Abstract
Most steps on the biogenesis of the mitochondrial ribosome (mitoribosome) occur near the mitochondrial DNA nucleoid, in RNA granules, which contain dedicated RNA metabolism and mitoribosome assembly factors. Here, analysis of the RNA granule proteome identified the presence of a set of small GTPases that belong to conserved families of ribosome assembly factors. We show that GTPBP10, a member of the conserved Obg family of P-loop small G proteins, is a mitochondrial protein and have used gene-editing technologies to create a HEK293T cell line KO for GTPBP10. The absence of GTPBP10 leads to attenuated mtLSU and mtSSU levels and the virtual absence of the 55S monosome, which entirely prevents mitochondrial protein synthesis. We show that a fraction of GTPBP10 cosediments with the large mitoribosome subunit and the monosome. GTPBP10 physically interacts with the 16S rRNA, but not with the 12S rRNA, and crosslinks with several mtLSU proteins. Additionally, GTPBP10 is indirectly required for efficient processing of the 12S-16S rRNA precursor transcript, which could explain the mtSSU accumulation defect. We propose that GTPBP10 primarily ensures proper mtLSU maturation and ultimately serves to coordinate mtSSU and mtLSU accumulation then providing a quality control check-point function during mtLSU assembly that minimizes premature subunit joining.
INTRODUCTION
Ribosome biogenesis involves the function of non-ribosomal proteins, globally known as ribosome assembly factors, which act at every step of the process. They promote processing, modification, and stability of rRNAs and ribosomal proteins, and their coming together to form the ribonucleoprotein particle. Whereas there is significant knowledge of the assembly factors involved in the biogenesis of bacterial and eukaryotic ribosomes, the available information is scarcer regarding the assembly of the mitochondrial ribosomes (mitoribosomes).
Mitoribosomes are made of components of dual genetic origin. In mammals, all mitoribosome proteins (MRPs) are encoded in the nuclear genome, whereas their three RNA components are encoded in the mtDNA. Mammalian mitoribosomes sediment as 55S particles and consist of a 28S small subunit (mtSSU), formed by a 12S rRNA and 30 MRPs, and a 39S large subunit (mtLSU), formed by a 16S rRNA, 52 MRPs and a structural tRNA (Val in human, Phe in porcine mtLSU) located in the central protuberance (1–4). Mitoribosomes are specialized in the synthesis of a small set of hydrophobic proteins encoded in the mitochondrial DNA (mtDNA). In mammals, the mtDNA encodes 13 proteins, all subunits of the oxidative phosphorylation (OXPHOS) system enzymes, which are essential for aerobic energy production. Hence, the dysfunction of mitochondrial protein synthesis leads to human diseases, mostly cardio- and encephalo-myopathies and contributes to the altered metabolism of cancer cells (5,6).
Knowledge of the mitoribosome assembly process and extra-ribosomal factors involved is starting to emerge. Recent reports have provided an overall view of the assembly landscapes of yeast (7) and human (8) mitoribosomes, which generally coincided to reveal co-operative assembly of protein sets forming structural clusters and, at least in yeast, also preassembled modules. Mitoribosome biogenesis is confined to specific compartments within the mitochondrial matrix. Early steps of human mitoribosome assembly occur at or near the nucleoids (8,9), probably in a co-transcriptional manner. The bulk of mitoribosome biogenesis occurs subsequently in the mitochondrial RNA granule, a membrane-less compartment that condenses and progressively separates from the nucleoids upon transcription (10). Mitochondrial RNA granules contain mitoribosome proteins and assembly factors and accumulate newly synthesized mRNAs and factors that regulate their processing, storage, sorting or translation (11–15).
To date, the proteins known to facilitate mitoribosome biogenesis, include rRNA modification enzymes and their co-factors, kinases, DEAD box helicases and GTPases. Attempts to characterize the RNA granule proteome have contributed to the identification of a subset of these factors (11–14). They include several conserved GTPases whose specific functions remain in most cases to be fully understood. Three conserved ribosome-assembly GTPases were initially identified in Saccharomyces cerevisiae mitochondria, the mtLSU assembly factors Mtg1 (16) and Mtg2 (17) as well as the mtSSU assembly factor Mtg3 (18). Their respective human homologs, MTG1 (19,20), MTG2 (19,21) and C4orf14 or NOA1 (22,23), also participate in mitoribosome assembly. Each of these proteins belongs to a different GTPase subfamily.
