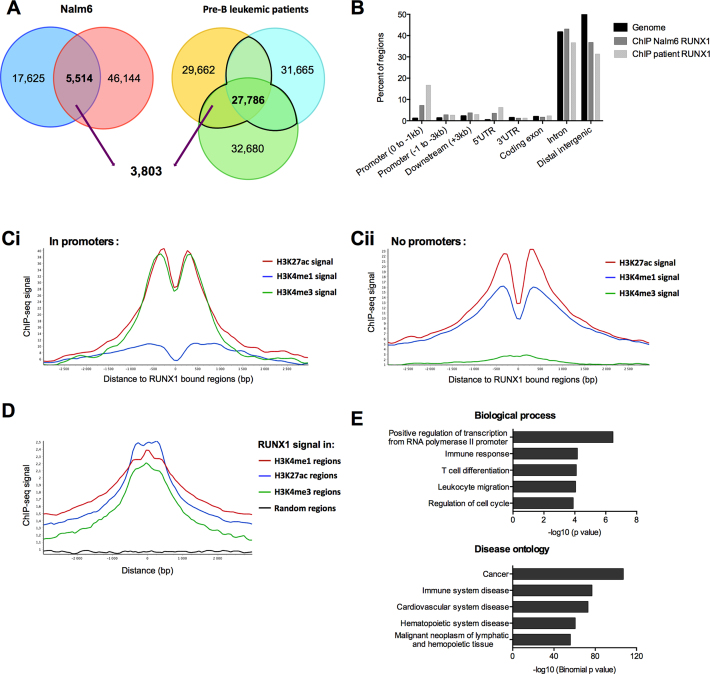
Replicates of RUNX1 ChIP-Seq in Nalm6 cells retrieved 5514 common peaks, 69% (3803 peaks) of which are also found in RUNX1 ChIP-Seq peaks in at least two out of three pre-B leukemic patients (Figure 1A). Those 3803 peaks were assigned to 2651 genes (Supplementary Table S3). RUNX1-bound regions were highly enriched in promoters (–1 kb from the TSS) and in 5′UTR regions (Figure 1B). RUNX1-bound regions were associated with enriched H3K4me3 and H3K27ac signals in promoters, and with H3K4me1 and H3K27ac signals in genomic regions excluding promoters (no-promoter regions) (Figure 1C). Finally, the average RUNX1 signal peaked at regions positive for H3K4me3, H3K4me1 and H3K27ac but not at random regions (Figure 1D). Ontology analyses of these genes reveal an enrichment in biological processes related to transcription regulation, cell cycle regulation and a variety of hematologic processes (Figure 1E and Supplementary Table S4). Cancer, cardiovascular disease, and hematopoietic disease are also enriched in disease ontology analysis (Figure 1E and Supplementary Table S4). Altogether, our data show that RUNX1 localizes on chromatin in transcriptionally active chromatin regions mainly in promoter and enhancer regions in human pre-B cells.
Figure 1.
RUNX1 ChIP-Seq signals are enriched within transcriptionally active chromatin regions. (A) Venn-diagrams presenting the intersection between RUNX1 binding regions in both replicates in Nalm6 cells, and bone marrow mononuclear cells isolated from three pre-B ALL patients. (B) Analyses of RUNX1 ChIP-Seq-enriched signals for pre-B Nalm6 cells and bone marrow mononuclear cells isolated from a pre-B acute lymphoblastic leukemia patient. ChIP-Seq reads were aligned to the reference human genome version GRCh37 (hg19). Analyses with the cis-regulatory Element Annotation System (CEAS) (47) software of RUNX1 ChIP-Seq provided RUNX1-bound regions enrichment in different genomic regions compared to the whole genome. (Ci and Cii) Read density plots for H3K4me1, H3K4me3 and H3K27ac enrichment centered around RUNX1 binding sites over a 3 kb window from RUNX1 Chip-Seq in Nalm6 cells, in promoter regions ([–4000 +1000] from TSS of annotated genes) (Ci) or without promoter regions (Cii). (D) Read density plots for RUNX1 signal around H3K4me1, H3K27ac, H3K4me3 regions over a 3 kb window and random regions of similar lengths in Nalm6 cells. The graph shows that the average RUNX1 signal peaked at regions positive for H3K4me3, H3K4me1 and H3K27ac. (E) Annotation enrichments of the 2651 potential RUNX1 target genes identified with DAVID (81) for biological process and GREAT (48) for disease ontology were shown. We selected 5 non-redundant annotations among the 10 most significant.
FUBP1 protein interacts with RUNX1 in hematopoietic lineages
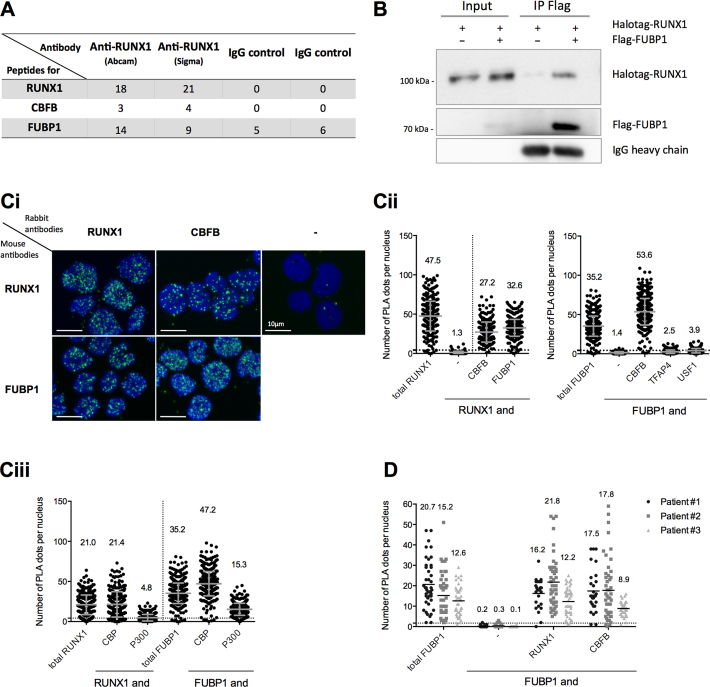
To identify RUNX1 co-factors able to modulate its transcription activity, we performed immunoprecipitations of endogenous RUNX1 protein in pre-B Nalm6 cells with two different antibodies followed by mass spectrometry (RIME) (41) (Figure 2A). We identified 20 RUNX1-associated proteins (Supplementary Table S5). RUNX1 was recovered with the highest number of peptides (18–21 peptides). CBFB, a well-known transcriptional cooperating factor responsible for RUNX1 stabilization on chromatin (14), was also recovered with 3–4 peptides. Together, these results validated the RUNX1-RIME approach. Interestingly, 9–14 peptides from the Far Upstream Element Binding Protein 1 (FUBP1) were identified, although FUBP1 peptides were also enriched with control IgGs but to a lower extent (Figure 2A). Given that RUNX1 and FUBP1 share common roles in hematopoietic lineages, a potential functional interaction between these two proteins was further investigated.
Figure 2.
