Abstract
Enhancers are distal cis-regulatory elements that modulate gene expression. They are depleted of nucleosomes and enriched in specific histone modifications; thus, calling DNase-seq and histone mark ChIP-seq peaks can predict enhancers. We evaluated nine peak-calling algorithms for predicting enhancers validated by transgenic mouse assays. DNase and H3K27ac peaks were consistently more predictive than H3K4me1/2/3 and H3K9ac peaks. DFilter and Hotspot2 were the best DNase peak callers, while HOMER, MUSIC, MACS2, DFilter and F-seq were the best H3K27ac peak callers. We observed that the differential DNase or H3K27ac signals between two distant tissues increased the area under the precision-recall curve (PR-AUC) of DNase peaks by 17.5–166.7% and that of H3K27ac peaks by 7.1–22.2%. We further improved this differential signal method using multiple contrast tissues. Evaluated using a blind test, the differential H3K27ac signal method substantially improved PR-AUC from 0.48 to 0.75 for predicting heart enhancers. We further validated our approach using postnatal retina and cerebral cortex enhancers identified by massively parallel reporter assays, and observed improvements for both tissues. In summary, we compared nine peak callers and devised a superior method for predicting tissue-specific mouse developmental enhancers by reranking the called peaks.
INTRODUCTION
In metazoans, enhancers are a major class of regulatory elements that drive cell-type-specific and time-restricted patterns of gene expression (1,2). Population-scale genetic studies indicate that many sequence variants associated with human diseases reside in enhancers (3,4). Thus, genome-wide maps of enhancers and their activity patterns across cell and tissue types can provide tremendous insights into mechanisms of gene regulation and disease etiology.
In recent years, epigenetic approaches have greatly advanced genome-wide identification of enhancers (5). Enhancers are enriched in several histone modifications, including monomethylation of histone H3 at lysine residue 4 (H3K4me1) and acetylation of H3 at lysine 27 (H3K27ac) (6,7). Chromatin immunoprecipitation of histone modifications followed by sequencing (ChIP-seq) is a powerful technique for mapping histone marks genome-wide. A related approach is to perform ChIP-seq on the histone acetyltransferases P300 or CBP (8). Because most enhancers are in open chromatin regions, another approach is to treat chromatin with Deoxyribonuclease I (DNase I) and sequence the liberated DNA (DNase-seq) (2,9). ATAC-seq is another technique for detecting open chromatin regions that offers some practical advantages over DNase-seq (10). The CpG dinucleotides in enhancers remain unmethylated so whole-genome bisulfite sequencing is another type of high-throughput data that can be used for identifying enhancers (11).
A number of supervised machine-learning algorithms have been developed to integrate ChIP-seq, DNase-seq, ATAC-seq and DNA methylation data along with annotations such as evolutionary conservation and sequence motifs to predict enhancers (12–17). Some of these methods, e.g. REPTILE (12), RFECS (16) and LEG (18), combine multiple types of epigenetic signals in a particular cell or tissue type to predict enhancers in the same cell or tissue type, while other algorithms, e.g. PEDLA (13) and EnhancerFinder (14), use thousands of features, including features from distant cell types. Being supervised, these algorithms are trained using known enhancers. There are also unsupervised algorithms that integrate multiple types of epigenetic signals in a cell type to define a set of chromatin states, including enhancers (19–21).
Despite the success of these algorithms, many studies continue to use just one or two types of epigenetic signal to define enhancers in the specified cell type. This is because it is costly to perform multiple assays, especially when many samples are studied. For example, Pennacchio and colleagues used ChIP-seq datasets of H3K27ac and P300 in human and mouse heart tissues to build a compendium of heart enhancers (22). Sun et al. used H3K27ac ChIP-seq to define enhancers in the brain tissues of a large cohort of autism spectrum disorder patients and controls (23). To call enhancers based on DNase-seq, ATAC-seq, or ChIP-seq data, one first identifies the genomic regions with significantly high signal, called peaks. (DNase peaks are commonly known as DNase hypersensitive sites or DHSs. Here, we also call them DNase peaks for convenience.) Computational algorithms devised for this purpose are peak-calling algorithms or peak callers.
More than thirty peak callers have been developed for ChIP-seq data over the past decade (see review (24)). Some of these algorithms are designed for finding punctate peaks such as those of transcription factors (25), while others identify broad domains of histone marks such as H3K9me3 (26,27). The widths of DNase-seq and ATAC-seq peaks lie midway between punctate and broad, and these peaks have characteristic shapes that differ from TF and histone mark ChIP-seq peaks. Thus, a subset of algorithms designed for both punctate and broad ChIP-seq signals can also be configured to work for DNase-seq and ATAC-seq data.
Several benchmarking studies have compared algorithms for their consistency in finding histone mark peaks (28–31) and DNase peaks (32), with several studies also evaluating these peaks’ efficacy for identifying promoters. However, to the best of our knowledge, there is not yet a study to compare the performance of peak callers for finding enhancers. As described above, the utility of enhancer-focused peak calling is apparent: these peaks can guide the identification of enhancers in a cell or tissue type at a developmental stage, and the predicted enhancers can be experimentally tested (22) and can be used for studying the genetic basis of gene regulation in diseases (23,33).
Pennacchio and colleagues have established transgenic assays to test enhancer function in embryonic day 11.5 (e11.5) mice (34); their results for over 2500 genomic regions are provided in the VISTA Enhancer Database (35). Using these VISTA regions as the gold standard, we evaluated the peaks called by nine algorithms for the DNase-seq and histone mark ChIP-seq data of e11.5 mouse tissues: MACS2 (36), F-seq (37), HOMER (38), Hotspot (39) and its newer version Hotspot2, MOSAiCS (40), RSEG (26), BCP (41), DFilter (42) and MUSIC (43). Among these algorithms, Hotspot was designed for DNase-seq data (39), DFilter, HOMER, MACS2 and F-seq were designed for both DNase-seq and ChIP-seq data, and the remaining algorithms were designed only for ChIP-seq data. We sampled these methods as they were representative of the various known methodological and statistical approaches and could potentially work well on DNase-seq data and ChIP-seq data for the five histone marks that have been shown to be predictive of enhancers. Besides the aforementioned H3K4me1 and H3K27ac, three additional histone marks were analyzed: H3K4me2, enriched in promoters and some enhancers (1,44); H3K4me3, enriched in promoters and present at enhancers (6,45,46) and H3K9ac, enriched in actively transcribed promoters and active enhancers (47,48).
Using tissue-specific VISTA enhancers as the gold standard, we found DHSs and H3K27ac peaks to be more predictive of enhancers than the peaks of the other histone marks (H3K4me1, H3K4me2, H3K4me3 and H3K9ac). We found that DNase peaks called by DFilter and Hotspot2 outperformed those called by MACS2 and HOMER, and H3K27ac ChIP-seq peaks called by HOMER, MUSIC punctate, MACS2, F-seq and Dfilter outperformed the peaks by the other algorithms we tested. Furthermore, the DHSs and H3K27ac peaks called by these algorithms showed a high concordance between biological replicates and were most stable upon downsampling of reads to simulate a lower sequencing depth.
Motivated by the knowledge that enhancers are highly tissue-specific, we asked whether contrasting the DNase and H3K27ac signals between different tissues would lead to improved prediction performance. Indeed, we found that contrasting DNase and H3K27ac signals in one tissue against another tissue with a substantially different regulatory landscape (e.g. midbrain versus limb) led to a drastic improvement in predicting VISTA enhancers, especially for DNase peaks. For DFilter-called DNase peaks, the areas under the precision-recall curves (PR-AUC) were improved by 17.5–33.3% in four tissues and 166.7% in craniofacial prominence (abbreviated as face). The same approach also improved the performance of H3K27ac peaks, albeit to a lesser extent (7.1–22.2% improvement). We further explored using combinations of tissues as the background, and found that a panel of tissues, weighted by the distances between their regulatory landscapes and that of the tissue under investigation, led to the best performance.
We validated our differential-signal approach in two ways. First, we conducted a blind test of the prediction of mouse e12.5 enhancers from the VISTA database, finding that differential H3K27ac signal improved the PR-AUC from 0.48 to 0.75 for heart enhancers. Second, we evaluated our approach on a different benchmark, roughly 3,500 DNase hypersensitive sites tested using massively parallel reporter assays (MPRA) by ex vivo plasmid electroporation into the mouse retina and in vivo adeno-associated virus (AAV) injection into the mouse cerebral cortex (49). Differential DNase signal improved the PR-AUC of predicting retinal enhancers from 0.28 to 0.42, and a modest improvement was also observed for predicting cerebral cortex enhancers.
In summary, we have evaluated and enhanced the general approach of using DNase-seq or histone mark ChIP-seq data for identifying tissue-specific enhancers. We found that computing the differential signal in a given tissue against a panel of background tissues led to the best performance. Furthermore, because our refined approach is general, it can be applied to the tens of thousands of publicly-available DNase-seq and H3K27ac ChIP-seq datasets (49).
MATERIALS AND METHODS
Datasets
We used all histone mark ChIP-seq, DNase-seq and RNA-seq datasets generated by the ENCODE Consortium for e11.5 mouse tissues. We downloaded the alignment files (in BAM format) and gene expression tables of these datasets from the ENCODE Portal (http://encodeproject.org). The H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac ChIP-seq data were produced by the Ren lab for eight e11.5 mouse tissues (forebrain, midbrain, hindbrain, neural tube, craniofacial prominence, liver, limb, heart) and the DNase-seq data were produced by the Stamatoyannopoulos lab for five e11.5 mouse tissues (midbrain, hindbrain, neural tube, craniofacial prominence, and limb). We also downloaded the e12.5 H3K27ac ChIP-seq data in forebrain, heart and limb from the ENCODE portal to perform a blind test of our differential signal method (see the penultimate section of Methods). These data were also generated by the Ren lab. The ENCODE accession numbers for all above datasets are listed in Supplementary Table S1A. The craniofacial prominence tissue is abbreviated as face throughout this work. We used the mouse genome assembly mm10 for all analyses.
Genomic regions tested by transgenic mouse assays using e11.5 tissues were downloaded from the VISTA enhancer browser (35) on 6 November 2016 (https://enhancer.lbl.gov/). The database contained 2581 regions with coordinates in mouse genome assembly mm9 or human genome assembly hg19, among which 2556 regions could be lifted over to the mouse assembly mm10. These VISTA regions are listed in Supplementary Table S2. On 6 November 2016, there were no e12.5 enhancers at the VISTA browser. Subsequently, 150 regions were tested for their activities in e12.5 tissues (see the penultimate section of Methods) and we used these regions for a blind test of our methods.