The Obg subfamily, which includes MTG2, is a class of highly conserved small monomeric P-loop GTPases principally involved in LSU assembly. In Bacillus subtilis, Obg associates with the ribosome (24), possibly through an interaction with L13 (25). Escherichia coli ObgE or CgtA also associates with the LSU, interacts with the 23S rRNA and several ribosomal proteins (26,27) and their GTPase activity is essential for LSU biogenesis (28,29). ObgE deletion in E. coli leads to the accumulation of LSU assembly intermediates containing reduced levels of L16, L33, and L34, further linking Obg proteins to roles in LSU biogenesis (30). Furthermore, ObgE genetically interacts with the 23S rRNA methyltransferase RrmJ since ObgE overexpression suppresses the LSU 50S assembly defect observed in rrmJ mutant cells (31). However, ObgE cofractionates not only with 50S but also with 30S ribosomal subunits, co-purifies with components of the two subunits and is required for efficient pre-16S rRNA processing (26), suggesting a more global role in ribogenesis. In S. cerevisiae, the nucleolar Obg protein Nog1 is important for pre-60S particle assembly (32,33). Within mitochondria, the Obg ortholog Mtg2 was initially identified in S. cerevisiae as a suppressor of mtLSU 21S rRNA methyltransferase mutant mrm2, suggesting a role during mtLSU biogenesis (17). In mammals, two Obg proteins exist in mitochondria, MTG2 (alias OBGH1 or GTPBP5) and GTPBP10 (alias OBGH2), but their molecular roles remain unknown. MTG2 was shown to specifically associate with the mtLSU (21), but partial MTG2 silencing did not produce any apparent phenotype (19,21). Regarding GTPBP10, it was localized to the nucleolus by immunofluorescence (21). However, it is predicted to have a mitochondrial targeting sequence (MTS) and is listed in the MitoCarta inventory of mammalian mitochondrial proteins (34).
In the present study, we have focused on clarifying the subcellular location of GTPBP10 and characterizing its function. We report that GTPBP10 is a mitochondrial RNA granule component that plays an essential role in mitoribosome biogenesis primarily by fueling late steps in the maturation of the 39S mtLSU subunit. This role is secondarily required for efficient 12S-16S rRNA precursor transcript processing and thus also for the normal biogenesis of the 28S mtSSU subunit. In this way, GTPBP10 coordinates the availability of matured 39S and 28S subunits and assembly into functional 55S mitoribosomes.
MATERIALS AND METHODS
Human cell lines and culture conditions
HEK293T embryonic kidney cells (CRL-3216), and 143B osteosarcoma cells (CRL-8303) were obtained from ATCC. The 143B.TK− rho0 derivative (143B206) was obtained from Dr. M. King (35). The three cell lines were cultured at 37°C under 5% CO2 in high-glucose DMEM (Dulbecco's Modified Eagle Medium) supplemented with 10% FBS (fetal bovine serum), 1 mM pyruvate, and 50 μg/ml uridine (complete DMEM medium). Analysis for mycoplasma contamination was routinely performed.
Key reagents
Tables presenting the list of antibodies, recombinant DNAs, oligonucleotides and siRNA oligoribonucleotides used in this study are included in the supplementary material.
Analysis of the DDX28 interactome
A stable pool of HEK293T cells expressing HA-6xHis-tagged DDX28 was established by puromycin (2 μg/ml) selection for 3 weeks after transfection with the construct DDX28- HA-6xHis/pIRESpuro2. Two mg of mitochondria isolated from this cell line or from HEK293T cells were extracted in 500 μl of extraction buffer (20 mM HEPES, pH 7.4, 0.5 mM PMSF, EDTA-free protease inhibitor cocktail (Roche), 1% digitonin, and 10 mM MgCl2) containing 12.5, 150 or 300 mM KCl. Mitochondrial proteins were incubated with α-IgG (control), or α-HA-conjugated Dynabeads (Life Technologies) at room temperature for 4h. The supernatant containing unbound material was subsequently collected and the beads were washed 3 times with low-salt NET-2 buffer (50 mM Tris–HCl pH 7.4, 12.5 mM NaCl, 0.05% NP-40, EDTA-free protease inhibitor cocktail (Roche)), twice with high-salt NET-2 buffer (with 150 mM NaCl) and once with 1× PBS. The bound proteins were eluted using 50 μl of dye-less Laemmli buffer, precipitated with methanol/chloroform and analyzed by mass spectrometry at the Keck Biotechnology Resource Laboratory (Yale University School of Medicine, New Haven, CT, USA).