FUBP1 protein interacts with RUNX1 in hematopoietic lineages. (A) Number of peptides from RUNX1 (49 kDa), CBFB (22 kDa) and FUBP1 (68 kDa) proteins identified by RIME (rapid immunoprecipitation mass spectrometry of endogenous proteins) with RUNX1 or immunoglobulin G (IgG) control in pre-B lymphoblastic Nalm6 cells. The lysates were incubated with 10 μg of anti-RUNX1 (ab23980, Abcam), 5 μg of anti-RUNX1 (HPA004176, Sigma Aldrich) or with 10 μg of normal anti-IgG (sc2027, Santa Cruz Biotechnology). (B) Co-immunoprecipitation (IP) using anti-Flag antibody (M2 clone) in HEK293 cells expressing Halotag-RUNX1 and/or Flag-FUBP1. Western blots were performed with RUNX1 and FUBP1 antibodies. Similar levels of IgG heavy chains in both IP-Flag lanes were used as loading control. (Ci) Representative images of a Proximity Ligation Assay (PLA) in Nalm6 cells. Four pairs of antibodies were used as indicated (references of the antibodies are listed in the Supplementary Table S1). Nuclei were stained by DAPI and appeared in blue. Colocalization of proteins, visualized by PLA dots, appeared in green. (Cii and Ciii) Quantitation of protein co-localization per nucleus–visualized by PLA dots- in Nalm6 cells, presented with the mean values ± S.D. The value of the mean is indicated at the top of each scatter dot plot. Quantitation of PLA dots was performed by an automatic counting with ImageJ as published by Debaize et al. (42). Positive controls (named total RUNX1 or total FUBP1, where primary antibodies against two different epitopes of RUNX1 or FUBP1 were used) and negative controls (either anti-RUNX1 alone or anti-FUBP1 alone) were included. Here, the positive threshold value represented by the dotted line (set at two S.D over the background signal) is 4.3 dots. One representative experiment of at least two independent experiments is shown. (Ciii) CBP and P300 are co-activators that are well known to interact with RUNX1. (D) Quantitation of protein colocalization per nucleus (visualized by PLA dots) in human mononuclear cells from bone marrow of three pre-B acute lymphoblastic leukemia patients, presented with the mean values. The value of the mean is indicated at the top of each scatter dot plot. Here, the positive threshold value represented by the dotted line is 1.7 dots.
We confirmed the RIME-based results using a targeted approach. Flag-tagged FUBP1 and Halotag-tagged RUNX1 were co-expressed in HEK293 cells. We found that Halotag-RUNX1 co-immunoprecipitated with Flag-FUBP1 using an anti-Flag antibody (Figure 2B) and conversely, Flag-FUBP1 co-immunoprecipitated with Halotag-RUNX1 using an anti-Halotag antibody (Supplementary Figure S1C). We then interrogated whether RUNX1 and FUBP1 colocalize under physiological conditions in human pre-B cells. To that aim, we used the Proximity Ligation Assay (PLA), which is a powerful technique to detect endogenous protein–protein spatial proximity (Figure 2C and D) (42). As expected (13,14), PLA dots were observed with RUNX1 and CBFB proteins in Nalm6 cells, thereby validating the approach. Importantly, we detected a close proximity between RUNX1 and FUBP1 proteins in the nuclear compartment (Figure 2C) in accordance with their transcriptional function (52,53). Interestingly, we also detected a close molecular proximity between FUBP1 and CBFB proteins, suggesting that RUNX1, FUBP1 and CBFB associate in one or several regulatory complexes. The absence of any detectable PLA signal between FUBP1 and other transcription factors (TFAP4, USF1) strengthened the specificity of the interactions observed between FUBP1, RUNX1 and CBFB (Figure 2C). Additional PLA analyses in Nalm6 cells demonstrated that FUBP1 is in close molecular proximity with CBP and P300, two transcriptional co-activators known to be part of RUNX1 complexes (54) (Figure 2C) suggesting that FUBP1 and RUNX1 can colocalize in a same transcriptional complex. Importantly, the proximity between FUBP1, RUNX1 and CBFB were confirmed in bone marrow cells from all three tested pre-B leukemic patients (Figure 2D). Finally, comparable results for FUBP1 and RUNX1 proximity were obtained in other pre-B cell lines (BJAB and REH cells) (Supplementary Figure S1D). Interestingly, we demonstrated a proximity between FUBP1 and the fusion protein ETV6-RUNX1 in REH cells (Supplementary Figure S1D). Moreover, we evidenced a spatial proximity between FUBP1 and RUNX1 in human CD34+ hematopoietic stem and progenitor cells (Supplementary Figure S1E). Altogether, these data indicate that FUBP1 is likely to influence RUNX1 transcriptional activity in blood cells.
FUBP1 and RUNX1 regulate c-KIT transcription by binding to a common intronic enhancer in the c-KIT gene
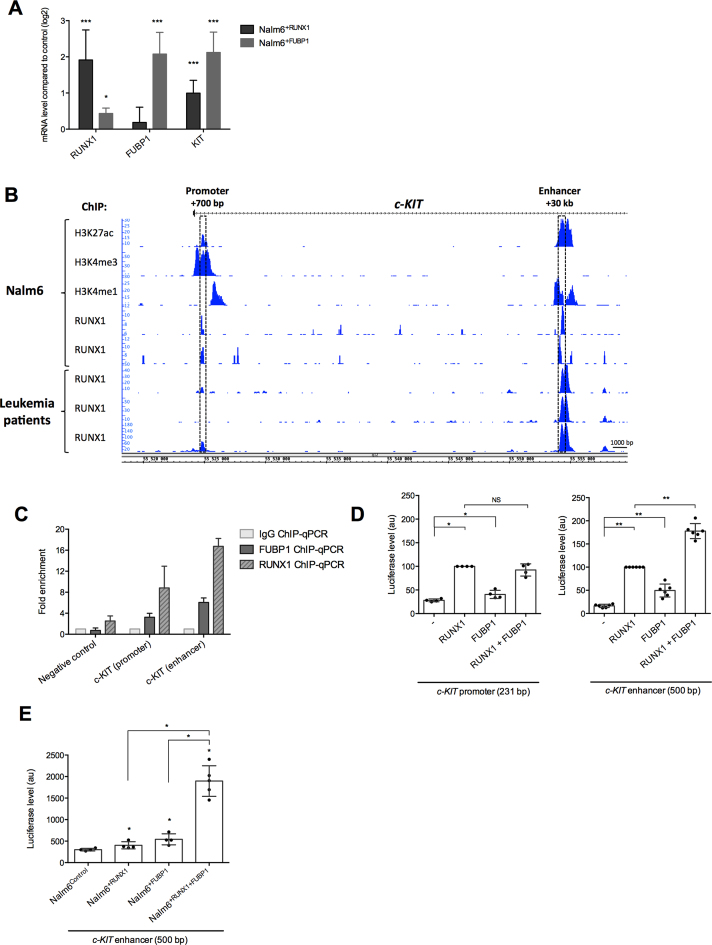
Among the 2651 genes putatively regulated by RUNX1 was c-KIT. Since c-KIT oncogene plays a central role in cell growth control and stimulation of proliferation of hematopoietic stem cells and early committed hematopoietic lineage cells (38,55), we decided to work further on the regulation of this gene by RUNX1. Upregulation of RUNX1 (in Nalm6+RUNX1) modulated positively c-KIT mRNA expression (Figure 3A, Supplementary Figure S2A). Interestingly, enforced expression of FUBP1 (in Nalm6+FUBP1) strongly upregulated c-KIT expression with an opposite profile upon depletion of FUBP1, suggesting that RUNX1 and FUBP1 could participate in c-KIT transcription (Figure 3A, Supplementary Figure S2A and B).
Figure 3.
FUBP1 and RUNX1 regulate c-KIT transcription by binding to a common intronic enhancer in the c-KIT gene. (A) Expression data analyses of c-KIT mRNA. RT-qPCR were performed in biological triplicate in Nalm6 cells overexpressing FUBP1 or RUNX1 (see Supplementary Figure S2A) and expression data are given compared to Nalm6control cells after normalizing with GAPDH mRNA (error bars are S.D.). *P < 0.05, **P < 0.01, ***P < 0.001 in Mann–Whitney tests. (B) Genomic tracks displayed ChIP-Seq data for RUNX1 and the histones H3K4me3, H3K27ac and H3K4me1 from Nalm6 cells (2 replicates) and bone marrow mononuclear cells isolated from three pre-B acute lymphoblastic leukemia patient across the first 35 kb of the human c-KIT gene (NM_000222). ChIP-Seq data were acquired by Illumina sequencing and visualized with Integrated Genome Browser 9.0.0 (82). ChIP-Seq reads were aligned to the reference human genome version GRCh37 (hg19). Both genomic regions of c-KIT gene that were associated with active histone marks are located within the first intron, one in a promoter region (+700 pb from TSS) and the another in an enhancer region (+30 kb from TSS) and are indicated by boxes. (C) ChIP-qPCR performed on samples from Nalm6 cells using a specific anti-RUNX1 (n = 3), anti-FUBP1 (n = 3) or a non-specific control IgG. ChIP-qPCR shows fold enrichment for c-KIT promoter (+700 pb) and enhancer (+30 kb) regulatory regions. Histogram indicates the means with S.D. of the fold enrichment of peaks relative to IgG. (D) Luciferase assays with a vector containing the c-KIT promoter or enhancer regions downstream a minimal promoter and a luciferase ORF, in presence of RUNX1 and FUBP1 expressing vectors in HEK293shFUBP1 cells (see Supplementary Figure S2E). Note that the HEK293shFUBP1 cells do not express RUNX1 and that the FUBP1 expression vector is resistant to shFUBP1. Luciferase levels (firefly luciferase/Renilla luciferase) are represented using a scatter dot plot indicating the means and S.D. of at least 4 independent experiments. NS: non-significant, * < 0.05 in Mann–Whitney tests. (E) Luciferase assays with a vector containing the c-KIT enhancer regions downstream a minimal promoter and a luciferase ORF, in presence of RUNX1 and FUBP1 expressing vectors in Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1 and Nalm6+RUNX1+FUBP1 cells. Luciferase levels are represented using a scatter dot plot indicating the means and S.D. of at least four independent experiments (i.e. each dot represents an independent stable cell line). The expression level of RUNX1 is similar in Nalm6+RUNX1 and Nalm6+RUNX1+FUBP1. The expression level of FUBP1 is similar in Nalm6+FUBP1 and Nalm6+RUNX1+FUBP1 (refer to Figure 6A, Supplementary Figure S3B). NS: non-significant, *P < 0.05 in Mann–Whitney tests.