Peak-calling algorithms
Among the large number of available peak callers, we sampled nine algorithms that are easy to use and are representative of the key methodological and statistical approaches (Supplementary Methods): MACS v2.1.0 (36) (called MACS2), F-Seq v1.8.5 (37), HOMER v4.7.2 (38), RSEG v0.4.9 (26), MOSAiCS v2.4.0 (40), BCP v1.1 (41), DFilter v1.5 (42), MUSIC (43) and Hotspot v5 (39). We applied the first eight algorithms for histone ChIP-seq data. Four of these algorithms can also be applied to DNase-seq data: DFilter, MACS2, HOMER and F-seq. However, F-Seq failed to run on several DNase-seq datasets, so we only evaluated the other three algorithms for DNase-seq data. In addition, we tested Hotspot v5 (39) and Hotspot2 for DNase-seq data. As recommended by the Hotspot developers, we evaluated the peaks called by Hotspot, which are all 151 bp long, and the DHSs called by Hotspot2 (these DHSs are also named hotspots in the program's outputs), which are ∼300 bp long. For simplicity, we refer to Hotspot peaks and Hotspot2 DHSs as DNase peaks henceforth.
We ran these algorithms using their recommended parameters for DNase-seq and histone mark ChIP-seq data respectively, and for specific histone marks if provided. The programs and their parameter settings are listed in Supplementary Tables S3. We did not tune the parameters, because Micsinai et al. performed extensive parameter tuning for a number of algorithms and concluded that the recommended parameters worked well for most of these algorithms (31). For MUSIC, we called ‘punctate peaks’ and ‘broad peaks’ and evaluated these two sets of peaks separately. For MACS2, we only evaluated narrow peaks as the P-values of its broad peaks and gapped peaks yielded poor performance. The peak lists were ranked as recommended by each algorithm: MACS2, Hotspot, and Hotspot2 peaks were ranked by P-value then signal, DFilter by P-value then max score, F-Seq by signal, MUSIC by P-value, HOMER by normalized tag count, MOSAiCS by average log P-value, BCP by posterior mean and RSEG by domain score.
Six of the algorithms (MACS2, BCP, F-Seq, HOMER, MUSIC and RSEG) required the average length of the sequenced DNA fragments as a parameter. All reads from the datasets were single-ended; we used the SPP algorithm (50) to estimate the average fragment length for each dataset using 5 million randomly-sampled reads.
Resizing peaks and excluding TSS-proximal peaks
The peaks called by different algorithms had different widths. To achieve a fair comparison of performance for predicting VISTA enhancers, motif enrichment analysis, and gene expression analysis (see below), we resized each peak in the final peak list produced by the uniform processing pipeline to a fixed length (2 kb for histone mark peaks and 300 bp for DHSs) centered on its summit. The summit of a peak is defined as the position in the original peak with the highest H3K27ac ChIP-seq or DNase-seq signal. Some of the originally called H3K27ac peaks or DNase might be shorter than 2 kb or 300 bp; thus, after resizing, they might overlap each other. We merged the overlapping peaks, averaged the coordinates of their summits, and resized again to 2k or 300 bp centered on the new summit. The merged and twice-resized peak was assigned the best rank of its constituent original peaks.
Because the VISTA enhancer team has intentionally avoided testing regions that are near annotated transcription start sites (TSSs), we excluded all H3K27ac and DNase peaks that were within 2 kb of a GENCODE-annotated TSS (Version M8) from our analysis henceforth, namely VISTA enhancer prediction, distance to the expressed genes and motif enrichment, as described in the remaining sections of Materials and Methods.
Metric for evaluating enhancer predictions
We evaluated how accurately a final, resized, and TSS-distal peak list called using a histone mark ChIP-seq or a DNase-seq dataset could predict enhancers, using the genomic regions downloaded from the VISTA database (34,51) as of Nov, 2016 as the gold standard. These VISTA regions had been experimentally tested for enhancer activity in e11.5 transgenic mouse assays; a VISTA region is deemed a VISTA enhancer for a particular tissue if the transgenic mice show reproducible reporter activity for this tissue in multiple mouse embryos, regardless of the results for the other tissues. For example, a VISTA enhancer that has reporter activity for both limb and heart is considered a positive enhancer for limb and heart but negative for all other tissues. Supplementary Table S2 shows the VISTA enhancers used in our study and the number of positives and negatives in each tissue.
We computed the area under Precision-Recall curve (PR-AUC) to evaluate how well the ranked lists of the final, resized, TSS-distal DNase or histone-mark peaks could predict VISTA enhancers. If a DNase or histone mark peak overlapped a VISTA positive (or negative) region by at least 1 bp, it was considered a true positive (or false positive) prediction. Precision is the fraction of true positives (i.e. DNase or histone mark peaks that were ranked above a certain threshold and overlapped a VISTA enhancer in a particular tissue) among a set of predicted regions (i.e. DNase or histone mark peaks that were ranked above the threshold and overlapped a VISTA region). Recall (equivalent to sensitivity) is the fraction of true positives out of all positives (all VISTA enhancers in a particular tissue). We calculated the PR-AUC using the auc function in the flux package in R. If a peak list overlaps all positive VISTA enhancers, its PR curve would extend all the way to the 100% recall. Note that for any PR curve, the precision at the 100% recall is simply the percentage of positives in the test set, independent of the performance of the predictions. Because different peak lists may overlap different numbers of positive VISTA enhancers in a given tissue, the PR curves of these peak lists may end at various locations before the 100% recall. For their PR-AUCs to be comparable, we include the last point (percentage of positives at 100% recall), which is common to all peak lists, in the PR-AUC calculation.
Enrichment of transcription factor binding
As shown in Results, the top 2000 DNase peaks called by DFilter did not overlap many of the top 2000 peaks of Hotspot2, MACS2 or HOMER, while the top peaks of the latter three algorithms overlapped substantially. We assessed whether these peaks were enriched in known sequence motifs of transcriptional factors. We first constructed two sets of peaks with equal numbers, one set called by DFilter only, and the other set by two or all three of the other algorithms but not by DFilter. We then searched for enriched motifs in these two sets of peaks using the findMotifsGenome.pl script in the HOMER suite of tools (38), which is independent of HOMER’s peak caller being evaluated in this study. We used the script to compute a P-value for each motif from a motif library provided by HOMER. We identified the motifs with a P-value lower than 0.01 in at least one peak set and then used a paired Wilcoxon signed-rank test to compare the -log10P-values of all such motifs between the two peak sets. Reciprocally, we performed de novo motif finding on the top 2000 DNase peaks shared by the Hotspot2, MACS2 and HOMER peak callers. To investigate how many of the top 2000 DNase peaks were bound by CTCF, we overlapped these peaks in e11.5 hindbrain and midbrain with postnatal 0 day CTCF peaks in hindbrain (ENCSR150RGT) and midbrain (ENCSR985ZTV) using the intersect function in BEDTools (52).
Using differential signal levels of DHSs and H3K27ac peaks between two tissue types to predict enhancers
We reranked the DNase peaks called by DFilter or Hotspot2 and the H3K27ac peaks called by HOMER by the differential signal in the tissue of interest (X) against another tissue (called the contrast tissue, C). We tested three metrics for evaluating the difference in signals: (i) signal (S) defined as control-normalized read counts computed by MACS2 (called ‘fold change over control’), averaged over all positions in a peak: S(X) – S(C); (ii) the signal defined as per (i) and further log transformed;: log(S(X)) – log (S(C)) and (iii) the signal defined as per (ii) and then transformed into a Z-score: Z(log(S(X))) – Z(log(S(C))). Note that the distribution of log-signal at peaks is approximately Gaussian, hence justifying the Z-score computation.
We also evaluated which contrast tissue could lead to the best enhancer prediction performance. First, for each set of peaks called in a tissue under investigation, we calculated the log of the read counts falling within the peaks for each contrast tissue. Then, to estimate the relative distance between each contrast tissue and the tissue under investigation, we applied principal component analysis (PCA) on all tissues based on the 500 peaks with the highest variances across tissues, using the ‘plotPCA’ function in Bioconductor package DESeq2 (53). The distance between two tissues is defined as the Euclidean distance between first two principal components (PCs). We used the top two PCs for most calculations, except for DNase data from 24 tissues where the top two PCs did not capture sufficient variation in the data; for these tissues, we used the top six PCs. The tissues were weighted proportionally to their Euclidean distances to the tissue under investigation, with the weights scaled to sum to 1. The datasets for all contrast tissues are listed in Supplementary Table S1B and C.
We performed the same motif enrichment analysis described in the previous section on the top 2000 final, resized, TSS-distal peaks (DFilter or Hotspot2 peaks for DNase and HOMER peaks for H3K27ac) and the top 2000 peaks ranked by differential signals, and tested whether these two sets of peaks differed in motif enrichment.
Differential expression analysis of the neighboring genes
To estimate the gene activation potential of DNase and H3K27ac peaks, we investigated the tissue specificity of neighboring genes among eight e11.5 tissues. First, we identified differentially expressed genes in each tissue compared with tissues of different origins. The eight e11.5 tissues fell into two groups—brain-related tissues (forebrain, midbrain, hindbrain and neural tubes) and non-brain-related tissues (face, limb, heart, and liver). We used DESeq2 (53) to perform differential expression analysis between each tissue and the four tissues in the other group. We then identified genes that were expressed at ≥1 FPKM in at least one of the eight e11.5 tissues and showed significant upregulation (adjusted P-value ≤ 0.05) in each tissue vs. the tissues in the other group. Finally, we selected the top 2000 promoter-distal peaks in each peak list and summarized the log2 fold change values of their upregulated neighboring genes. The neighboring genes of a DNase or an H3K27ac peak are defined as the genes with a GENCODE-annotated TSS that is located within 50 kb of the peak, identified using the window function in BEDTools (52). A Wilcoxon rank sum test was used to compare the upregulated neighboring genes’ log2 fold change levels between the top 2000 DFilter, Hotspot2 or HOMER peaks ranked by differential signals and the top 2000 peaks originally ranked by the corresponding algorithms.
A blind test of the differential H3K27ac method for predicting e12.5 enhancers
We tested the performance of our differential signal method formulated using e11.5 VISTA enhancers for predicting VISTA enhancers in the e12.5 forebrain, heart and limb. Note that when we downloaded the VISTA enhancers in November 2016, all of them were for e11.5, and e12.5 VISTA enhancers became available only later. First, ENCODE H3K27ac peaks were downloaded from the ENCODE Portal. The peaks were processed with the ENCODE uniform processing pipeline, which uses MACS2 as a peak caller and ranks peaks by P-value (signal is used to break ties). Second, we called peaks with the HOMER algorithm, using the same parameters as used for predicting e11.5 enhancers. Third, we ranked the HOMER peaks by contrasting H3K27ac signals in the e12.5 forebrain, heart or limb against the other 16 tissues (eight e11.5 tissues, the other seven e12.5 tissues, and postnatal day 0 or P0 intestine). We submitted predictions from our differential signal method to the ENCODE Portal on 21 March 2017 (accession numbers: ENCFF091DKM, ENCFF635WWK and ENCFF760OKN), before the Pennacchio lab released the experimental results at the VISTA database on 2 May 2017. The experimental method of transgenic mouse assays and results for these regions are described in a preprint (BioRxiv: https://doi.org/10.1101/166652). These e12.5 VISTA regions are provided in Supplementary Table S4.