Generation of GTPBP10-edited cell lines and plasmid transfection
To create a stable human GTPBP10 knockout (KO) line in HEK293T cells, we used a pair of TALEN constructs obtained from Invitrogen and CRISPR-CAS9 constructs obtained from Santa Cruz Biotechnology. The left and right TALENs for the pair and the guide RNAs for the CRISPR system were designed to bind the sequences shown in Supplemental Figure S1A. HEK293T cells grown in a six-well plate at 30% confluency were transfected with 2 μg of each right and left TALEN plasmid as a pair using 5 μl of Lipofectamine 2000 pre-incubated in 300 μl of Opti-MEM (ThermoFisher). After 8 h of incubation, the media was changed to complete DMEM medium. After five times of repeated transfections every three days, cells were collected and single-cell sorted by FACs in multiple 96 well plates. Upon growth, each clone was tested by immunoblotting to assess the steady-state levels of GTPBP10, as well as the mtDNA-encoded COX2 protein as a surrogate of mitochondrial protein synthesis capacity. Clones that had attenuated GTPBP10 and COX2 levels were further analyzed by genotyping. Similarly, HEK293T cells grown in a six-well plate at 30% confluency were transfected with 2 μg of GTPBP10 CRISPR/Cas9 KO plasmid comprising of a pool of three guided RNAs targeting each to Exon 2, Exon 4 and Exon 6 respectively along with Homology-Directed Repair (HDR) Plasmid. After 8 h of incubation, the media was changed to complete DMEM medium. On day 2, the cells were kept under 2 μg/ml of puromycin selection. On day 5, cells were collected and by serial dilution single cell were plated in multiple 96 well plates. Upon growth, the clones were tested by immunoblotting similar to the clones generated by TALEN mentioned above.
To establish stable lines expressing FLAG-tagged or untagged versions of GTPBP10 cloned into a mammalian vector with hygromycin as the selection marker, 2 μg of each construct or an empty vector was transfected to HEK293T cells using a standard Lipofectamine 2000 protocol. Two days after transfection, the medium was supplemented with 200 μg/ml hygromycin for three weeks. Two vectors were used, pCMV6, in which GTPBP10 expression was placed under the control of a standard human cytomegalovirus (CMV) intermediate early enhancer/promoter, and a modified vector with an attenuated CMV promoter (Δ5), generated by a deletion that eliminates most transcription factor binding sites (36).
Pulse labeling of mitochondrial translation products
Mitochondrial protein synthesis was assayed by pulse-labeling 60-70% confluent human wild-type (WT) HEK293T cells and GTPBP10-edited derivatives in the presence of 100 μg/ml emetine to inhibit cytoplasmic protein synthesis as reported (37,38) and described in the supplementary methods.
Isolation of mitochondria
Mitochondria from WT HEK293T cells, GTPBP10-knockout cells (GTPBP0-KO) or GTPBP0-KO cells stably expressing GTPBP10-FLAG were isolated as described previously (39).
Sucrose gradient analysis
The sedimentation properties of GTPBP10, mitoribosomal proteins, and assembly factors were analyzed by sedimentation in sucrose gradients collected using a Brandel fractionator, essentially as reported (14) and described in the supplementary methods.
RNA analysis
Total RNA was prepared from whole cells by using Trizol (Invitrogen) following the manufacturer instructions for northern-blot and quantitative qPCR analyses. Experimental details are described in the supplementary methods.
GTPBP10 interactome analysis
To study the GTPBP10 protein interactome, we used modified versions of co-immunoprecipitation analysis of protein-protein interaction in the presence or absence of crosslinkers. We also used modified versions of PAR-CLIP assays to study protein-RNA interactions (40). Experimental details are described in the supplementary methods.
Statistical analysis
All of the experiments were done in triplicates or otherwise indicated. Data in X-ray films were digitalized and analyzed using the ImageJ software. Statistical analyses were performed using the Prism-5/6 software and Microsoft Excel. The data are presented as the means ± S.D. or mean ± S.E.M. of absolute values or percentages of control. The values obtained for WT and the GTPBP10-KO strains for the different parameters studied were compared using a Student's two-tailed unpaired t-test for comparison of two groups. For comparison of multiple groups, we performed one-way analysis of variance (ANOVA) followed by Tukey's post hoc test for all the groups of the experiment (*P < 0.05; **P < 0.01; ***P < 0.001).