We then analyzed ChIP-Seq data for RUNX1 and histones marks along the c-KIT gene in pre-B Nalm6 and bone marrow cells from leukemic patients. We identified two strong RUNX1 binding regions in the first intron of c-KIT: one at +700 bp and another one at +30 kb from TSS, that overlapped respectively with H3K4me3 and H3K4me1 marks and displayed H3K27ac marks, indicating active chromatin regions (Figure 3B).
To investigate the binding of FUBP1 on those two chromatin regions, FUBP1 antibody was first validated for IP in HEK293+FUBP1 (Supplementary Figure S2C) and tested for ChIP-qPCR on CDKN1A and MYC loci. Very few target regions, including CDKN1A and MYC loci, are described as bound by FUBP1, with results varying with the models (19,20,56–58). We found that FUBP1 binds to CDKN1A promoter in Nalm6control compared to Nalm6shFUBP1, but not to the –1.5kb MYC promoter (Supplementary Figure S2D). These results validate FUBP1 antibody for ChIP. Bindings of RUNX1 and FUBP1 to both c-KIT intronic regions were validated by ChIP-qPCR and showed higher fold enrichment at the +30 kb enhancer compared to the +700 bp site (Figure 3C). Importantly, the +30 kb enhancer conferred an additive responsiveness to transcription activation upon RUNX1 and FUBP1 transfection in luciferase reporter assays performed in HEK293shFUBP1 cells (Figure 3D), in which endogenous FUBP1 was depleted (Supplementary Figure S2E). Conversely, the promoter region (+700 bp) mostly responded to RUNX1 expression (Figure 3D). We therefore focused on the +30 kb enhancer region (500-nucleotide-long). Stable RUNX1 or FUBP1 overexpression in Nalm6 cells demonstrated the responsiveness of the +30 kb enhancer of c-KIT to transcription activation upon RUNX1 or FUBP1 expression. Moreover, the combination of RUNX1 and FUBP1 shows synergetic effect on the c-KIT +30 kb enhancer (Figure 3E). Altogether, these data demonstrate that the c-KIT enhancer at +30 kb is controlled by RUNX1, FUBP1 and more importantly RUNX1 together with FUBP1 in pre-B lymphoblasts, suggesting a cooperativity between those two transcription regulators.
RUNX1 and FUBP1 cooperate for the regulation of the +30 kb enhancer of c-KIT
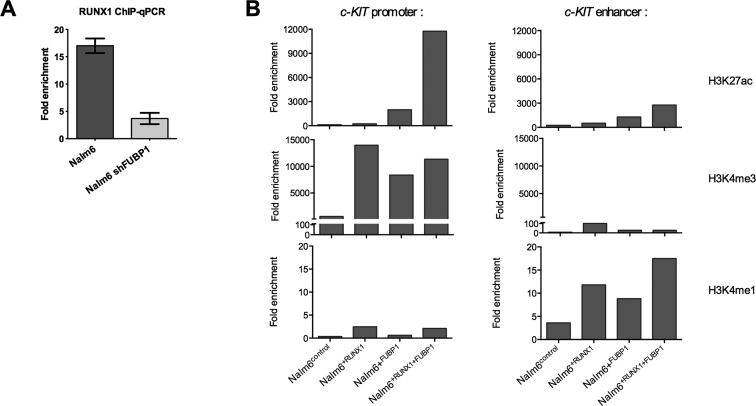
To address the question of the cooperativity between RUNX1 and FUBP1 for the regulation of c-KIT expression (Figure 4A), we conducted RUNX1 ChIP-qPCR on +30 kb enhancer upon presence (in Nalm6control) or depletion of FUBP1 (in Nalm6shFUBP1). We showed that there is ∼4.5-fold more RUNX1 bound to the +30 kb enhancer in presence of FUBP1 than upon depletion of FUBP1.
Figure 4.
FUBP1 is a facilitator of the binding of RUNX1 on the +30kb enhancer of c-KIT. (A) ChIP-qPCR performed on samples from Nalm6control and Nalm6shFUBP1 cells using a specific anti-RUNX1 (n = 3) or a non-specific control IgG (n = 3) at the c-KIT enhancer (+30 kb) regulatory regions. Histogram indicates the means with S.D. of the fold enrichment of peaks relative to IgG. (B) ChIP-qPCR performed on samples from Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1 and Nalm6+RUNX1+FUBP1 cells using specific anti-histones H3K4me3, H3K27ac and H3K4me1 at the c-KIT promoter (+700 bp) and enhancer (+30 kb) regulatory regions. Histogram indicates the fold enrichment of peaks relative to IgG.
We next investigated the impact of RUNX1 and FUBP1 modulation on histones marks at the promoter and the enhancer. We showed that enforced expression of RUNX1, FUBP1 or both RUNX1 with FUBP1 increases H3K4me1 and H3K27ac signal at the +30 kb enhancer, and H3K4me3 and H3K27ac at the promoter (Figure 4B). Importantly, the strong increase of H3K27ac signal when both RUNX1 and FUBP1 are overexpressed indicates a strong activation of the gene transcription (Figure 4B). Those data demonstrate that RUNX1 and FUBP1 cooperate at the +30 kb c-KIT enhancer and modulate the chromatin state at this region.
Identification of RUNX1 and FUBP1 DNA binding motifs within the +30 kb enhancer of c-KIT
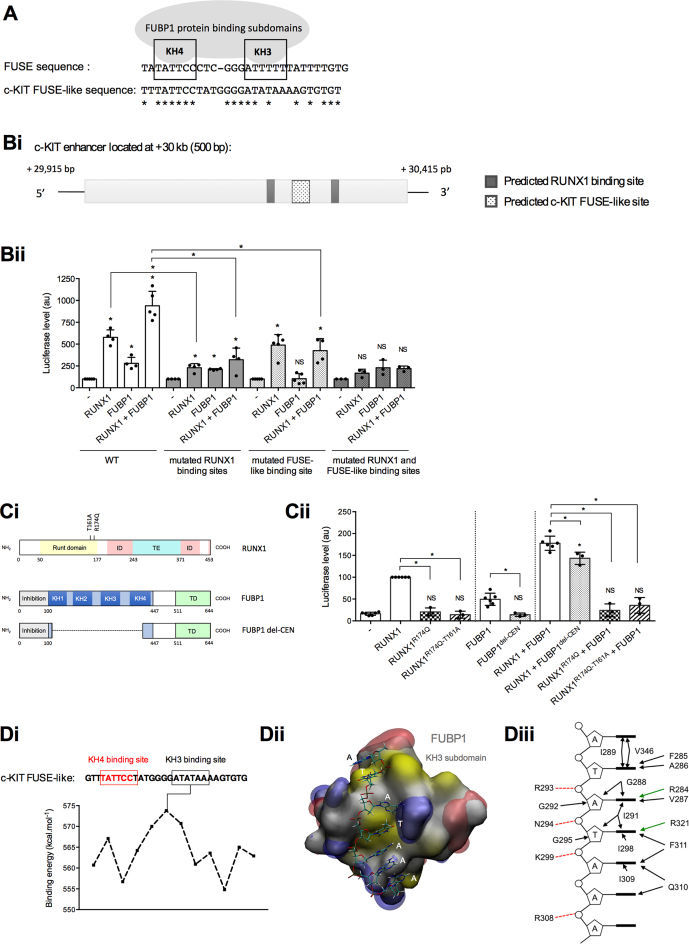
Analysis of the +30 kb c-KIT enhancer (500-nucleotide-long) revealed the presence a 29-nucleotide-long FUSE-like sequence homologous to the sole FUBP1-binding motif described so far and called the FUSE sequence (18,59) (Figure 5A). We also predicted two RUNX1 binding sites that displayed high sequence conservation with the RUNX1 consensus motif 5′-TGTGGT-3′ (60,61). To investigate the importance of these sequences in RUNX1- and FUBP1-dependent transcriptional activation, we mutated RUNX1- and FUBP1-predicted binding sites in +30 kb enhancer luciferase reporter vector. Mutations of those predicted binding sequences resulted in the abrogation of the effects of RUNX1 and FUBP1, indicating that identified RUNX1- and FUBP1 FUSE-like binding sites are functional and that each element is required for the transcriptional activation of the +30 kb enhancer of c-KIT (Figure 5B).