Prediction of MPRA CRE-seq enhancers in retina and cerebral cortex
We further tested the performance of our differential signal method for predicting enhancers identified by massively parallel reporter assays (MPRA) in postnatal day 0 retina and adult cerebral cortex (54). We selected ‘high expression’ constructs as positive enhancer regions and ‘not high expression’ constructs as negative regions, as defined by (54). Overlapping constructs were merged to form a contiguous region, yielding 320 positive and 3019 negative regions in the retina, and 110 positive and 3272 negative regions in the cerebral cortex (Supplementary Table S5). We then downloaded DNase-seq datasets in retina postnatal day 7 and forebrain postnatal day 0 (ENCODE accessions: ENCSR000CNU and ENCSR791AJY), and H3K27ac ChIP-seq datasets in forebrain postnatal day 0 (ENCODE accession: ENCSR094TTT) from the ENCODE portal. We re-ranked DNase peaks by contrasting the retina or forebrain DNase signals against 23 others tissues (Supplementary Table S1B), and we re-ranked the H3K27ac peaks by contrasting the H3K27ac signals in forebrain against 17 other tissues (Supplementary Table S1C) prior to prediction.
RESULTS
Comparison of peak callers
We compared nine peak callers: DFilter, HOMER, Hotspot/Hotspot2 and MACS2 for DNase-seq data; and BCP, DFilter, F-seq, HOMER, MACS2, MOSAiCS, MUSIC and RSEG for histone mark ChIP-seq data (see Supplementary Methods for a brief description of the key features of these algorithms and our rationale of choosing them). We first compared the numbers and lengths of the peaks called by these algorithms on the same DNase-seq or histone-mark ChIP-seq data (Supplementary Results; Supplementary Figure S1A and B; Supplementary Table S6). We further assessed the consistency of the peaks after halving or quartering sequencing depth (Supplementary Results; Supplementary Figures S1C, D, S2 and S3). We found the DNase-seq algorithms to differ in the peaks they call, consistency between biological replicates, and robustness against downsampling, and several H3K27ac algorithms achieved high consistency and robustness. We implemented the ENCODE uniform processing pipeline (Supplementary Figure S4) to filter out peaks that were not consistently called between biological replicates (Supplementary Results; Supplementary Figure S5–S10; Supplementary Table S7). After comprehensive comparison of the peak callers, we decided to perform subsequent analyses using the final, resized DNase and histone-mark peaks produced by the uniform processing pipeline with each peak caller.
The performance of DNase and histone mark peaks for predicting VISTA enhancers
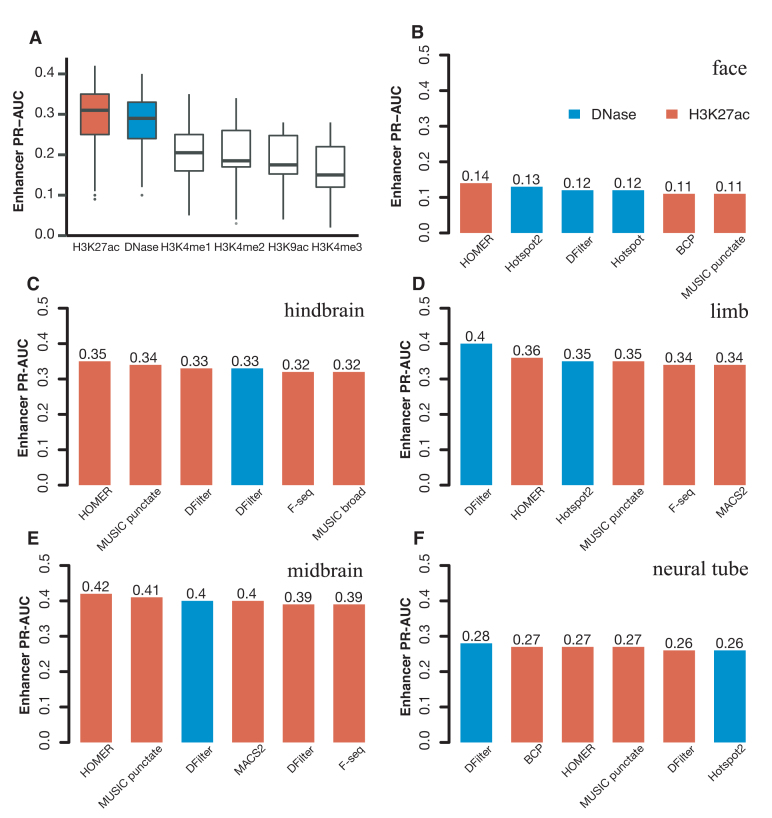
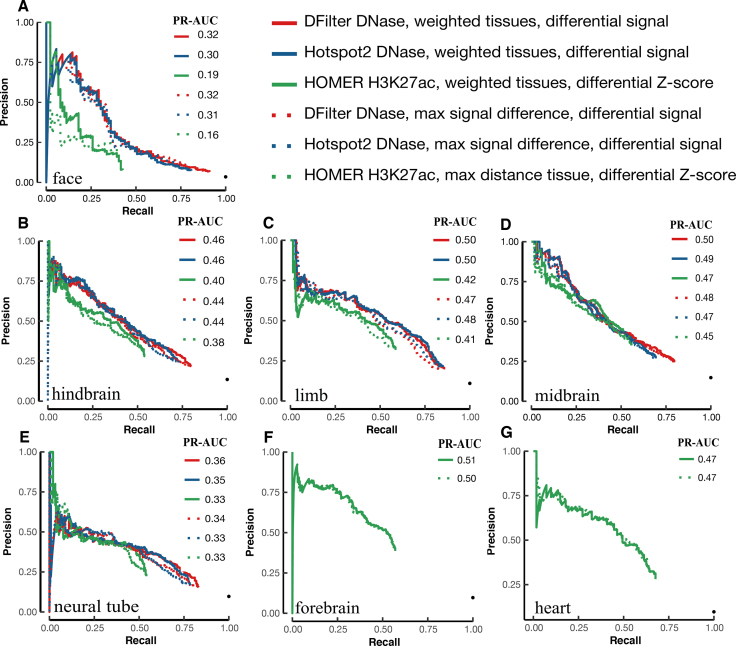
We compared the performance of the final, resized, TSS-distal peaks of DNase and histone marks for predicting VISTA enhancers (Methods). We used five tissues for testing both DNase and histone mark data and two additional tissues for testing histone mark data alone—there were no DNase-seq data for the e11.5 forebrain or heart. Despite having histone mark ChIP-seq data, liver has only eight positive VISTA enhancers, so we left it out of the PR-AUC evaluation. Figure 1A shows that H3K27ac and DNase achieved substantially higher PR-AUCs than the other four histone marks: the median PR-AUCs across five tissues were 0.35, 0.33, 0.25, 0.20, 0.19, 0.17 for H3K27ac, DNase, H3K4me1, H3K4me2, H3K9ac and H3K4me3 respectively when the best algorithm for each tissue was used. H3K27ac also outperformed the other four histone marks by similar margins in forebrain and heart (Supplementary Figures S11). Because our goal was to search for the best method for predicting enhancers, we focused on DNase and H3K27ac for the rest of this study. For completeness, the PR-AUCs for individual algorithms on DNase and all five histone marks are provided in Supplementary Tables S8).
Figure 1.
The performance of H3K27ac and DNase-seq peaks for predicting VISTA enhancers in the corresponding tissue, assessed by PR-AUC values. Final, resized, TSS-distal peaks were used for comparing the peak calling algorithms. (A) Boxplots of the comparison of DNase peaks (four algorithms in five tissues) with the peaks of five histone marks (eight algorithms in the same five tissues). (B–F) Barplots of the top six methods for DNase peaks (blue) and H3K27ac peaks (red) in (B) face, (C) hindbrain, (D) limb, (E) midbrain and (F) neural tube.
Among the five tissues with both DNase and H3K27ac data, DNase performed better for limb and neural tube, while H3K27ac performed better for face, hindbrain, and midbrain (Figure 1B–F for the top six algorithms and Supplementary Tables S8A-B for all algorithms). Note that the performance of the top algorithms differed among the tissues because of varying numbers of VISTA enhancers in these tissues: 3.5% VISTA regions were active in face, 13.5% in hindbrain, 11.0% in limb, 15.0% in midbrain and 9.7% in neural tube, and these fractions are highly correlated with the average PR-AUC of the top six methods in each tissue (correlation coefficient R2 = 0.92). Nevertheless, the performance of different algorithms in the same tissue can be directly compared. For DNase, DFilter was the most predictive, with Hotspot2 being a close second (mean PR-AUCs across five tissues = 0.31 and 0.29 respectively). Hotspot2 outperformed Hotspot for all five tissues. Among the histone mark peak callers, HOMER achieved the highest PR-AUC, followed by MUSIC-punctate, MACS2, F-seq and DFilter (mean PR-AUCs across seven tissues = 0.34, 0.33, 0.31, 0.31 and 0.31 respectively). For MUSIC, the punctate mode outperformed the broad mode for all seven tissues. Although the absolute differences of PR-AUC among the algorithms were small, the relative rankings of the algorithms were consistent across the tissues.
The top DNase peaks called by DFilter were mostly distinct from the DNase peaks called by Hotspot2, MACS2 and HOMER
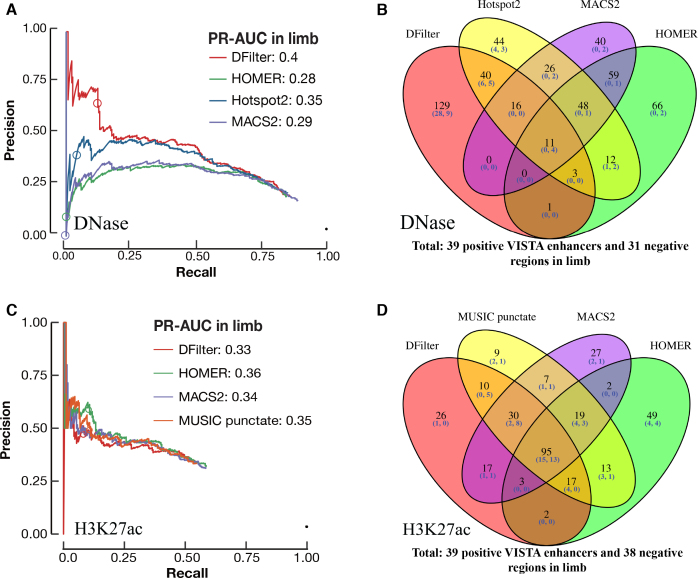
For limb DNase-seq data, DFilter peaks achieved a higher PR-AUC (0.40) than the other peak callers. The top DFilter peaks had a much higher precision (65% for the top 200 peaks) than the top peaks of the other algorithms (39% for Hotspot2, the next best algorithm; Figure 2A). We omit Hotspot henceforth because Hotspot2 was nearly always superior. The PR curve for DFilter lay above the curves of the other algorithms throughout the entire range of recall even though the differences became smaller when more predictions were included. The top-ranked DNase peaks by DFilter also achieved a higher precision than the top-ranking H3K27ac peaks called by HOMER (Figure 2A versus C).