RESULTS AND DISCUSSION
A set of conserved ribosome assembly GTPases co-purify with mitochondrial RNA granules
To extend the list of mitochondrial RNA granule protein components and the strength of their interactions, we used the bona fide granule component DDX28 as bait for co-immunoprecipitation studies. The strength of their interactions was probed by using HEK293T mitochondrial extracts prepared in the presence of increasing salt concentrations, from 12.5 to 300 mM KCl, which did not affect the efficiency of DDX28 immunoprecipitation (not shown), and the samples were subsequently analyzed by mass spectrometry (see datasets in the Supplementary Table S1). Proteins with MASCOT score >65 and found specifically in the DDX28-precipitated sample but not in the control IgG-precipitated sample are presented in Figure 1A.
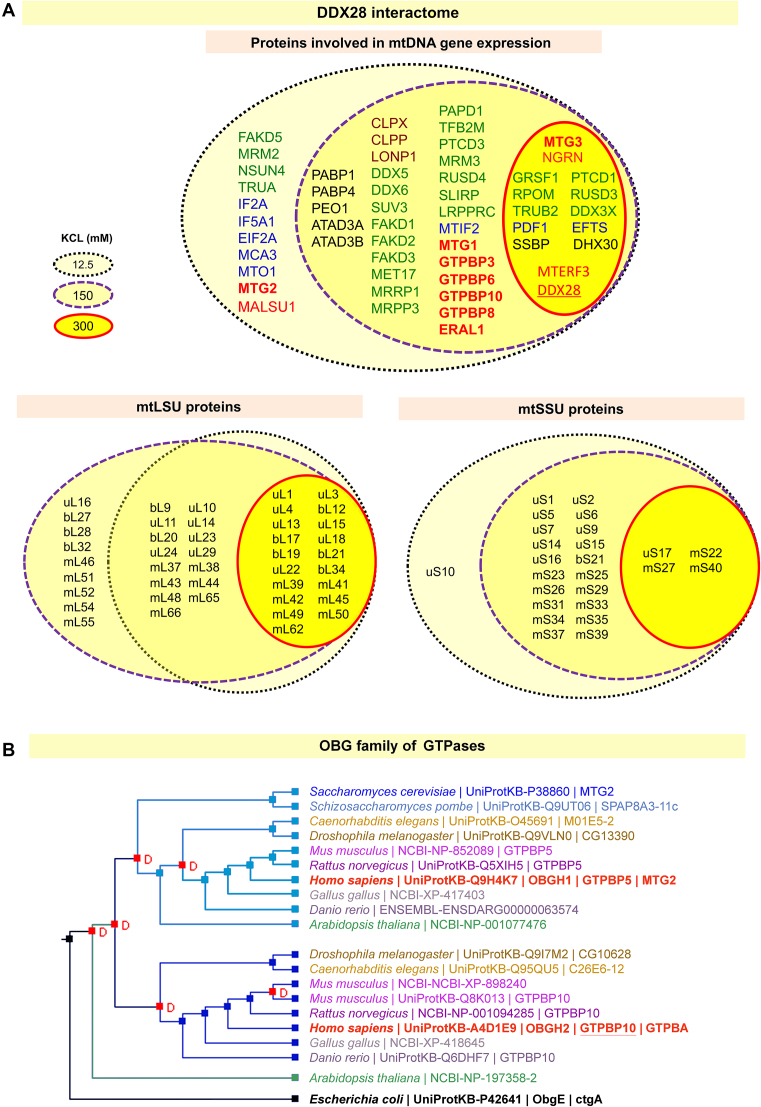
Figure 1.
The mitochondrial RNA granules contain members of several families of GTPases. (A) DDX28 interacting proteome analyzed by immunoprecipitation of endogenous DDX28 from HEK293T mitochondria extracted using conditions of increasing salt stringency and an anti-DDX28-specific antibody and protein A magnetic beads. IgG was used as a control. The full list of proteins is present in Supplemental Table S1. Proteins co-immunoprecitated with DDX28 are displayed confined by black dotted (12.5 mM KCl), purple dashed (150 mM KCl) and red solid (300 mM KCl) Venn diagrams. The diagram in the upper panel list proteins within five categories: RNA metabolism factors (green), ribosome assembly factors (red, with GTPases in bold), translational factors (blue), proteases (brown) and mtDNA nucleoids (black). The diagrams in the lower panel list mtSSU and mtLSU mitoribosome proteins. (B) Cluster analysis of Obg proteins, highlighting the clustering of yeast Mtg2 and animal GTPBP5 (human OBGH1) and the lack of yeast homolog for animal GTPBP10 (human OBGH2), obtained using the Princeton Protein Orthology Database (P-POD). A Notug 2.6 graph is presented (http://ppod.princeton.edu) (56).