Figure 5.
Identification of RUNX1 and FUBP1 DNA binding motifs within the +30 kb enhancer of c-KIT. (A) Schematic representation of two functional subdomains (KH3 and KH4) of FUBP1 required for its binding to the FUSE sequence located –1.5 kb upstream the c-MYC P1 promoter (20,59). Potential c-KIT FUSE-like sequence within the +30 kb c-KIT intronic enhancer (identified by in silico analysis) is provided. Nucleic acid homology is represented by asterisks (*). The c-KIT FUSE-like sequence showed 48.2% homology compared to the FUSE sequence according to CLUSTAL 2.1 Multiple Sequence Alignments software (83). (Bi) Scheme of the human c-KIT enhancer. Predicted RUNX1 binding sites with more than 80% sequence conservation from Jaspar RUNX1 matrix (MA0002.1) have been selected with Jaspar software (61). (Bii) Luciferase assays with a vector full-length or mutated variants of the +30 kb c-KIT enhancer downstream a minimal promoter and a luciferase ORF, in presence of RUNX1 and FUBP1 expressing vectors were performed in HEK293shFUBP1 cells. In the mutated variants, RUNX1 predicted binding sites 5′-GGTGTTGTG-3′ and 5′-ATTGTGGTTA-3′ were site-directed mutated or the predicted FUBP1 FUSE-like binding sequence was deleted. Luciferase levels (firefly luciferase/Renilla luciferase) are represented using a bar chart-scatter dot plot indicating the means with S.D. of at least four independent experiments. Luciferase activities are normalized to the control (–). NS: non-significant, *P < 0.05 in Mann–Whitney tests. (Ci) Scheme of RUNX1 and FUBP1 proteins with indication of the position of mutations for RUNX1, and truncated FUBP1. ID: inhibition domain, TE/TD: transactivation domain. (Cii) Luciferase assays with a vector containing the +30 kb c-KIT enhancer downstream a minimal promoter and a luciferase ORF, in presence of RUNX1 mutants and truncated FUBP1 expressing vectors were performed in HEK293shFUBP1 cells. Luciferase levels are represented as in Bii.(Di) Binding energies (kcal.mol-1) of the KH3 subdomain of FUBP1 protein with the c-KIT FUSE-like sequence. Red framed sequence corresponds to the positioning of KH4 subdomain by analogy to the FUSE sequence retrieved on c-MYC promoter associated to the solution NMR structure of KH4 (PDB ID: 1J4W). Maximum binding energy of KH3 is computed for 5′-ATATAAA-3′. (Dii) Structural model for the most stable complex formed by the KH3 subdomain with the c-KIT +30 kb enhancer sequence. Protein is shown as a molecular surface colored according to hydrophobicity (84) (white for hydrophilic, yellow for hydrophobic), electrostatic potentials are represented as iso-surfaces (–50kTe in red and +50kTe in blue). Tilt observed between second (dT) and third (dA) bases originates from the position of the KH3 first alpha-helix, as it was observed in the experimental structure used as structural template for modeling. (Diii) Schematic representation of the amino-acids of KH3 subdomain of FUBP1 involved in the binding to the FUSE sequence.
Additionally, DNA-binding mutants of RUNX1 protein (RUNX1R174Q and RUNX1T161A,R174Q) (62,63) and truncation of FUBP1 central DNA-binding domain encompassing four K-Homology (KH) subdomains (50) reduced the transcription activity of the +30kb enhancer in luciferase reporter assays (Figure 5C). Those results demonstrated that RUNX1- and FUBP1-DNA-binding domains are necessary for a maximal transcriptional activation of the +30 kb enhancer of c-KIT.
Since FUBP1 requires two functional subdomains (KH3 and KH4) to bind DNA (18,59,64,65) (Figure 5A), and because FUBP1-DNA binding sequences are documented only for a few genes (19,64), we investigated the binding energies of the KH3 and KH4 subdomains on the +30 kb c-KIT enhancer (Figure 5D). We generated 12 structural models of KH3 binding to single-stranded-DNA sliding sequences in 3′ from the theoretical positioning of the KH4 subdomain on the 5′-TATTCC-3′ sequence. A maximal binding energy was measured for the AT-rich sequence 5′-ATATAAA-3′ located in 3′ at 7-nucleotide-apart from the KH4-bound sequence. This novel KH3-bound sequence is coherent with the previously described AU-rich FUBP1 RNA-bound sequence (66) and its position relative to the KH4-bound motif is fully consistent with the inter-domain distance suggested by previous structural studies (59). The structural model of KH3 binding to 5′-ATATAAA-3′ sequence indicates that stabilization of this complex is mainly driven through hydrophobic contacts with the ssDNA nucleobases and electrostatic interactions with the ssDNA sugar phosphate backbone of amino-acids located in a groove (Figure 5D).
Hence, we have identified the RUNX1 binding sequences and a new FUBP1 FUSE-like binding sequence, where KH4 and KH3 FUBP1-subdomains bind on the intronic enhancer of c-KIT, which functionally regulate c-KIT expression.