Figure 2.
Identifying VISTA enhancers in limb using DNase-seq and H3K27ac peaks. Final, resized, TSS-distal peaks were used for comparison. The circle in each PR curve indicates the 200th prediction. (A) PR curves for DNase peak callers. PR-AUC values are indicated in the legend. (B) Venn diagram of the top 200 peaks by each DNase peak callers. Among the top 200 peaks called by DFilter, 129 were not called by the other three algorithms. (C) PR curves for four H3K27ac peak callers. PR-AUC values are indicated in the legend. (D) Venn diagram of the top 200 peaks by each of the four H3K27ac peak callers.
We asked how much the top predictions of the four algorithms differed. Figure 2B shows the overlap among the top 200 peaks of the four algorithms in limb—only 11 peaks were ranked in the top 200 by all four algorithms; of these, four overlapped with VISTA regions but none of the four were active in limb. Among DFilter's top 200 peaks, 129 were unique to DFilter; among these, 37 overlapped VISTA regions and 28 were positive in limb (75.7% precision). The other three algorithms called fewer unique peaks (44, 40, and 66 for Hotspot2, MACS2, and HOMER), and few of their unique peaks overlapped with VISTA regions (7, 2 and 2). Those that did overlap achieved lower precisions than DFilter-only peaks (57%, 0% and 0%).
We manually inspected the top 20 peaks called by each algorithm in limb with the UCSC genome browser (Supplementary Figures S12–S15) to identify features that might differentiate the algorithms. Of the top 20 DFilter peaks, nine (DFilter ranks 7–12, 16 and 18–19) were not ranked in the top 200 by any of the other algorithms. Among these, four peaks (7th, 9th, 10th and 12th) overlapped positive VISTA enhancers and only one peak (18th) overlapped negative VISTA regions in limb. One common feature shared among the top-ranked DFilter peaks was their broad and multimodal signals, suggesting the presence of multiple evicted nucleosomes at these open chromatin regions. The DNase signal climaxes at the trough of the surrounding H3K27ac signal, which indicate the positions of the remaining nucleosomes (Supplementary Figure S12). In contrast, most of the top 20 peaks called by the other three algorithms showed sharp and unimodal signals, suggesting a single evicted nucleosome (Supplementary Figures S13–S15). Most of these unimodal, high-signal peaks were ranked poorly by DFilter.
For midbrain, the H3K27ac peaks outperformed the best set of DNase peaks (Figure 1D); nevertheless, DFilter remained the best DNase peak caller (Supplementary Figure S16A). Among the top 200 peaks called by DFilter in midbrain, 120 were not shared by the other algorithms; 23 of these peaks overlapped VISTA regions and 16 were tested positive (precision = 69.6%; Supplementary Figure S16B). The PR curves and the four-way Venn diagrams for the other three tissues are shown in Supplementary Figures S17 and S18.
We also compared the top 2000 DNase peaks called by the four algorithms in each tissue and observed similar results as described above: DFilter peaks were unique (Supplementary Figure S19) and tended to be broad and multimodal (UCSC genome browser shots not shown).
DFilter-only and shared DNase peaks differ in TF motifs and TF binding
To provide more quantitative comparison between the DFilter-only peaks and the peaks shared by the other three algorithms, we evaluated the motif enrichment of these two sets of peaks (see Materials and Methods). In all five tissues, the DFilter-only peaks were significantly more enriched in known transcription factor motifs than the peaks shared by the other three algorithms but not called by DFilter (Supplementary Figure S20). In particular, two of the most enriched motifs correspond to the Pitx1 transcription factor regulates genes involved in limb morphogenesis (55) and the Sox family of transcription factors has been assigned important roles in the specification and differentiation of the neuronal lineage (56). The Pitx1 motif was more enriched in the DFilter-only peaks than in the peaks shared by the other three algorithms in limb and face, while the Sox3 motif was more enriched in the DFilter-only peaks than in the shared peaks in midbrain, hindbrain and neural tube (Supplementary Figure S20, Supplementary Table S9).
For comparison, we performed de novo motif discovery on the top 2000 peaks shared by Hotspot2, MACS2, and HOMER but not by DFilter. The three most enriched motifs were all related to the insulator binding protein CTCF (57) in all five tissues, while these CTCF motifs were much less enriched among DFilter-only peaks (Supplementary Figure S20, Supplementary Table S9).
We wanted to confirm the results of our motif analysis using ChIP-seq data of transcription factors in the same tissues, but we only found CTCF ChIP-seq data on P0 midbrain and hindbrain tissues (Materials and Methods). Thus we compared the top 2000 DNase peaks with the CTCF ChIP-seq peaks in the corresponding tissues, and found that 91% and 95% of the DNase peaks shared by Hotspot2, MACS2 and HOMER (among the top 2000 peaks called by each algorithm) overlapped CTCF ChIP-seq peaks in midbrain and hindbrain, respectively, while much smaller overlaps were observed for DFilter-only peaks (28% and 31%).
In conclusion, DFilter preferentially identifies broad DNase peaks with multiple evicted nucleosomes, which are likely bound by tissue-specific transcription factors. In comparison, Hotspot2, MACS2 and HOMER preferentially identify narrow DNase peaks with a single evicted nucleosome, which are bound by CTCF. These CTCF-bound DNase peaks shared by the three algorithms are likely to function as insulators or regulate chromatin structures during development.
The top H3K27ac peaks called by the best-performing algorithms mostly overlap
Averaged over seven tissues, HOMER achieved 1–3% higher PR-AUCs than the four next best H3K27ac algorithms, MUSIC-punctate, MACS2, DFilter, and F-seq (Figure 1, Supplementary Table S8B). We compared the top peaks by HOMER, MUSIC-punctate, MACS2 and DFilter but omitted F-seq for further comparison because, as mentioned above, F-seq called many peaks and the low-ranked F-seq peaks were not reproducible between biological replicates. Figure 2C and Supplementary Figure S16C show the PR curves of these algorithms for limb and midbrain. In contrast to the DNase results, the four H3K27ac algorithms shared many of their top 200 peaks—95 and 102 of these peaks were called by all four algorithms for limb and midbrain, respectively (Figure 2D and Supplementary Figure S16D). The H3K27ac peaks called by all four algorithms overlapped 28 and 33 VISTA regions and achieved 53.6% and 66.7% precision, respectively, higher than the DNase peaks called by all four algorithms. The results for other tissues are in Supplementary Figures S21–S22 and the results for the top 2,000 peaks in all tissues are in Supplementary Figure S23. The conclusions remain the same—the top H3K27ac peaks called by the top algorithms were largely shared, and these shared peaks achieved high precisions in predicting VISTA enhancers in the respective tissues. We tested whether using only these H3K27ac peaks called by all four algorithms could lead to a better performance, but it did not improve the average PR-AUC (0.32) beyond that of HOMER (0.34).
Contrasting the DNase or the H3K27ac signal in one tissue against a distant tissue substantially improved the performance of enhancer prediction
Thus far, we have used DNase peaks or H3K27ac peaks called from a particular tissue for predicting enhancers in the same tissue. Given the tissue specificity of many enhancers, we hypothesized that peaks having significantly higher signal in a target tissue as compared to a contrast tissue were more likely to be active enhancers in the target tissue. Thus, we next explored whether we could improve the accuracy of enhancer prediction using differential signals between different tissues. For DNase, we focused on peaks called by DFilter and Hotspot2 because they performed well individually but did not share many of their top peaks. For H3K27ac peaks, we focused on those called by HOMER because, as described in the previous section, four H3K27ac algorithms called highly similar peaks and among them, HOMER had the highest average PR-AUC.
Our differential signal approach was first tested in neural tube. We called DNase peaks in neural tube and then reranked these peaks according to the differential DNase signal between neural tube and a contrast tissue. We tested three metrics for computing differential DNase signals: sequencing-depth normalized signal level (abbreviated as ‘signal’ in figure legends); sequencing-depth normalized signal level followed by log transformation (abbreviated as ‘log’); and sequencing-depth normalized signal level followed by Z-score transformation (abbreviated as ‘Z-score’). We used each of the other four tissues with DNase data as the contrast tissue and obtained different results: using face or limb increased the PR-AUC, while using hindbrain or midbrain decreased the PR-AUC. Thus, we concluded that the contrast tissue needs to be ‘distant’ from the tissue under investigation in order for the differential signal to be predictive of enhancers.
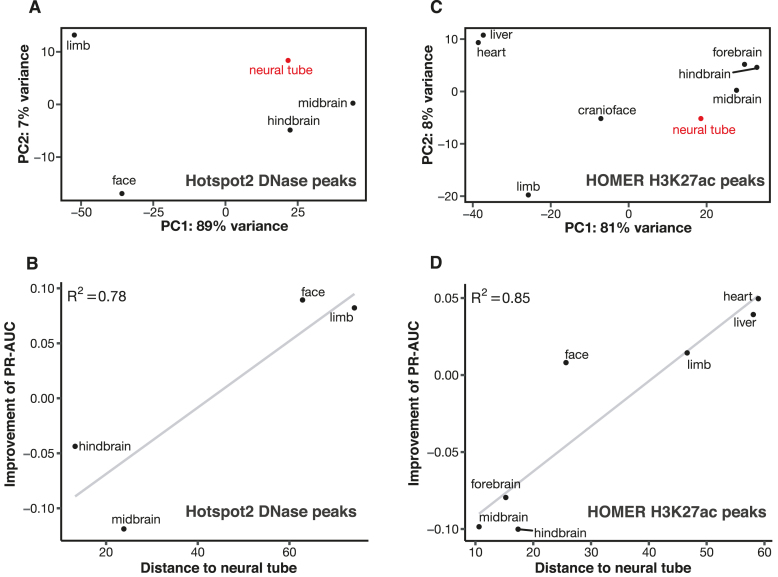
To formulate a method for quantifying the distance between two tissues, we performed principal component analysis (PCA) on the DNase signal profiles of the five tissues computed on the neural tube DNase peaks (Figure 3A). All ChIP-seq data used in this study were generated by the same ENCODE production center (led by Bing Ren) and all DNase-seq data were generated in the same production center (led by John Stamatoyannopoulos; Materials and Methods), and we did not observe batch effects upon the PCA. The first two principal components accounted for 96% of the variance in the data; thus, we defined the distance using the Euclidean distance defined by the first two principal components (i.e. the straight-line distance between the dot for each tissue and the dot for neural tube in Figure 3A). We observed a high Pearson correlation (R2 = 0.78) between the change in PR-AUC for predicting enhancers and the distance between the two tissues (Figure 3B). Face and limb are distant from neural tube, and using either one of them as the contrast tissue for neural tube led to an increase in PR-AUC of 0.09. On the other hand, midbrain and hindbrain are close to neural tube; using either one of them as the contrast tissue led to a decrease in PR-AUC of 0.04 or 0.12, respectively. Similar results were observed for predicting enhancers in the other four tissues using DFilter or Hotspot2 peaks (Supplementary Figures S24 and S25). Among the three metrics for computing differential DNase signals, the signal metric worked the best (Supplementary Figure S26).