The proteins were broadly classified into five categories according to their functions. (i) Mitoribosome proteins; (ii) Mitoribosome assembly factors; (iii) Mitochondrial translational factors; (iv) RNA metabolism factors, including transcription factors, RNA processing and modifying enzymes, and RNA stability and degradosome proteins; and (v) Proteins previously identified as mtDNA nucleoid components. As expected, we observed an inverse correlation between the stringency of the extraction conditions and the number of proteins identified. Notably, the RNA helicase DHX30 associates with DDX28 even in the 300 mM-KCl extracts, stringency enough to disrupt some protein-protein interactions within the ribosome (14). DHX30 was initially identified at the central core region of mitochondrial nucleoids where it has been proposed that proteins related to mitochondrial replication and transcription accumulate (41). More recent studies have localized DHX30 to the mitochondrial RNA granules and shown to be important for mitoribosome LSU assembly (12). These observations confirm that RNA granules and mtDNA nucleoids are located in close proximity and a portion of them dynamically overlap (42) to attend the necessities of newly transcribed RNAs. Our data also confirm the role of mitochondrial RNA granules in compartmentalization of mitoribosome assembly, a process proposed to initiate in granule-nucleoid overlapping foci (8,9) to end within the granule environment (12,14,15).
Focusing on mitoribosome assembly factors, our datasets include a collection of seven GTPases, four of them previously identified as mitoribosome assembly factors (MTG1, MTG2, MTG3 or NOA1, ERAL1) and three other that remain poorly characterized (GTPBP6, GTPBP8 and GTPBP10).
From these proteins, MTG2 (GTPBP5 or OBGH1) and GTPBP10 (OBGH2) belong to the family of Obg proteins whose Escherichia coli founder is the ribosome assembly factor ObgE (ctgA) (see cluster analysis presented in Figure 1B), suggesting a similar kind of function for the two human OBG proteins. However, the finding of GTPBP10 in the mitochondrial DDX28 interactome differs from findings in a single report that identified the protein as part of the nucleolus (21). Therefore, we devoted efforts to further clarify the location and characterize the function of GTPBP10.
GTPBP10 is a mitochondrial matrix protein that interacts with the mitoribosome subunits
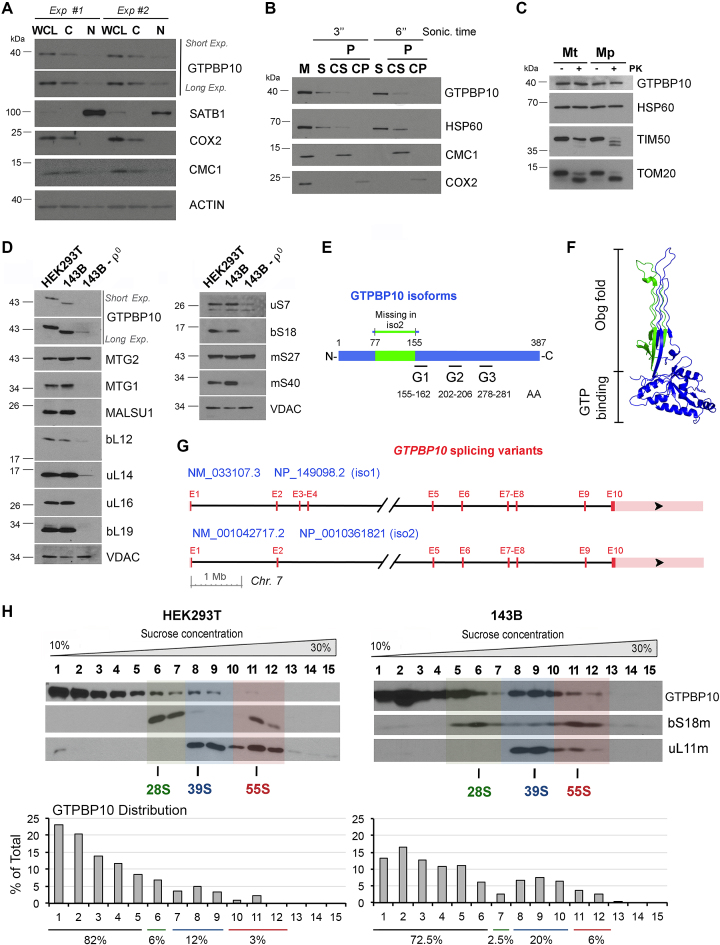
Analyses of nuclear and cytoplasmic fractions obtained from HEK293T cells revealed that endogenous GTPBP10 is a mitochondrial protein, although traces were detected in the nuclear fraction (Figure 2A) as previously shown for the mitoribosome assembly helicase DDX28 (14). Using brief sonication, alkaline extraction (pH 11.5), and proteinase protection assays in mitochondria and mitoplasts, the ∼42-kDa GTPBP10 protein was sub-localized as a soluble mitochondrial matrix protein (Figure 2B–C). Our results are in agreement with a recent publication in which GTBP10 was fully extracted with sodium carbonate pH 11.5 but only partially when the pH was 10.8 (43), which brought the authors to suggest that a fraction of GTPBP10 is peripherally interacting with the inner membrane. Our data, instead, contrast with the previously reported exclusive nucleolar localization of a GFP-tagged version of the protein (21).