FUBP1 and RUNX1 exacerbate the c-KIT pathway and contribute to cell proliferation
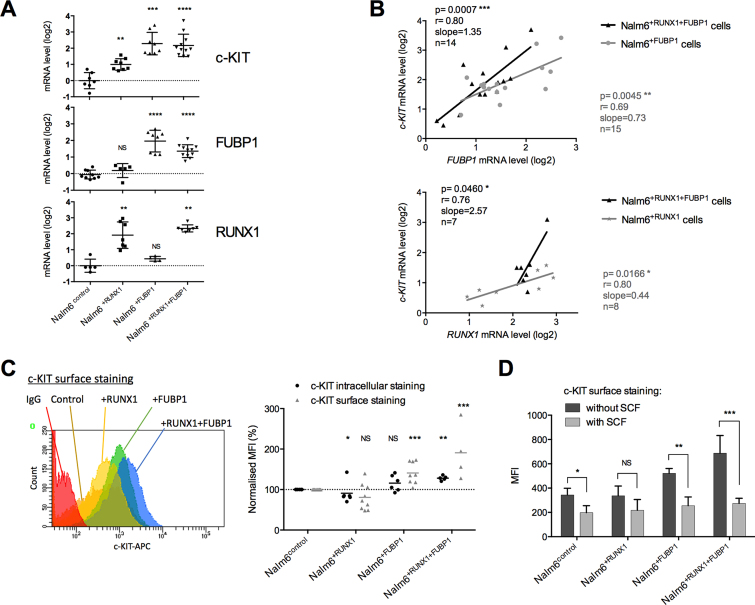
To further investigate the cooperative role of FUBP1 and RUNX1 in c-KIT regulation, we compared c-KIT level and functionality in pre-B Nalm6 cells overexpressing FUBP1 and/or RUNX1. c-KIT transcript levels were elevated in Nalm6+RUNX1, Nalm6+FUBP1 and in Nalm6+RUNX1+FUBP1 stable cell lines (Figure 6A). Furthermore, we identified a positive correlation between FUBP1 and c-KIT mRNA levels as well as between RUNX1 and c-KIT mRNAs (Figure 6B). Comparable results were obtained in REH cells and ETV6-RUNX1 leukemia patient biopsies (data from St. Jude Children's Research Hospital—Pediatric Cancer Data Portal) (67) (Supplementary Figure S3A). Importantly, the presence of RUNX1 reinforces the regulation of c-KIT by FUBP1 (Figure 6B). The c-KIT protein steady-state levels were also elevated (Supplementary Figure S3B). This phenotype was more pronounced for both intracellular and surface c-KIT in Nalm6 cells overexpressing FUBP1 or FUBP1+RUNX1 as shown by FACS (Figure 6C). Upon binding of the c-KIT ligand, the Stem Cell Factor (SCF), c-KIT dimerizes and autophosphorylates allowing the activation of its downstream pathways, including the ERK MAP-kinase pathway (68). Hence, to investigate the functionality of c-KIT, we next analyzed phospho-c-KIT in Nalm6control and Nalm6+FUBP1 cells treated with SCF. Nalm6+FUBP1 cells showed a faster response to SCF than control cells with higher levels of phospho-c-KIT and phospho-ERK1/2 after 5 minutes of treatment (Supplementary Figure S3C). In addition, SCF induces c-KIT internalization and degradation, one of the hallmarks of c-KIT functionality (69). Following SCF stimulation, c-KIT was more intensively internalized in Nalm6+FUBP1 and especially in Nalm6+RUNX1+FUBP1 than in Nalm6control cells. These results suggest that a stronger biological response to SCF is elicited by overexpressing RUNX1 and FUBP1 (Figure 6D).
Figure 6.
FUBP1 and RUNX1 exacerbate the c-KIT pathway. (A) mRNA levels analyzed by RT-qPCR of c-KIT, FUBP1, and RUNX1 in Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1, and Nalm6+RUNX1+FUBP1 cells. Results are presented in terms of a fold change after normalizing with GAPDH mRNA. Each dot represents a biological replicate. NS: non-significant, **P < 0.01, ***P < 0.001, ****P < 0.0001 in Mann–Whitney tests. (B) Correlation between FUBP1 and KIT mRNA levels from different transduced Nalm6+FUBP1 and Nalm6+RUNX1+FUBP1 stable cell lines, and correlation between RUNX1 and KIT mRNA levels in Nalm6+RUNX1 and Nalm6+RUNX1+FUBP1 cells are presented. P-value (p), Pearson correlation coefficient (r), slope of the correlation curve (slope), and number of biological replicates analyzed (n) are indicated. (C) FACS analyses following c-KIT (CD117) intracellular and surface staining in Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1, or Nalm6+RUNX1+FUBP1 cells. The c-KIT expression was assessed in comparison with the control isotype staining. A representative analysis of the KIT surface staining is shown on the left. Percentage of the mean fluorescence intensity (MFI) is presented on the right panel (n = 4–8 per group) after normalizing with Nalm6control cells. Statistical significance was assessed compared to Nalm6control cells by Mann–Whitney tests (NS: non-significant, *P < 0.05, **P < 0.01, ***P < 0 .001). (D) Mean fluorescence intensity (MFI) analyzed by FACS from Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1, and Nalm6+RUNX1+FUBP1 cells following SCF treatment (0 or 100 ng/ml of SCF were added in the culture media and incubated 5 min at 37°C) and c-KIT surface staining. SCF induces c-KIT internalization and degradation, one of the hallmarks of c-KIT functionality (69). Statistical significance was assessed compared to untreated cells (without SCF) by Mann–Whitney tests (NS: non-significant, *P < 0.05, **P < 0.01, ***P < 0 .001).
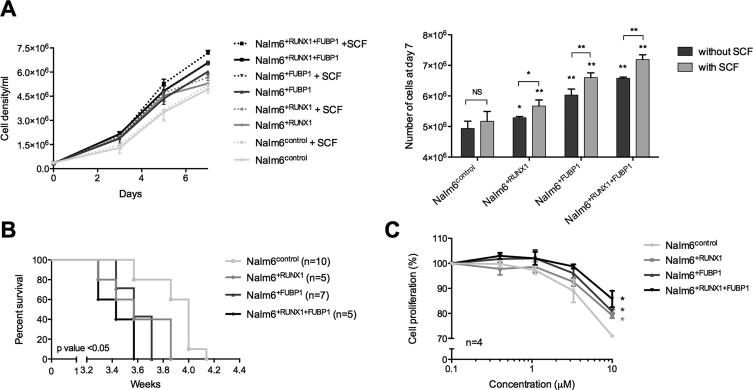
Having demonstrated that FUBP1 and RUNX1 regulate c-KIT and that c-KIT is properly functional, we next addressed the question of the physiological consequence of this regulation. Nalm6+FUBP1, Nalm6+RUNX1 and Nalm6+RUNX1+FUBP1 cells presented a higher proliferation rate than Nalm6control cells (Figure 7A). This result was consistent with the proliferation arrest induced by FUBP1 loss-of-function (32,70). The gain-in-proliferation phenotype was slightly exacerbated in presence of SCF (Figure 7A) as anticipated from the surface membrane c-KIT internalization (Figure 6D), even if Nalm6 cells are not fully dependent on SCF for proliferation. Apoptosis and necrosis were minimal in all those cell lines (Supplementary Figure S3D). A BrdU pulse-chase assay allowed us to correlate this increase in proliferation rate to an acceleration of cell cycle in Nalm6+FUBP1, Nalm6+RUNX1 and Nalm6+RUNX1+FUBP1 cells compared to Nalm6control cells (Supplementary Figure S3E). Finally, mice xenografted with Nalm6+FUBP1, Nalm6+RUNX1 or Nalm6+RUNX1+FUBP1 cells presented a reduced survival compared to mice injected with Nalm6control cells (Figure 7B). Altogether, these data demonstrate that enforced expression of FUBP1 and RUNX1 regulate c-KIT expression and subsequent cell proliferation.
Figure 7.
FUBP1 and RUNX1 contribute to c-KIT-dependent cell proliferation. (A) Proliferation curves from Nalm6control (n = 5), Nalm6+RUNX1 (n = 4), Nalm6+FUBP1 (n = 5), Nalm6+RUNX1+FUBP1 (n = 5) cells exposed to 0 or 100 ng/ml of SCF the first day (day 0). Histogram (on the right) represents the number (means with S.D.) of cells at day 7. Statistical significance was assessed compared to Nalm6control by Mann–Whitney tests (NS: non-significant, *P < 0.05, **P < 0.01). (B) Survival curves from immunodeficient NOD/scid IL2 Rgnull mice xenografted with 100 000 cells: Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1, or Nalm6+RUNX1+FUBP1 human cell lines (n = 5–10 per group). The general condition of mice was monitored daily until experiment ended. Kaplan-Meier survival curves were plotted. P-value was assessed with Mantel-Cox tests compared to mice xenografted with Nalm6control. (C) Inhibition curves from Nalm6control, Nalm6+RUNX1, Nalm6+FUBP1, and Nalm6+RUNX1+FUBP1 cell lines treated with 100 ng/ml of SCF and with growing concentrations of imatinib mesylate (0–10 μM) for 48h. Each point is the mean of four biological replicates ± S.E.M. Statistical significance was assessed at 10 μM compared to Nalm6control by Mann–Whitney tests (*P < 0.05).
We next evaluated Nalm6 cells responsiveness to a c-KIT inhibitor, the imatinib mesylate (71). While Nalm6control, Nalm6+FUBP1, Nalm6+RUNX1 and Nalm6+RUNX1+FUBP1 cells were sensitive to imatinib mesylate at a standard concentration of 1 μM as shown by the decreased phosphorylation of c-KIT and of downstream targets such as ERK and AKT (Supplementary Figure S3F), cells overexpressing FUBP1, RUNX1, alone or together, were less sensitive to the inhibitor in a proliferation assay beyond the dose of 1 μM (Figure 7C). This result demonstrates that FUBP1 and RUNX1 levels can impact on the sensitivity of the cells to a c-KIT inhibitor.