Figure 3.
Enhancer prediction accuracy is positively correlated with the distance between the contrast tissue and the tissue under investigation. (A) Principal component analysis (PCA) for DNase signal profiles in DNase peaks called by Hotspot2 in neural tube. The distance between two tissues in the scatter plot is computed using the first two principal components (PCs) of the signal profiles in these two tissues. Neural tube—the tissue under investigation—is in red. (B) Correlation between the distance of the contrast tissue and the improvement of enhancer PR-AUC for the Hotspot2 DNase peaks in neural tube. (C) PCA for the signal profiles of the HOMER H3K27ac peaks in neural tube. (D) Correlation between the distance of the contrast tissue and the improvement of enhancer PR-AUC for the HOMER H3K27ac peaks in neural tube.
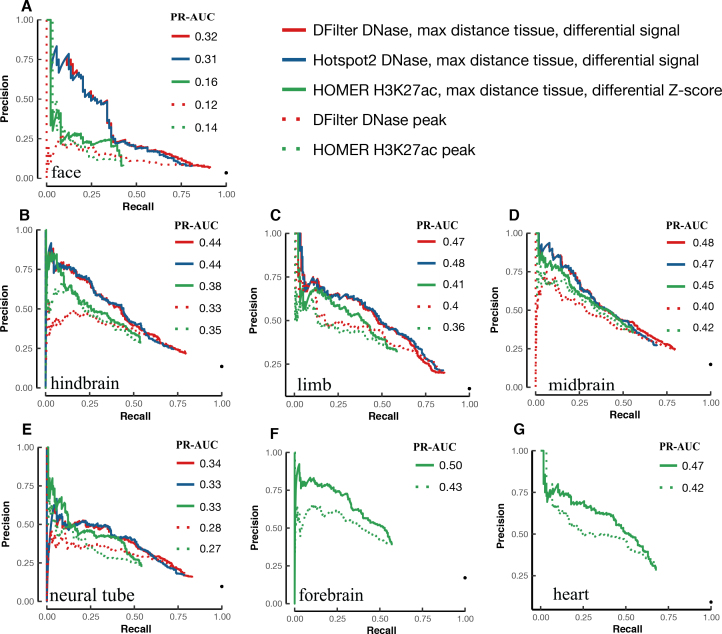
Figure 4 shows the PR curves of the DNase peaks called by DFilter or Hotspot2 (solid red and blue lines, respectively), using the difference in DNase signals between the tissue under investigation and the most distant tissue. It is apparent that using differential DNase signals substantially improved PR-AUCs over using DNase signal from the tissue under investigation alone (dotted red line). The improvement is large for all five tissues (0.06–0.20) and most dramatic for face, which improved from a PR-AUC of 0.12 to 0.32 for DFilter peaks (a 167% increase). The improvements arise mostly from increased precision in the top-ranked peaks—the peaks that have the greatest impact on guiding experimental testing. For example, top DFilter peaks as ranked by DNase signal in face alone had <25% precision when overlapped with VISTA regions (i.e. <25% of these peaks were active in face according to VISTA), but the top ranked DFilter peaks according to the differential DNase signal between face and midbrain achieved 75% precision (comparing the solid red line with the dashed red line in Figure 4A).
Figure 4.
Improvement in VISTA enhancer prediction achieved by differential DNase or H3K27ac signals between two distant tissues. The PR curves of the DFilter or Hotspot2 DNase peaks and the HOMER H3K27ac peaks, ranked by differential signals (solid lines) or original algorithms (dotted lines), were evaluated using VISTA enhancers in (A) face, (B) hindbrain, (C) limb, (D) midbrain, (E) neural tube, (F) forebrain and (G) heart. PR-AUC values are indicated in the legend of each panel.
The similarity of the PR curves in Figure 4 for DFilter and Hotspot2 may seem surprising because, as noted previously, the top 2000 peaks called and ranked by these algorithms were largely non-overlapping. Indeed, only 32.7–54.2% of the top 2000 original DFilter and Hotspot2 peaks overlap but 91.4–96.9% of the top 2000 reranked DFilter and Hotspot2 peaks overlap (Supplementary Figure S27). These results highlight that DFilter and Hotspot2 call a similar overall set of peaks, but the two algorithms rank their peaks quite differently.
We performed the same analysis using differential H3K27ac signals between the eight tissues with H3K27ac ChIP-seq data. We included liver as a possible contrast tissue but did not compute its PR-AUC because, as mentioned above, there are only eight validated VISTA enhancers in liver. Of our three metrics, differential Z-scores of H3K27ac signals most precisely predicted enhancers (Supplementary Figure S28). Figure 3C shows the projection of the eight tissues in the first two principal components, revealing that hindbrain, midbrain and forebrain are the three closest tissues to neural tube, while heart and liver are the two most distant. As for DNase, we observed a strong correlation between the change in PR-AUC and the distance between the two tissues (Figure 3D). Using heart or liver as the contrast tissue for neural tube led to a 0.04–0.05 improvement in PR-AUC, while using any of the three brain regions resulted in a 0.08–0.10 decrease in PR-AUC. Similar results were observed for using differential H3K27ac signals to predict enhancers in the other six tissues (Supplementary Figure S29). The tissue distances computed using H3K27ac signals at H3K27ac peaks are strongly correlated with those computed using DNase signals at DFilter DNase peaks (R2 = 0.80–0.96; Supplementary Figure S30) or DNase signals at Hotspot2 DNase peaks (R2 = 0.79–0.98; Supplementary Figure S31), indicating that our approach to computing differential signals is generally applicable.
Figure 4 also includes the PR curves for the HOMER H3K27ac peaks ranked according to the differential H3K27ac Z-scores between a tissue under investigation and its most distant tissue (solid green lines). These PR curves exhibit improvement over the H3K27ac peaks ranked by HOMER in the tissue under investigation alone (dotted green lines), with increases in PR-AUC from 0.02 to 0.07 seen across seven tissues. The improvement was largest for forebrain and across the entire range of recall.
Contrasting DNase signals or H3K27ac signals in one tissue against a group of tissues weighted by their distances led to further improvements
For the results shown in the previous section, contrasting against the most distant tissue typically produced the largest increase in performance, although there were a few exceptions. For example, for Hotspot2 peaks called in limb, midbrain was the most distant tissue from limb, but the second most distant tissue—neural tube—led to the greatest improvement (a 0.02 PR-AUC difference; Supplementary Figure S25C). Thus, we explored the approach of contrasting the target tissue against a large group of tissues, weighting each contrast tissue by its distance to the tissue under investigation. We tested this approach using the existing five tissues for DNase and eight tissues for H3K27ac. We observed slight improvements for H3K27ac, but slightly worse results for DNase, suggesting this approach may require a sufficiently large number of tissues to be effective.
To expand our tissue coverage, we took all 24 DNase datasets on mouse tissues from ENCODE that were mapped to the mm10-minimal mouse genome build at the time of our study. For H3K27ac, we included eight tissues at e11.5 and e12.5. We also added the intestine from P0, which we had previously found to be the most distant from all other tissues by hierarchical clustering of gene expression profiles. These datasets are listed in Supplementary Tables S1B-C.
We performed PCA and calculated Euclidean distances as described in the previous section. We then computed differential DNase signals between the tissue under investigation and the weighted sum of the DNase signals in the other 23 tissues, with the weights proportional to the distances and summing to one. For H3K27ac, we calculated the difference in the Z-scores between the tissue under investigation and the weighted sum of the H3K27ac Z-scores in the other 16 tissues. We continued to observe a positive correlation between the improvement in PR-AUC and the distances from the individual contrast tissues (Supplementary Figures S32–S34). Figure 5 compares the results using the larger group of weighted tissues (solid lines) with the results using the most distant tissue among the smaller group of tissues (dotted lines, which correspond to the solid lines in Figure 4). Using weighted tissues over the single most distant tissue led to 0.02–0.03 increases in PR-AUC for DNase peaks (for both DFilter peaks and Hotspot2 peaks) in four out of five tissues, with face being the exception, showing a decrease of 0.01 for Hotspot2 peaks. For the H3K27ac peaks called by HOMER, a slight increase in PR-AUC was observed in five tissues (0.01–0.03) while neural tube and heart showed no change. In summary, for the larger groups of tissues (24 for DNase and 17 for H3K27ac), using weighted tissues was consistently, albeit slightly, better than using the most distant tissue alone (Supplementary Figure S35). Despite its large distance from all other tissues, the addition of the P0 intestine did not significantly change our results (data not shown).
Figure 5.
Additional improvement in enhancer prediction achieved by contrasting against a group of distance-weighted tissues. We used 24 tissues for DNase and 17 tissues for H3K27ac, weighted proportionally to their distances from the tissue under investigation. The PR curves using weighted tissues are drawn with solid lines, and the PR curves using the most distant tissue alone are drawn in dotted lines of the corresponding colors for comparison. VISTA enhancers in the tissue of interest were used for evaluating the precision and recall in (A) face, (B) hindbrain, (C) limb, (D) midbrain, (E) neural tube, (F) forebrain and (G) heart. PR-AUC values are indicated in the legend in each panel.
To further investigate the difference in performance between signal and differential signal, we compared the ranks for the top 2000 peaks as determined by these two metrics (Supplementary Figures S36-S38). Differential signal assigns poor ranks to many of the top 2000 peaks as ranked by the original algorithms, more so for DNase peaks than for H3K27ac peaks (compare Supplementary Figures S36–S37 with S38). Accordingly, the differential signal approach led to a greater improvement in enhancer prediction for DNase peaks (especially Hotspot2 peaks) than for H3K27ac peaks. This difference may also be attributable to the greater abundance of called DNase peaks versus H3K27ac peaks (17 fold more on average), which results in many of the called DNase peaks being non-enhancer regulatory elements or false positives. Indeed, when we examined the top 2000 DNase peaks by DFilter or Hotspot2, 11.5–51.3% and 23.4–70.3% of them were ranked poorly (beyond 20k) by the differential signal metric, suggesting many of these might be false positives. In contrast, only 13.8–27.6% of the top 2000 H3K27ac peaks were ranked beyond 5k, indicating a smaller fraction of false positives.
We also performed motif enrichment analysis (see Methods) to compare the top 2000 peaks called and ranked by each algorithm (DFilter or Hotspot2 for DNase and HOMER for H3K27ac) with the top 2000 peaks called by the corresponding algorithm but reranked by differential signals. The reranked Hotspot2 DNase peaks were significantly more enriched in known transcription factor motifs than the peaks originally ranked by Hotspot2 in all five tissues (Supplementary Figure S40), and for four out of five tissues when considering DFilter peaks (except neural tube; Supplementary Figure S39). Reranked HOMER H3K27ac peaks were significantly more enriched in motifs than the peaks originally ranked by HOMER in all seven tissues (Supplementary Figure S41). In particular, we observed a higher enrichment for the Pitx1 motif in the reranked DNase and H3K27ac peaks than in the original peaks in limb and face and a higher enrichment for the Sox3 motif in the reranked DNase and H3K27ac peaks than in the corresponding original peaks in most of the brain tissues (Supplementary Figure S39–S41), consistent with the known function of Pitx1 in limb development and Sox3 in neural development (55,56).