Figure 2.
GTPBP10 is a mitochondrial protein that sediments with mitoribosome large and small subunits. (A) Immunoblot analyses of GTPBP10 levels in HEK293T whole cell lysate (WCL), cytoplasmic (C) and nuclear (N) fractions. Antibodies against organelle-specific proteins were used as controls. (B) Mitochondria (M) isolated from HEK293T cells were first fractionated into soluble (S) and membrane-bound (P) proteins by brief sonication and centrifugation. The pellet was submitted to alkaline carbonate extraction (pH: 11.5) to allow the separation of the extrinsic proteins present in the supernatant (CS) from the intrinsic proteins in the pellet (CP). Equivalent volumes of each fraction were analyzed by immunoblotting using antibodies against GTPBP10, the matrix-soluble protein HSP60, the extrinsic membrane-associated protein CMC1 and the inner membrane intrinsic protein COX2. (C) Proteinase K protection assay in mitochondria (Mt) and mitoplasts (Mp) prepared by hypotonic swelling of mitochondria. The samples were analyzed by immunoblotting using antibodies against GTPBP10, the matrix protein HSP60, the inner membrane protein TIM50, and the outer membrane protein TOM20. (D) Immunoblot analyses of the steady-state levels of mitoribosome LSU, SSU proteins and assembly factors in mitochondria from HEK293T, 143B and 143B-206 rho0 (ρ0) cells. VDAC used as a loading control. (E) Scheme of the GTPBP10 protein indicating the region truncated in the isoform 2, and the location of the GTP binding domains (G1-3). (F) Structure of the B. subtilis Obg GTP-binding protein (PDB 1LNZ) (44). The conserved Obg fold (glycine-rich domain) and GTP binding domain are indicated. The protein segment absent in GTPBP10 isoform 2 is labeled in green color. (G) Scheme of the GTPBP10 locus on human chromosome 7 depicting the two known splicing variants. (H) Sucrose gradient sedimentation analysis of GTPBP10 and mitoribosomal proteins in extracts from wild-type HEK293T or 143B mitochondria prepared in the presence of the 20 mM MgCl2. The fractions were analyzed by immunoblotting using Abs against the indicated proteins. Transparent green, blue and red colors mark the fractions where the 28S mtSSU, 39S mtLSU and 55S monosome sediment, respectively. GTPBP10 levels in each fraction was quantified by densitometric integration of the bands using the histogram panel of Adobe Photoshop and plotted as the percentage of the total signal in the bottom graphs.
To test whether GTPBP10 stability depends on the presence of mtDNA and mitochondrial ribosomes, we assessed GTPBP10 steady-state levels in a derivative of an osteosarcoma 143B cell line devoid of mtDNA (rho° cells) and therefore of all mtRNAs including rRNAs. Several ribosome proteins tested (uL16, uL19, bS18) did not accumulate in rho° cells and other (mS27) were markedly decreased (Figure 2D), whereas only traces of GTPBP10 were detected. We noticed that the migration pattern of GTPBP10 was different in HEK293T and 143B cells and derivatives. The GTPBP10 gene, located on chromosome 7, contains 6 exons. According to NCBI, two transcript variants exist. Variant #1 (NM_033107.3 → NP_149098.2) includes all the exons and yields a protein (isoform 1) of 387 aa and an estimated MW of ∼43 kDa (Figure 2E–G). Variant #2 lacks alternate in-frame exons 3 and 4 in the 5′ coding region, compared to variant 1. The resulting protein (isoform 2) has 308 aa and an estimated MW of ∼35 kDa (Figure 2E–G). Our data indicate that HEK293T (embryonic kidney) and 143B (bone osteosarcoma) predominantly express one or the other isoform (Figure 2D). Members of the Obg family contain two highly conserved domains, a C-terminal GTP binding domain and an N-terminal glycine-rich domain (Figure 2F). The two GTPBP10 isoforms retain the three conserved G protein elements (G1-G3) that form the C-terminal GTP binding domain of the protein. Isoform 2, however, has a shorter N-terminal glycine-rich Obg domain (Figure 2E). The Obg fold (residues 1-158 in the B. subtilis protein; residues 1-155 in GTPBP10) is found in all known Obg nucleotide-binding proteins (44) and contains 21 conserved glycines, 9 of which are lost in GTPBP10 isoform 2. The functional relevance of the Obg fold has been supported by the isolation of temperature-sensitive obg alleles in B. subtilis (45) and E. coli (46). Specifically, mutations in several residues within the N-terminal glycine-rich domain were found to disrupt Obg function, indicating that the integrity of the Obg fold is required for function in vivo. The mutations include G79E affecting the conserved Gly79 residue (45), which in GTPBP10 lies within the fragment missing in isoform 2. The architecture of the Obg glycine-rich domain has suggested a platform or scaffolding role (44). We speculate that existence of two GTPBP10 isoforms in human mitochondria may obey to tissue-specific requirements or to facilitate/prevent protein-protein interactions to adapt to changing environmental conditions, as it has been shown for E. coli ObgE (28). These queries are out of the scope of this manuscript but warrant future investigations.