DISCUSSION
In our search for identifying transcriptional co-factors of RUNX1 and deciphering its function, we demonstrated that RUNX1 and FUBP1 co-immunoprecipitated and are physically located close to each other on active chromatin of human hematopoietic cells.
Examination of RUNX1 genome-wide distribution together with gene expression data led us to propose that the oncogene c-KIT is commonly activated by RUNX1 and FUBP1. Moreover, we identified two regulatory regions in the first intron of c-KIT (the +700 bp promoter and the +30 kb enhancer regions) that are bound by RUNX1, FUBP1 and present active histone marks. Interestingly, a very recent study in AML1-ETO acute myeloid leukemia spotted identical regulatory regions in c-KIT gene bound by RUNX1 (or AML1) and the fusion protein AML1-ETO (72). The role of the transcriptional regulators RUNX1 and FUBP1 in a potential interplay between c-KIT enhancer and promoter has not been addressed yet. However, Tian et al. identified a DNA-loop between the promoter and the +30 kb enhancer of c-KIT that plays a significant role in transcriptional regulation (72). Since FUBP1 induces a similar promoter-enhancer loop in the c-MYC gene through interaction with TFIIH (73), we propose that FUBP1 could trigger the formation of such a DNA-loop at the c-KIT locus.
Optimal FUBP1 binding sequences are still a matter of debate (19,22,59,64,74). Here, we benefit from the functional c-KIT enhancer to bring new insights on novel FUBP1 DNA-binding sequences. Analysis of the +30 kb c-KIT enhancer, by molecular biology and structural modeling, allows us to report a c-KIT FUSE-like sequence encompassing the KH4 binding sequence (retrieved from the c-MYC promoter (18,59)), a 7-nucleotide-long spacer that may not interact with FUBP1 as described previously (59), and an AT-rich sequence bound by the KH3 subdomain consistently with FUBP1-bound AU-rich sequences described previously (66).
c-KIT encodes a tyrosine kinase receptor and is expressed in hematopoietic stem and progenitor cells (38,75). Binding of SCF to the c-KIT receptor triggers a cascade of signaling pathways resulting in the regulation of apoptosis, proliferation and differentiation (68). Inappropriate c-KIT expression bears oncogenic consequences. c-KIT gene is found overexpressed in human leukemia from myeloid and some lymphoid lineages (67,68,76,77) and constitutive c-KIT activity triggers lymphoblastic leukemia in mouse model (78). Our findings suggest a new mechanism for the control of cell proliferation via the fine-tuned regulation of c-KIT by FUBP1 and to a lesser extent RUNX1. Importantly, we demonstrated that FUBP1-induced c-KIT expression produces a functional protein with increased tyrosine phosphorylation of its substrates and faster internalization following SCF stimulation. The phenotype on proliferation associated to c-KIT pathway activation is reinforced when both FUBP1 and RUNX1 are overexpressed. FUBP1 mRNA level was shown to be significantly elevated in leukemia stem cell-enriched cell populations and in some lymphoblastic leukemia subtypes (data from St. Jude Children's Research Hospital) (67,79) as well as RUNX1 (80). Here, we proposed for the first time a rationale for oncogenic function of FUBP1 in leukemia. Therefore, since RUNX1, FUBP1 and c-KIT are overexpressed in some subtypes of leukemia, abnormalities in this regulatory network could participate in the onset or the maintenance of RUNX1-related leukemia. We hypothesize that the c-KIT pathway may be impaired in those models and draw attention to the possible decrease in sensitivity under c-KIT inhibitor treatment.
In conclusion, we demonstrate a regulatory network involving two transcriptional regulators, RUNX1 and FUBP1, in the control of the +30 kb c-KIT enhancer. FUBP1 and RUNX1 colocalize on active chromatin, cooperate for regulating c-KIT transcription, and promote cell cycle progression. Although our study focuses on the role of the cooperativity between RUNX1 and FUBP1 in the regulation of c-KIT, we can envision that this partnership most likely impacts on other target genes. Deregulation of this regulatory mechanism may explain some oncogenic function of RUNX1 and FUBP1.
DATA AVAILABILITY
All ChIP-seq data have been deposited to GEO with the accession number of GSE109377 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109377).
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of Gene Expression and Oncogenesis team for insightful discussions and for providing scientific expertise. We are most grateful to patients and patients’ families from Rennes University Hospital, CHRU Rennes. We thank the following core facilities from BIOSIT SFR UMS CNRS 3480 – INSERM 018: MRic-Photonics (Stéphanie Dutertre), Flow cytometry and cell sorting (Laurent Deleurme, Gersende Lacombe), ImPACcell (Rémy Le Guével), ARCHE animal housing (Laurence Bernard-Touami), Human and Environmental Genomics of Rennes facilities (GEH) (Marc Aubry).
Author Contributions: L.D. performed experiments, analyzed and interpreted the data. H.J. and J.G. performed experiments and analyzed the data. S.A. performed and interpreted Chip-Seq bioinformatics analyses. A.G.R. assisted with experiments. A.S. performed RIME and some Chip-Seq experiments. L.Do performed CD34+ cell sorting. F.C. performed bioinformatics analyses. J.S.C. collaborates for RIME and Chip-Seq experiments. M.Z. and M.A.R. provided CD34+ sorted cells and interpreted the data. O.D. performed modeling studies, analyzed and interpreted those data. G.S., M.D.G. and V.G. analyzed and interpreted the data. V.G. provided patients samples. M.B.T. designed the study, analyzed and interpreted the data. M.B.T. and L.D. wrote the manuscript with consultation and contribution from all coauthors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ligue Régionale contre le cancer [comité 22,35,56,79,41 to M.B.T., L.D., V.G.]; SFR Biosit UMS CNRS 3480-INSERM 018 (to M.B.T.); Région Bretagne (to L.D., M.B.T.); The Société Française d’Hématologie (to L.D.); Rennes Métropole (to M.B.T.); French Research Ministry (to H.J.); FHU CAMIN (to J.G., V.G.); the société française de lutte contre les cancers et les leucémies de l’enfant et de l’adolescent and the Fédération Enfants et Santé (to M.B.T.); a private donator Mrs M-Dominique Blanc-Bert (to M.B.T.); Cancéropole Grand Ouest (to L.D.); The Société Française de Biochimie et Biologie Moléculaire (to H.J.); CNRS; Université de Rennes 1 and the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme [FP7/2007-2013] under REA grant agreement [291851 to M.B.T.]. Funding for open access charge: People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sood R., Kamikubo Y., Liu P.. Role of RUNX1 in hematological malignancies. Blood. 2017; 129:2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yzaguirre A.D., de Bruijn M.F.T.R., Speck N.A.. The role of Runx1 in embryonic blood cell formation. Adv. Exp. Med. Biol. 2017; 962:47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brady G., Elgueta Karstegl C., Farrell P.J.. Novel function of the unique N-terminal region of RUNX1c in B cell growth regulation. Nucleic Acids Res. 2013; 41:1555–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayashi K., Natsume W., Watanabe T., Abe N., Iwai N., Okada H., Ito Y., Asano M., Iwakura Y., Habu S. et al. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J. Immunol. 2000; 165:6816–6824. [DOI] [PubMed] [Google Scholar]
- 5. Huang G., Zhang P., Hirai H., Elf S., Yan X., Chen Z., Koschmieder S., Okuno Y., Dayaram T., Growney J.D. et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 2008; 40:51–60. [DOI] [PubMed] [Google Scholar]
- 6. Ichikawa M., Asai T., Chiba S., Kurokawa M., Ogawa S.. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle. 2004; 3:722–724. [PubMed] [Google Scholar]
- 7. Ichikawa M., Yoshimi A., Nakagawa M., Nishimoto N., Watanabe-Okochi N., Kurokawa M.. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int. J. Hematol. 2013; 97:726–734. [DOI] [PubMed] [Google Scholar]
- 8. Lorsbach R.B., Moore J., Ang S.O., Sun W., Lenny N., Downing J.R.. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004; 103:2522–2529. [DOI] [PubMed] [Google Scholar]
- 9. Okuda T., van Deursen J., Hiebert S.W., Grosveld G., Downing J.R.. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996; 84:321–330. [DOI] [PubMed] [Google Scholar]