DNase and H3K27ac peaks with the highest differential signals are near differentially expressed genes
We asked whether the top TSS-distal DNase or H3K27ac peaks (≥2 kb away from any GENCODE-annotated TSS) were near genes specifically expressed in the same tissue, hypothesizing that the peaks near tissue-specific genes were more likely bona fide tissue-specific enhancers. We analyzed the top 2000 DNase peaks (called by DFilter or Hotspot2) and the top 2000 H3K27ac peaks (called by HOMER), ranked by the default approach of the peak callers or by differential signals against weighted tissues as described in the previous section. To quantify tissue-specific genes, we used RNA-seq data to compute differential expression (quantified as log fold change), retaining only the genes with positive log fold changes in the corresponding tissue against contrast tissues (see Materials and Methods). If the tissue under investigation was brain-related (forebrain, midbrain, hindbrain or neural tube), we computed differential expression against the other four non-brain tissues (face, limb, heart, and liver). Conversely, if the tissue under investigation was not brain-related, we instead computed differential expression against the four brain tissues. We identified 1747 upregulated genes in face, 2068 in limb, 988 in midbrain, 1071 in hindbrain, 988 in neural tube with the adjusted P-value ≤ 0.05. Supplementary Figure S42A illustrates that the neighboring genes of the top 2000 DFilter peaks identified using differential DNase signals were more differentially expressed in the corresponding tissue than the neighboring genes of the top 2000 DFilter peaks identified by the default metric provided by the DFilter algorithm, and the differences were significant in two out of five tissues (P-value < 0.05). The differences were significant in three tissues for Hotspot2 DNase peaks and significant in all seven tissues for HOMER H3K27ac peaks (Supplementary Figure S42B and C).
Differential H3K27ac signals led to substantial improvement in e12.5 heart enhancer prediction
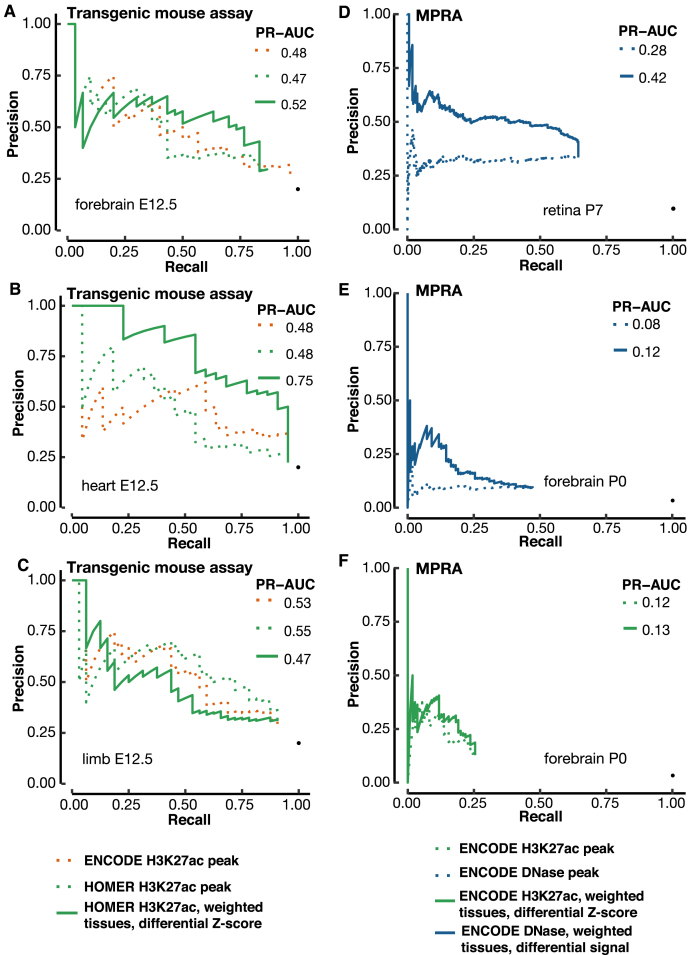
As part of the ENCODE consortium, the Pennacchio lab used transgenic mouse assays to test 150 regions of the mouse genome for enhancer activity, identified from 50 H3K27ac peaks each from the e12.5 forebrain, heart, and limb tissues. Due to the nature of transgenic mouse assays, the enhancer activities of these regions were tested in all mouse tissues, not just the tissues in which the peaks were identified. Predictions were solicited to rank these regions before the release of the experimental data, i.e. this was a blind test of the computational methods. We ranked the 150 regions for each tissue using differential H3K27ac signals and submitted the predictions to the ENCODE Portal on 21 March 2017, 1.5 months before the Pennacchio lab released the experimental results at the VISTA database (Supplementary Table S4). The transgenic mouse data on these 150 regions are described in a preprint (BioRxiv: https://doi.org/10.1101/166652).
We compared the performance of three ranking methods (See Methods): H3K27ac peak ranks downloaded from the ENCODE Portal, which were called and ranked using MACS2, H3K27ac peaks called and ranked by HOMER, and H3K27ac peaks called using HOMER but ranked by contrasting H3K27ac signals in the e12.5 forebrain, heart, or limb against the other 16 tissues as described above. We did not test the approach using differential DNase signals because no DNase-seq data were available for these three tissues at e12.5. The PR-AUC values for the three methods are comparable for forebrain and limb, but the differential H3K27ac method is substantially better for heart (PR-AUC = 0.75, while ENCODE and HOMER tied at 0.48), with the improvement spanning the entire range of recall (Figure 6B).
Figure 6.
Validation of our differential signal approaches with a blind test and MPRA enhancers. (A–C) A blind test of the differential H3K27ac signal method for predicting e12.5 transgenic mouse enhancers in (A) forebrain, (B) heart and (C) limb. We used 17 contrast tissues as in Figure 5, weighted proportionally to their distances from the tissue under investigation. VISTA enhancers in the tissue of interest were used for evaluating the precision and recall. The predictions were submitted to the ENCODE Portal before the release of experimental results, and hence were a blind test. PR-AUC values are indicated in the legend in each panel. (D, E) Using the differential DNase signal for predicting MPRA CRE-seq enhancers in retina (D) and (E) cerebral cortex. (F) Using the differential H3K27ac signal for predicting MPRA CRE-seq enhancers in cerebral cortex. We used the same set of contrast tissues as in Figure 5, weighted proportionally to their distances from the tissue under investigation.
We were surprised that the differential H3K27ac method did not outperform the other two methods for the e12.5 limb enhancers, in contrast with the results for e11.5 enhancers (Figures 4 and 5). One reason for this is that few of the e12.5 limb enhancers are limb-specific, whereas many of the e11.5 limb enhancers are. Among the 150 regions, 32 are active in the e12.5 limb, but only two of these regions (6.25%) are exclusively active in limb at this stage. In sharp contrast, 146 of the 280 (52.14%) e11.5 limb enhancers are exclusively active in limb (Chi-square P-value = 2.15E–6). The difference between e12.5 and e11.5 is smaller for forebrain and heart. Seven of the 30 (23.3%) e12.5 forebrain enhancers are exclusively active in forebrain, compared with 146 of the 437 (33.4%) e11.5 forebrain enhancers (P-value = 0.35). Twelve of the 22 (54.5%) e12.5 heart enhancers are exclusively active in heart, compared with 187 of the 235 (79.7%) e11.5 heart enhancers (P-value = 0.016).
To investigate whether the differential H3K27ac approach is particularly effective in identifying tissue-specific enhancers, we examined the 280 e11.5 limb enhancers in greater detail. We separated these 280 enhancers into two groups: 146 (52.14%) e11.5 limb enhancers that are exclusively active in limb, and the remaining 134 enhancers active in limb and another e11.5 tissue. We computed PR-AUC separately for these two groups of enhancers against the 2276 VISTA regions that are inactive in the limb. Note that the baseline performance as measured by PR-AUC is the percentage of positives in the test set, which would be 146/2,276 or 0.064 for limb-specific enhancers and 0.059 for the enhancers that are active in limb and another tissue. Compared with this baseline performance, HOMER H2K27ac achieved PR-AUC of 0.22 and 0.24 respectively, while the weighted H3K27ac Z-score achieved PR-AUC of 0.30 and 0.27. Indeed, the differential H3K27ac approach is more effective in predicting limb-specific enhancers than enhancers active in limb and other tissues (PR-AUC improved by 0.08 and 0.03 respectively); yet, the differential H3K27ac approach shows an improvement for both groups of enhancers. We observed similar results for the differential DNase approach and in other tissues.
Further validation with MPRA CRE-seq enhancers
So far, our performance evaluations have been based on VISTA regions. We further tested the differential signal approach using a completely different gold standard—enhancers identified by massively parallel reporter (MPRA) CRE-seq assays performed on mouse P0 retina and adult cerebral cortex (54). We used the DNase peaks of P7 retina and the DNase peaks and H3K27ac peaks of P0 forebrain, the best matching biosamples with ENCODE data. For retina DNase peaks, ranking by differential signal showed a significant improvement over ranking by the original algorithm, with PR-AUC increasing from 0.28 to 0.42 (Figure 6D). Forebrain DNase peaks and H3K27ac peaks also showed modest improvements, with PR-AUCs increasing from 0.08 to 0.12 and 0.12 to 0.13 respectively (Figure 6E and F). Overall, these PR-AUC values are lower than those for VISTA enhancers, partially because of the lack of a perfect match in developmental time points between the biosamples with epigenetic and MPRA data. Nevertheless, these results validate the differential signal approach.
DISCUSSION
Chromatin accessibility (DNase-seq or ATAC-seq) and histone mark ChIP-seq data have been widely used to predict enhancers in specific cell types. Although sophisticated algorithms have been developed to integrate multiple types of data in multiple cell and tissue types to predict enhancers, calling peaks using a single dataset remains a widely used approach for predicting enhancers in the corresponding cell type (22,23,33). In this study, we systematically compared nine algorithms for calling peaks on DNase or ChIP-seq signals with the goal of predicting enhancers. We found that for the same data, different algorithms predicted widely varying numbers of peaks, with systematically different widths and boundaries. We implemented the ENCODE uniform processing pipeline for all nine algorithms we tested in order to filter peaks not reproducible between the two biological replicates, and observed a 30–50% reduction in the peaks called by most algorithms. The uniform processing pipeline also substantially decreased the variations among the final numbers of peaks across the algorithms.
The algorithms we tested were representative of published algorithms applicable to DNase-seq and histone mark ChIP-seq data. Earlier efforts have been made to benchmark peak calling performance, but because of the large number of peak callers available such efforts are far from complete. Thomas et al. (58) selected six out of 30 available peak callers for ChIP-seq data and compared their performance on ChIP-seq data for H3K4me3 and H3K36me3, as well as various transcription factors. They found BCP and MUSIC performed best on these two types of histone ChIP-seq data, assessed by actively transcribed genes. Koohy et al. (32) compared F-seq, Hotspot, MACS2 and ZINBA on DNase-seq data, using transcription factor ChIP-seq peaks in the same cell type as the gold standard. They found F-seq to be slightly more sensitive than Hotspot and MACS2 and ZINBA to be the least sensitive. Our work complements these earlier benchmarking efforts.