However, since both HEK293T and 143B cells are well known to have a sound mitochondrial protein synthesis and metabolism for ex vivo standards, we have explored a possible association of each GTPBP10 isoform with the mitoribosome. For that purpose, we used sucrose gradient sedimentation analysis of mitochondrial extracts prepared in the presence of 20 mM MgCl2 to stabilize the association of ribosome subunits. In both cell lines, a similar portion of GTPBP10 (∼10% of total) co-sediments with mtSSU and mtLSU markers and a smaller fraction (∼1-2%) co-sediments with the 55S monosome whereas the rest accumulates in lighter fractions (Figure 2H). The interaction with the monosome must be very transient or labile since it is not consistently detected throughout the multiple gradients that we have performed. Our data is reminiscent of the properties of E. coli ObgE, which cofractionates with 30S and 50S ribosomal subunits to facilitate their maturation, although affinity of ObgE to the 30S subunit was shown to be weaker (30). Here, GTPBP10 may play roles in mtSSU and mtLSU biogenesis that extend until the formation of the monosome.
GTPBP10 is essential for mitochondrial protein synthesis
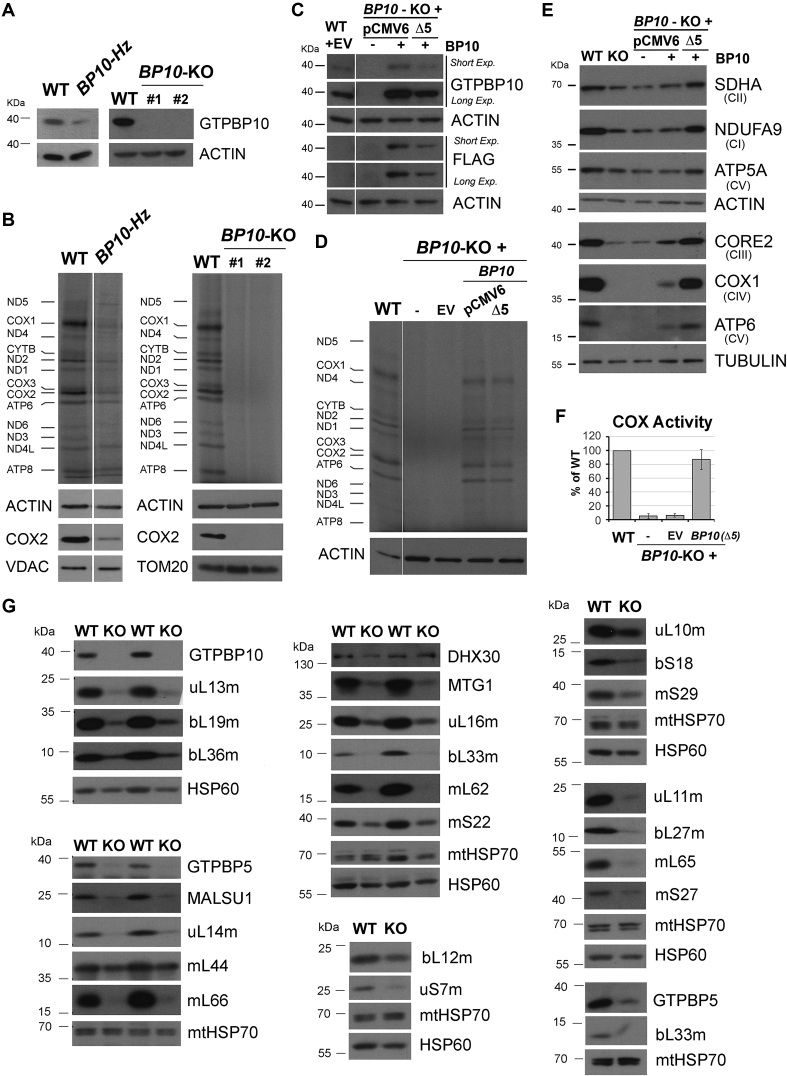
To gain insight into the possible role of GTPBP10 in mitochondrial translation and mitoribosome assembly, we attempted to knockout (KO) GTPBP10 in HEK293T cells using transcription activator-like effector nucleases (TALENs) (Supplemental Figure S1A). Although several transfection strategies were used and nearly 200 clones were screened by immunoblot against GTPBP10 and subsequent genotyping, we identified several heterozygous (see GTPBP10-Hz example in Figure 3A) but did not identify any homozygous KO. Further GTPBP10 editing attempts involved the CRISPR/CAS9 technology using three different guide RNAs (Supplemental Figure S1A). Analysis of another set of ∼200 clones finally yield one carrying a full set of GTPBP10-KO alleles (see genotyping in Supplemental Figure S1B) and therefore did not express any GTPBP10 protein (Figure 3A). The KO clone grows poorly in glucose-rich medium, which becomes fast acidified even when supplemented with uridine, pyruvate, and formate to minimize metabolic consequences of a potential respiratory deficiency.
Figure 3.