- 10. Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A.. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonifer C., Levantini E., Kouskoff V., Lacaud G.. Runx1 structure and function in blood cell development. Adv. Exp. Med. Biol. 2017; 962:65–81. [DOI] [PubMed] [Google Scholar]
- 12. Ito Y., Bae S.-C., Chuang L.S.H.. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015; 15:81–95. [DOI] [PubMed] [Google Scholar]
- 13. Huang G., Shigesada K., Ito K., Wee H.-J., Yokomizo T., Ito Y.. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin–proteasome-mediated degradation. EMBO J. 2001; 20:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Bushweller J.H., Bories J.C., Alt F.W., Ryan G. et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996; 87:697–708. [DOI] [PubMed] [Google Scholar]
- 15. Draper J.E., Sroczynska P., Tsoulaki O., Leong H.S., Fadlullah M.Z.H., Miller C., Kouskoff V., Lacaud G.. RUNX1B Expression is highly heterogeneous and distinguishes megakaryocytic and erythroid lineage fate in adult mouse hematopoiesis. PLoS Genet. 2016; 12:e1005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niebuhr B., Kriebitzsch N., Fischer M., Behrens K., Gunther T., Alawi M., Bergholz U., Muller U., Roscher S., Ziegler M. et al. Runx1 is essential at two stages of early murine B-cell development. Blood. 2013; 122:413–423. [DOI] [PubMed] [Google Scholar]
- 17. Davis-Smyth T., Duncan R.C., Zheng T., Michelotti G., Levens D.. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996; 271:31679–31687. [DOI] [PubMed] [Google Scholar]
- 18. Duncan R., Bazar L., Michelotti G., Tomonaga T., Krutzsch H., Avigan M., Levens D.. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994; 8:465–480. [DOI] [PubMed] [Google Scholar]
- 19. Zhang J., Chen Q.M.. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013; 32:2907–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Avigan M.I., Strober B., Levens D.. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J. Biol. Chem. 1990; 265:18538–18545. [PubMed] [Google Scholar]
- 21. Malz M., Weber A., Singer S., Riehmer V., Bissinger M., Riener M.-O., Longerich T., Soll C., Vogel A., Angel P. et al. Overexpression of far upstream element binding proteins: A mechanism regulating proliferation and migration in liver cancer cells. Hepatology. 2009; 50:1130–1139. [DOI] [PubMed] [Google Scholar]
- 22. Rabenhorst U., Beinoraviciute-Kellner R., Brezniceanu M.-L., Joos S., Devens F., Lichter P., Rieker R.J., Trojan J., Chung H.-J., Levens D.L. et al. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology. 2009; 50:1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo L., Zaysteva O., Nie Z., Mitchell N.C., Amanda Lee J.E., Ware T., Parsons L., Luwor R., Poortinga G., Hannan R.D. et al. Defining the essential function of FBP/KSRP proteins: Drosophila Psi interacts with the mediator complex to modulate MYC transcription and tissue growth. Nucleic Acids Res. 2016; 44:7646–7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang P.-N., Lin J.-Y., Locker N., Kung Y.-A., Hung C.-T., Lin J.-Y., Huang H.-I., Li M.-L., Shih S.-R.. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 2011; 39:9633–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irwin N., Baekelandt V., Goritchenko L., Benowitz L.I.. Identification of two proteins that bind to a pyrimidine-rich sequence in the 3′-untranslated region of GAP-43 mRNA. Nucleic Acids Res. 1997; 25:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin J.-Y., Li M.-L., Shih S.-R.. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009; 37:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miro J., Laaref A.M., Rofidal V., Lagrafeuille R., Hem S., Thorel D., Mechin D., Mamchaoui K., Mouly V., Claustres M. et al. FUBP1: a new protagonist in splicing regulation of the DMD gene. Nucleic Acids Res. 2015; 43:2378–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bettegowda C., Agrawal N., Jiao Y., Sausen M., Wood L.D., Hruban R.H., Rodriguez F.J., Cahill D.P., McLendon R., Riggins G. et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011; 333:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singer S., Malz M., Herpel E., Warth A., Bissinger M., Keith M., Muley T., Meister M., Hoffmann H., Penzel R. et al. Coordinated expression of stathmin family members by far upstream sequence Element-Binding Protein-1 increases motility in Non–Small cell lung cancer. Cancer Res. 2009; 69:2234–2243. [DOI] [PubMed] [Google Scholar]
- 30. Weber A., Kristiansen I., Johannsen M., Oelrich B., Scholmann K., Gunia S., May M., Meyer H.-A., Behnke S., Moch H. et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer. 2008; 8:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang L., Zhu J., Zhang J., Bao B., Guan C., Yang X., Liu Y., Huang Y., Ni R., Ji L.. Far upstream element-binding protein 1 (FUBP1) is a potential c-Myc regulator in esophageal squamous cell carcinoma (ESCC) and its expression promotes ESCC progression. Tumor Biol. 2016; 37:4115–4126. [DOI] [PubMed] [Google Scholar]
- 32. Rabenhorst U., Thalheimer F.B., Gerlach K., Kijonka M., Böhm S., Krause D.S., Vauti F., Arnold H.-H., Schroeder T., Schnütgen F. et al. Single-Stranded DNA-Binding transcriptional regulator FUBP1 is essential for fetal and adult hematopoietic stem cell Self-Renewal. Cell Rep. 2015; 11:1847–1855. [DOI] [PubMed] [Google Scholar]
- 33. Zhou W., Chung Y.-J., Parrilla Castellar E.R., Zheng Y., Chung H.-J., Bandle R., Liu J., Tessarollo L., Batchelor E., Aplan P.D. et al. Far upstream element binding protein plays a crucial role in embryonic development, hematopoiesis, and stabilizing Myc expression levels. Am. J. Pathol. 2016; 186:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klener P., Fronkova E., Berkova A., Jaksa R., Lhotska H., Forsterova K., Soukup J., Kulvait V., Vargova J., Fiser K. et al. Mantle cell lymphoma-variant Richter syndrome: Detailed molecular-cytogenetic and backtracking analysis reveals slow evolution of a pre-MCL clone in parallel with CLL over several years: Mantle cell lymphoma-variant Richter syndrome. Int. J. Cancer. 2016; 139:2252–2260. [DOI] [PubMed] [Google Scholar]
- 35. Lindqvist C.M., Lundmark A., Nordlund J., Freyhult E., Ekman D., Almlöf J.C., Raine A., Övernäs E., Abrahamsson J., Frost B.-M. et al. Deep targeted sequencing in pediatric acute lymphoblastic leukemia unveils distinct mutational patterns between genetic subtypes and novel relapse-associated genes. Oncotarget. 2016; 7:64071–64088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Landau D.A., Tausch E., Taylor-Weiner A.N., Stewart C., Reiter J.G., Bahlo J., Kluth S., Bozic I., Lawrence M., Böttcher S. et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015; 526:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ashman L.K. The biology of stem cell factor and its receptor C-kit. Int. J. Biochem. Cell Biol. 1999; 31:1037–1051. [DOI] [PubMed] [Google Scholar]
- 38. Broudy V.C. Stem cell factor and hematopoiesis. Blood. 1997; 90:1345–1364. [PubMed] [Google Scholar]
- 39. Miettinen M., Lasota J.. KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl. Immunohistochem. Mol. Morphol. 2005; 13:205–220. [DOI] [PubMed] [Google Scholar]
- 40. Yarden Y., Kuang W.J., Yang-Feng T., Coussens L., Munemitsu S., Dull T.J., Chen E., Schlessinger J., Francke U., Ullrich A.. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987; 6:3341–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mohammed H., D’Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L.L. et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013; 3:342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Debaize L., Jakobczyk H., Rio A.-G., Gandemer V., Troadec M.-B.. Optimization of proximity ligation assay (PLA) for detection of protein interactions and fusion proteins in non-adherent cells: application to pre-B lymphocytes. Mol. Cytogenet. 2017; 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S.Y., Hahn W.C., Sharp P.A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003; 9:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arnaud M.-P., Vallée A., Robert G., Bonneau J., Leroy C., Varin-Blank N., Rio A.-G., Troadec M.-B., Galibert M.-D., Gandemer V.. CD9, a key actor in the dissemination of lymphoblastic leukemia, modulating CXCR4-mediated migration via RAC1 signaling. Blood. 2015; 126:1802–1812. [DOI] [PubMed] [Google Scholar]