Using VISTA enhancers as the gold standard, we evaluated the predictiveness of TSS-distal peaks (≥2 kb away from any GENCODE-annotated TSS) for DNase and five histone marks (H3K4me1/2/3 and H3K9/27ac) called by the various algorithms. H3K4me1, H3K4me2 and H3K9ac have all been shown to be enriched at known enhancers and were used to predict enhancers (6,59,60). We found TSS-distal DNase-seq and H3K27ac peaks to be substantially more accurate in identifying VISTA enhancers than TSS-distal H3K4me1/2/3 and H3K9ac peaks. Our results agree with an earlier study (12), which compared these ‘single epigenetic features’ using a smaller set of VISTA regions (they used 546 regions, while we used 2,556 regions). Another study showed that H2BK20ac was more predictive of enhancers than H3K27ac (61). We reanalyzed the ChIP-seq data in that work and confirmed their results; however, the ENCODE H3K27ac ChIP-seq data achieved higher PR-AUC than both the H2BK20ac and H3K27ac ChIP-seq data used by Kumar et al. (0.42 versus 0.37 and 0.32; all peaks called by HOMER).
For DNase data, we found DFilter to be the most accurate in predicting enhancers as well as the most consistent in ranking called peaks between pairs of biological replicates, followed by Hotspot2. The top DNase peaks called by DFilter were mostly not shared by the top peaks of the other algorithms; this corresponded with the distinctively higher performance of DFilter. DFilter uses a linear detection filter known as the Hotelling observer to perform windowed smoothing of the DNase signal to achieve optimal signal detection accuracy (42). This approach is likely to detect DNase peaks with distinct shape characteristics. Meanwhile, the other algorithms do not employ techniques for taking into account the shapes of peaks and appear more focused on the raw signal level. As a result, most of the top DFilter-unique DNase peaks were broad and multimodal, suggesting multiple consecutive evicted nucleosomes. On the other hand, most of the peaks called by other algorithms (but not DFilter) were of high signal and unimodal, suggesting a single evicted nucleosome. The two sets of peaks also differ in their enriched TF motifs, with DFilter-only peaks enriched in tissue-specific motifs while the peaks shared by the other algorithms enriched in the CTCF motif. Furthermore, a vast majority of the shared peaks overlap CTCF ChIP-seq peaks based on further analysis in midbrain and hindbrain. These results are consistent with our earlier finding that CTCF binding sites are flanked by multiple well-positioned nucleosomes (62), i.e. a single evicted nucleosome at the center. Thus, Hotspot2, MACS2, and HOMER, but not DFilter, preferentially identify DNase peaks with a single evicted nucleosome and bound by CTCF.
For H3K27ac ChIP-seq, HOMER was the most predictive algorithm, followed closely by MUSIC punctate, MACS2, DFilter and F-seq. In contrast to DNase peaks, many of the top H3K27ac peaks called by HOMER were shared by other top performing algorithms (MUSIC punctate, MACS2, and DFilter). Because the same algorithm was used by DFilter to identify DNase and H3K27ac peaks (42), this suggests that, in contrast to DNase peaks, existing algorithms did not identify multiple distinct classes of H3K27ac peaks that could be predictive of active enhancers. Note that DNase-seq identifies many types of regulatory regions, of which enhancers are a subset. As illustrated above, the DNase peaks that are preferentially identified by Hotspot2, MACS2 and HOMER are likely CTCF-bound insulators; such regions tend not to overlap enhancers with high H3K27ac signals. Furthermore, H3K27ac peaks are much wider than DNase peaks, and the H3K27ac peaks that contain a multi-evicted-nucleosome DNase peak in their troughs may not differ substantially from the H3K27ac peaks that contain a single-nucleosome DNase peak in their troughs.
We substantially improved the prediction of VISTA enhancers by reranking DNase and H3K27ac peaks with differential signals against a distant tissue. Moreover, the top peaks after reranking are significantly more enriched in known TF motifs than the top peaks originally ranked by the peak callers, and the genes closest to the top peaks after reranking were significantly more upregulated in the tissue of interest than the genes closest to the top peaks without reranking. These three lines of independent evidence support the validity of the differential signal approach. Furthermore, even though DFilter performed better than Hotspot2 in ranking peaks, reranking led to nearly identical performance for the peaks called by these two algorithms. This suggests that as long as a peak caller identifies true positives peaks in its collection, reranking using differential signals can push these true positive peaks to the top of the list.
The differential signal approach led to a more substantial improvement for DNase data than for H3K27ac data, which could be due to several reasons. First, many more peaks are called for the DNase datasets than for H3K27ac datasets in the same tissue (17-fold more averaged over the nine algorithms). This is because DNase hypersensitivity characterizes all classes of regulatory elements, including enhancers, promoters, silencers, insulators, boundary elements, and nuclear matrix attachment regions, while H3K27ac is predominantly enriched at active enhancers and promoters. A case in point is potential insulators bound by CTCF—over 90% of the top 2000 DNase peaks that are shared among Hotspot2, MACS and HOMER overlap CTCF ChIP-seq peaks, as mentioned above. Second, active enhancers are more tissue-specific than other types of regulatory elements, e.g., promoters, poised enhancers, unannotated promoters, insulators, and matrix attachment regions. Differential DNase signal would rank the other types of elements low, thus improving the power to predict active enhancers. One example is CTCF-bound DNase peaks, which are preferentially identified by Hotspot2, MACS2 and HOMER, as shown above. Such potential insulators tend to be ubiquitously active across multiple cell types (63), and would be ranked low by our differential signal approach. Indeed, 35%, 46% and 65% of the top 2000 DNase peaks identified by DFilter, Hotspot2 and HOMER overlapped CTCF ChIP-seq peaks in the corresponding tissue (percentages averaged between midbrain and hindbrain), while after reranking using our differential signal approach, only 11% of the top 2000 DNase peaks for all three algorithms overlapped CTCF peaks. In contrast, H3K27ac would not benefit as much from differential scoring since it is already specific to active enhancers. Third, the DNase-seq datasets we analyzed had higher sequencing depths than the H3K27ac datasets (196–334 M versus 11–22 M), and a higher sequencing depth corresponds to a higher power in detecting peaks. The third possibility does not seem to play a large role because when we downsampled the DNase datasets to 5 M reads we did not observe a decrease in the DNase peak number called by DFilter.
Several earlier studies reported that differential signals led to improved performance in enhancer prediction. Zeitlinger and colleagues performed differential H3K27ac analysis for identifying Drosophila dorsoventral enhancers (64). Several studies showed that many differentially DNA methylated regions overlap with enhancers (12). Monti et al. noted a DNase I enrichment score that contributed significantly to their model performance on predicting limb VISTA enhancers—the enrichment of DNase signal in the limb over a headless embryo (18). We tested the headless embryo as a contrast tissue for computing the differential DNase signal in limb, and indeed we observed improved PR-AUC of 0.43 and 0.44 for DFilter and Hotspot2 DNase peaks over the original ranking by DFilter or Hotspot2 (PR-AUC = 0.40 and 0.35). However, the headless embryo is much closer to the limb than the three brain tissues (midbrain, hindbrain, and neural tube). Indeed, using any of these three brain tissues as the contrast tissue for computing differential DNase signal achieved greater improvements (PR-AUC = 0.47, 0.49 and 0.48 for DFilter peaks and 0.48, 0.49 and 0.50 for Hotspot2 peaks respectively). Thus, our study corroborates the findings by previous studies, and we additionally show that the performance can be further improved by choosing a different contrast tissue.
We have extended previous work by thoroughly exploring the differential DNase and H3K27ac approach for predicting developmental enhancers in seven mouse tissues. One novelty of our study is that our results indicate that it is important to choose the most distant tissue as the contrast tissue (Supplementary Figures S32-S34). If the tissue under investigation is a brain tissue, it works well to contrast with a non-brain tissue, and vice versa (Supplementary Figures S32-S34); nevertheless, the exact choice of the contrast tissue still impacts the performance. Furthermore, the approach may be sensitive to the differential qualities of the two datasets being contrasted, although this was not an issue with the high-quality ENCODE data produced by the same labs. Indeed, PCA did not reveal batch effects in the DNase-seq and ChIP-seq data we used; however, batch correction may be necessary when datasets are from different sources or protocols. To simplify the process of choosing the contrast tissue, we extended our approach by contrasting against a broad group of tissues, weighted by their distances from the tissue under investigation. This extension improved the results moderately, but eliminated the reliance on a particular contrast tissue. We should be able to use the large panel of ENCODE data as contrast tissues for differential DNase and H3K27ac analysis in any future cell types. The good performance of our method in a blind test of e12.5 heart enhancers and in predicting MPRA enhancers in postnatal retina and cerebral cortex further supports the predictive power of our approach.
Monti et al. used machine learning approaches to predict limb VISTA enhancers (18). Using 50 chromatin features measured on the limb tissue and sequence features, they achieved a median PR-AUC of 0.545 upon 10-fold cross validation, higher than the highest PR-AUC we achieved using a single feature (PR-AUC = 0.50 for weighted differential DNase signal). Nevertheless, our single-feature approach achieves competitive performance with machine learning approaches that require many more input data types, which are not always available for the biological system under study. Note that Monti et al. also focused on TSS-distal regions and used two features called ‘DNase enrichment’, which allows a direct comparison of their performance with ours. Furthermore, our approach does not require training and thus is more likely to maintain its performance across different tissues, while machine learning models trained using the data in one tissue often do not perform as well in another tissue.
Although the differential signal approach achieves improved overall performance, it may distort the overall enhancer landscape and eliminate or demote the enhancers that are active in many cell types. Our comparison of VISTA enhancers that are limb-specific vs. those that are active in limb and other tissues indicates that the differential signal approach works well for both types of enhancers; nevertheless, the approach may not work well for ubiquitously active enhancers.
Clear demarcation of peaks is important for defining individual enhancers and quantifying their activities in specific cell types. The different algorithms predict peaks with widely different widths and accordingly different boundaries. Some algorithms do not call peaks with base-pair resolution (e.g., DFilter peaks have a resolution of 200 bp, which is too low for DNase peaks). Based on visualizing signal tracks using the UCSC genome browser (Supplementary Figures S12–S14), Hotspot2 DNase peaks frequently have more appropriate boundaries than the peaks of the other algorithms. Nevertheless, more systematic analyses are required. Our three gold standards—VISTA enhancers, motif enrichment, and tissue-specific genes—do not have sufficient spatial resolution for evaluating which peak sets have the most accurate widths and boundaries; therefore, we avoided performing such analyses in this study by resizing all peaks to the same width. Other types of functional data such as massively parallel reporter assays may have sufficient resolution.