- 45. Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O., Campisi J., Yaswen P., Cooper P.K., Kaufman P.D.. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009; 4:e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sérandour A.A., Avner S., Oger F., Bizot M., Percevault F., Lucchetti-Miganeh C., Palierne G., Gheeraert C., Barloy-Hubler F., Péron C.L. et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012; 40:8255–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin H., Liu T., Manrai A.K., Liu X.S.. CEAS: cis-regulatory element annotation system. Bioinforma. Oxf. Engl. 2009; 25:2605–2606. [DOI] [PubMed] [Google Scholar]
- 48. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nègre D., Mangeot P.E., Duisit G., Blanchard S., Vidalain P.O., Leissner P., Winter A.J., Rabourdin-Combe C., Mehtali M., Moullier P. et al. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 2000; 7:1613–1623. [DOI] [PubMed] [Google Scholar]
- 50. Zheng Y., Miskimins W.K.. Far upstream element binding protein 1 activates translation of p27Kip1 mRNA through its internal ribosomal entry site. Int. J. Biochem. Cell Biol. 2011; 43:1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krieger E., Vriend G.. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015; 36:996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He L., Weber A., Levens D.. Nuclear targeting determinants of the far upstream element binding protein, a c-myc transcription factor. Nucleic Acids Res. 2000; 28:4558–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lu J., Maruyama M., Satake M., Bae S.C., Ogawa E., Kagoshima H., Shigesada K., Ito Y.. Subcellular localization of the alpha and beta subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol. Cell. Biol. 1995; 15:1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zelent A., Greaves M., Enver T.. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004; 23:4275–4283. [DOI] [PubMed] [Google Scholar]
- 55. Melchers F. Checkpoints that control B cell development. J. Clin. Invest. 2015; 125:2203–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Atanassov B.S., Dent S.Y.R.. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011; 12:924–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rabenhorst U., Beinoraviciute-Kellner R., Brezniceanu M.-L., Joos S., Devens F., Lichter P., Rieker R.J., Trojan J., Chung H.-J., Levens D.L. et al. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatol. Baltim. Md. 2009; 50:1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Malz M., Bovet M., Samarin J., Rabenhorst U., Sticht C., Bissinger M., Roessler S., Bermejo J.L., Renner M., Calvisi D.F. et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014; 60:1241–1250. [DOI] [PubMed] [Google Scholar]
- 59. Braddock D.T., Louis J.M., Baber J.L., Levens D., Clore G.M.. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002; 415:1051–1056. [DOI] [PubMed] [Google Scholar]
- 60. Levanon D., Groner Y.. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004; 23:4211–4219. [DOI] [PubMed] [Google Scholar]
- 61. Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B.. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004; 32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matheny C.J., Speck M.E., Cushing P.R., Zhou Y., Corpora T., Regan M., Newman M., Roudaia L., Speck C.L., Gu T.-L. et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007; 26:1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang L., Li Z., Yan J., Pradhan P., Corpora T., Cheney M.D., Bravo J., Warren A.J., Bushweller J.H., Speck N.A.. Mutagenesis of the Runt domain defines two energetic hot spots for heterodimerization with the core binding factor beta subunit. J. Biol. Chem. 2003; 278:33097–33104. [DOI] [PubMed] [Google Scholar]
- 64. Benjamin L.R., Chung H.-J., Sanford S., Kouzine F., Liu J., Levens D.. Hierarchical mechanisms build the DNA-binding specificity of FUSE binding protein. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18296–18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Siomi H., Matunis M.J., Michael W.M., Dreyfuss G.. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993; 21:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sully G., Dean J.L.E., Wait R., Rawlinson L., Santalucia T., Saklatvala J., Clark A.R.. Structural and functional dissection of a conserved destabilizing element of cyclo-oxygenase-2 mRNA: evidence against the involvement of AUF-1 [AU-rich element/poly(U)-binding/degradation factor-1], AUF-2, tristetraprolin, HuR (Hu antigen R) or FBP1 (far-upstream-sequence-element-binding protein 1). Biochem. J. 2004; 377:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou X., Edmonson M.N., Wilkinson M.R., Patel A., Wu G., Liu Y., Li Y., Zhang Z., Rusch M.C., Parker M. et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016; 48:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lennartsson J., Jelacic T., Linnekin D., Shivakrupa R.. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005; 23:16–43. [DOI] [PubMed] [Google Scholar]
- 69. Yee N.S., Hsiau C.W., Serve H., Vosseller K., Besmer P.. Mechanism of down-regulation of c-kit receptor. Roles of receptor tyrosine kinase, phosphatidylinositol 3′-kinase, and protein kinase C. J. Biol. Chem. 1994; 269:31991–31998. [PubMed] [Google Scholar]
- 70. He L., Liu J., Collins I., Sanford S., O’Connell B., Benham C.J., Levens D.. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000; 19:1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cammenga J., Horn S., Bergholz U., Sommer G., Besmer P., Fiedler W., Stocking C.. Extracellular KIT receptor mutants, commonly found in core binding factor AML, are constitutively active and respond to imatinib mesylate. Blood. 2005; 106:3958–3961. [DOI] [PubMed] [Google Scholar]
- 72. Tian Y., Wang G., Hu Q., Xiao X., Chen S.. AML1/ETO trans-activates c-KIT expression through the long range interaction between promoter and intronic enhancer. J. Cell. Biochem. 2017; 119:3706–3715. [DOI] [PubMed] [Google Scholar]
- 73. Liu J., He L., Collins I., Ge H., Libutti D., Li J., Egly J.-M., Levens D.. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell. 2000; 5:331–341. [DOI] [PubMed] [Google Scholar]
- 74. Liu J., Chung H.-J., Vogt M., Jin Y., Malide D., He L., Dundr M., Levens D.. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011; 30:846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sykora K.W., Tomeczkowski J., Reiter A.. C-Kit receptors in childhood malignant lymphoblastic cells. Leuk. Lymphoma. 1997; 25:201–216. [DOI] [PubMed] [Google Scholar]
- 76. Andersson A., Olofsson T., Lindgren D., Nilsson B., Ritz C., Edén P., Lassen C., Råde J., Fontes M., Mörse H. et al. Molecular signatures in childhood acute leukemia and their correlations to expression patterns in normal hematopoietic subpopulations. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:19069–19074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ikeda H., Kanakura Y., Tamaki T., Kuriu A., Kitayama H., Ishikawa J., Kanayama Y., Yonezawa T., Tarui S., Griffin J.D.. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 1991; 78:2962–2968. [PubMed] [Google Scholar]
- 78. Weidemann R.R., Behrendt R., Schoedel K.B., Müller W., Roers A., Gerbaulet A.. Constitutive Kit activity triggers B-cell acute lymphoblastic leukemia-like disease in mice. Exp. Hematol. 2017; 45:45–55. [DOI] [PubMed] [Google Scholar]
- 79. Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J. et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011; 17:1086–1093. [DOI] [PubMed] [Google Scholar]
- 80. Gandemer V., Rio A.G., de Tayrac M., Sibut V., Mottier S., Ly Sunnaram B., Henry C., Monnier A., Berthou C., Le Gall E. et al. Five distinct biological processes and 14 differentially expressed genes characterize TEL/AML1-positive leukemia. BMC Genomics. 2007; 8:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A.. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012; 28:1805–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nicol J.W., Helt G.A., Blanchard S.G., Raja A., Loraine A.E.. The integrated genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinforma. Oxf. Engl. 2009; 25:2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D.. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Efremov R.G., Chugunov A.O., Pyrkov T.V., Priestle J.P., Arseniev A.S., Jacoby E.. Molecular lipophilicity in protein modeling and drug design. Curr. Med. Chem. 2007; 14:393–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ChIP-seq data have been deposited to GEO with the accession number of GSE109377 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109377).