We used VISTA enhancers as a gold standard for enhancer prediction; this has been done in previous studies (12,14). Although VISTA enhancers are the most extensively validated mammalian enhancers, they have several disadvantages as the gold standard. First, there are only ∼2,500 VISTA regions and their activities have only been measured during embryonic development. Although we evaluated DNase and H3K27ac data in the same tissue and developmental time point as VISTA enhancers, it is still likely that our results are limited due to the small number of VISTA enhancers and our conclusions are specific to mouse embryonic development. Second, VISTA regions have not been randomly chosen. The VISTA regions that were tested in early years were chosen because of high evolutionary conservation. More recently, regions were chosen from H3K27ac or EP300 ChIP-seq peaks called by MACS (36) and CCAT (65); both algorithms approximate windowed ChIP-seq reads with a Poisson model. Thus regions that are not called by these algorithms are underrepresented in the VISTA enhancer collection, and the PR-AUCs we obtained may not reflect the performance that would be obtained by testing randomly-chosen regions. Third, VISTA enhancers may represent the strongest developmental enhancers while weaker enhancers can still be important for development (66). Because our approach does not require any training and does not have any adjustable parameters, it should be less affected by the non-random selection of VISTA regions than supervised machine learning approaches with many adjustable parameters would be. Nevertheless, we further tested our differential signal approach using a different gold standard—postnatal retina and cerebral cortex enhancers identified by massively parallel reporter assays. Despite the mismatch in developmental time points between the input epigenetic data and the MPRA data, we observed improved performance by our differential signal approach over the original algorithms.
CONCLUSION
We performed extensive analysis in search of the best approach for using a single DNase-seq or histone mark ChIP-seq dataset to predict tissue-specific enhancers. We developed a method to contrast DNase or H3K27ac signal in one tissue against a panel of other tissues, resulting in a substantial improvement over existing algorithms, and the improvement is replicated in a blind test. Our approach does not require training and can be directly applied to predict enhancers in new cell types. Our method pertains to the situation when just one histone mark ChIP-seq or DNase-seq experiment is available in the cell or tissue type of interest. When multiple types of experiments are available, methods that integrate these data may achieve better performance.
DATA AVAILABILITY
Code for calling peaks, processing data, and plotting figures in this study are deposited in https://github.com/weng-lab/Diff_signal_pred.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the ENCODE Consortium for the DNase-seq, ChIP-seq, RNA-seq, and transgenic mouse assay data. We also thank members of the Weng Lab for insightful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Thousand Talents Plan funding from the Chinese government and Tongji University [to Z. W., S.F., Q.W., K.F., C.G.] (in part); National Institutes of Health [U24-HG009446 to Z. W., J.M., M.P., H.P., A.K.] (in part). National Natural Science Foundation of China [31571362, 31500626, 91640201 to A. L.]. Funding for open access charge: Thousand Talents Plan funding (to Z.W.) from the Chinese Goverment and Tongji University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bulger M., Groudine M.. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 2010; 339:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012; 337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calo E., Wysocka J.. Modification of enhancer Chromatin: What, how, and why. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 7. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009; 457:854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., Wirbelauer C., Oakeley E.J., Gaidatzis D., Tiwari V.K. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011; 16:6. [DOI] [PubMed] [Google Scholar]
- 12. He Y., Gorkin D.U., Dickel D.E., Nery J.R., Castanon R.G., Lee A.Y., Shen Y., Visel A., Pennacchio L.A., Ren B. et al. Improved regulatory element prediction based on tissue-specific local epigenomic signatures. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E1633–E1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu F., Li H., Ren C., Bo X., Shu W.. PEDLA: predicting enhancers with a deep learning-based algorithmic framework. Sci. Rep. 2016; 6:28517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erwin G.D., Oksenberg N., Truty R.M., Kostka D., Murphy K.K., Ahituv N., Pollard K.S., Capra J.A.. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput. Biol. 2014; 10:e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu Y., Qu W., Shan G., Zhang C.. DELTA: A distal enhancer locating tool based on adaboost algorithm and shape features of chromatin modifications. PLoS One. 2015; 10:e0130622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rajagopal N., Xie W., Li Y., Wagner U., Wang W., Stamatoyannopoulos J., Ernst J., Kellis M., Ren B.. RFECS: A Random-Forest based algorithm for enhancer identification from chromatin state. PLoS Comput. Biol. 2013; 9:e1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Firpi H.A., Ucar D., Tan K.. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics. 2010; 26:1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monti R., Barozzi I., Osterwalder M., Lee E., Kato M., Garvin T.H., Plajzer-Frick I., Pickle C.S., Akiyama J.A., Afzal V. et al. Limb-Enhancer Genie: An accessible resource of accurate enhancer predictions in the developing limb. PLoS Comput. Biol. 2017; 13:e1005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E. et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013; 41:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., An L., Yue F., Hardison R.C.. Jointly characterizing epigenetic dynamics across multiple human cell types. Nucleic Acids Res. 2016; 44:6721–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dickel D.E., Barozzi I., Zhu Y., Fukuda-Yuzawa Y., Osterwalder M., Mannion B.J., May D., Spurrell C.H., Plajzer-Frick I., Pickle C.S. et al. Genome-wide compendium and functional assessment of in vivo heart enhancers. Nat. Commun. 2016; 7:12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun W., Poschmann J., Cruz-Herrera Del Rosario R., Parikshak N.N., Hajan H.S., Kumar V., Ramasamy R., Belgard T.G., Elanggovan B., Wong C.C.Y. et al. Histone Acetylome-wide association study of autism spectrum disorder. Cell. 2016; 167:1385–1397. [DOI] [PubMed] [Google Scholar]
- 24. Nakato R., Shirahige K.. Recent advances in ChIP-seq analysis: from quality management to whole-genome annotation. Brief. Bioinform. 2017; 18:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bardet A.F., Steinmann J., Bafna S., Knoblich J.A., Zeitlinger J., Stark A.. Identification of transcription factor binding sites from ChIP-seq data at high resolution. Bioinformatics. 2013; 29:2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song Q., Smith A.D.. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics. 2011; 27:870–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu S., Grullon S., Ge K., Peng W.. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 2014; 1150:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laajala T.D., Raghav S., Tuomela S., Lahesmaa R., Aittokallio T., Elo L.L.. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009; 10:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilbanks E.G., Facciotti M.T.. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010; 5:e11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szalkowski A.M., Schmid C.D.. Rapid innovation in ChIP-seq peak-calling algorithms is outdistancing benchmarking efforts. Brief. Bioinform. 2011; 12:626–633. [DOI] [PubMed] [Google Scholar]
- 31. Micsinai M., Parisi F., Strino F., Asp P., Dynlacht B.D., Kluger Y.. Picking ChIP-seq peak detectors for analyzing chromatin modification experiments. Nucleic Acids Res. 2012; 40:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koohy H., Down T.A., Spivakov M., Hubbard T.. A comparison of peak callers used for DNase-Seq data. PLoS One. 2014; 9:e96303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016; 167:1398–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D. et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006; 444:499–502. [DOI] [PubMed] [Google Scholar]
- 35. Visel A., Minovitsky S., Dubchak I., Pennacchio L.A.. VISTA enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007; 35:D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boyle A.P., Guinney J., Crawford G.E., Furey T.S.. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008; 24:2537–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. John S., Sabo P.J., Thurman R.E., Sung M.-H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A.. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011; 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuan P.F., Chung D., Pan G., Thomson J.A., Stewart R., Keleş S.. A statistical framework for the analysis of ChIP-Seq Data. J. Am. Stat. Assoc. 2011; 106:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xing H., Liao W., Mo Y., Zhang M.Q.. A novel Bayesian change-point algorithm for genome-wide analysis of diverse ChIPseq data types. J.Visual. Exp.: JoVE. 2012; 70:e4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar V., Muratani M., Rayan N.A., Kraus P., Lufkin T., Ng H.H., Prabhakar S.. Uniform, optimal signal processing of mapped deep-sequencing data. Nat. Biotechnol. 2013; 31:1–11. [DOI] [PubMed] [Google Scholar]
- 43. Harmanci A., Rozowsky J., Gerstein M.. MUSIC: identification of enriched regions in ChIP-Seq experiments using a mappability-corrected multiscale signal processing framework. Genome Biol. 2014; 15:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pekowska A., Benoukraf T., Ferrier P., Spicuglia S.. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010; 20:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M.Q. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008; 40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Consortium E.P., Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M.. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu Y., Sun L., Chen Z., Whitaker J.W., Wang T., Wang W.. Predicting enhancer transcription and activity from chromatin modifications. Nucleic Acids Res. 2013; 41:10032–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L. et al. Cistrome data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017; 45:D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kharchenko P.V., Tolstorukov M.Y., Park P.J.. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008; 26:1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Visel A., Rubin E.M., Pennacchio L.A.. Genomic views of distant-acting enhancers. Nature. 2009; 461:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quinlan A.R. BEDTools: The Swiss-Army tool for genome feature analysis. Curr.Protoc. Bioinformatics. 2014; 47:doi:10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Love M.I., Huber W., Anders S., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shen S.Q., Myers C.A., Hughes A.E.O., Byrne L.C., Flannery J.G., Corbo J.C.. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 2016; 26:238–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Infante C.R., Park S., Mihala A.G., Kingsley D.M., Menke D.B.. Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev. Biol. 2013; 374:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., Muhr J.. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011; 25:2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ong C.-T., Corces V.G.. CTCF: an architectural protein bridging genome topology and function. Nat. Revi. Genetics. 2014; 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomas R., Thomas S., Holloway A.K., Pollard K.S.. Features that define the best ChIP-seq peak calling algorithms. Brief. Bioinform. 2017; 18:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J.-C., Ferrier P., Spicuglia S.. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011; 30:4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L.. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012; 13:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar V., Rayan N.A., Muratani M., Lim S., Elanggovan B., Xin L., Lu T., Makhija H., Poschmann J., Lufkin T. et al. Comprehensive benchmarking reveals H2BK20 acetylation as a distinctive signature of cell-state-specific enhancers and promoters. Genome Res. 2016; 26:612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fu Y., Sinha M., Peterson C.L., Weng Z.. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008; 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xi H., Shulha H.P., Lin J.M., Vales T.R., Fu Y., Bodine D.M., McKay R.D.G., Chenoweth J.G., Tesar P.J., Furey T.S. et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007; 3:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Koenecke N., Johnston J., Gaertner B., Natarajan M., Zeitlinger J.. Genome-wide identification of Drosophila dorso-ventral enhancers by differential histone acetylation analysis. Genome Biol. 2016; 17:aaa5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu H., Handoko L., Wei X., Ye C., Sheng J., Wei C.-L., Lin F., Sung W.-K.. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics. 2010; 26:1199–1204. [DOI] [PubMed] [Google Scholar]
- 66. Levo M., Segal E.. In pursuit of design principles of regulatory sequences. Nat. Rev. Genet. 2014; 15:453–468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code for calling peaks, processing data, and plotting figures in this study are deposited in https://github.com/weng-lab/Diff_signal_pred.