GTPBP10 is essential for mitochondrial protein synthesis. (A) Immunoblot analysis of the steady-state levels of GTPBP10 in HEK293T (WT), heterozygous knockout (BP10-Hz) and knockout (BP10-KO) GTPBP10 clones. KO cells collected at two different passages (#1 and #2) are presented. (B) Metabolic labeling with 35S-methionine of newly synthesized mitochondrial translation products in whole cells from the indicated lines during a 30-min pulse in the presence of emetine to inhibit cytoplasmic protein synthesis. Immunoblotting for ACTIN, VDAC and TOM20 were used as loading controls. Newly-synthesized polypeptides are identified on the left. (C) Immunoblot analysis of the steady-state levels of COX2 in HEK293T wild-type (WT) and in GTPBP10 knockout (BP10-KO) cells transfected with an empty vector (EV) or a construct expressing FLAG-tagged GTPBP10. GTPBP10-FLAG was expressed under the control of either a standard human cytomegalovirus (CMV) intermediate early enhancer/promoter (plasmid pCMV6) or an attenuated CMV promoter (Δ5), generated by a deletion that eliminates a large proportion (4/5) of the transcription factor binding sites (36). (D) Metabolic labeling as in panel (B) using GTPBP10 knockout (BP10-KO) cells reconstituted with a construct expressing FLAG-tagged GTPBP10 under the control of either a standard CMV promoter (pCMV6) or a truncated promoter (Δ5). (E) Immunoblot analysis of the steady-state levels of oxidative phosphorylation complex subunits in wild-type (WT) and GTPBP10-KO HEK293T cells and GTPBP10-KO cells reconstituted with a construct expressing GTPBP10 under the control of either a standard CMV promoter (pCMV6) or the CMVΔ5 truncated promoter. NDUFA9 is a subunit of complex I, SDHA of CII, CORE2 or CIII, COX1 of CIV, ATP5α and ATP6 of the F1Fo-ATP synthase or CV. Immunoblotting for β-ACTIN and β-TUBULIN were used as loading controls. (F) Measurement of the enzymatic activity of CIV or cytochrome c oxidase (COX), expressed as the percentage of WT. Data represent the mean ± SD of three independent repetitions. (G) Immunoblot analysis of the steady-state levels of mitoribosome proteins and assembly factors in mitochondrial extracts from HEK293T (WT) and GTPBP10 knockout (BP10-KO) cell lines. HSP60 and mtHSP70 were used as loading controls.
Following the observation that GTPBP10 co-sediments with the mitoribosome, we examined the mitochondrial translation capacity of GTPBP10-Hz and GTPBP10-KO cells by following the incorporation of [35S]-methionine into newly synthesized mitochondrial polypeptides in the presence of emetine to inhibit cytoplasmic protein synthesis. De novo mitochondrial protein synthesis rates were markedly decreased in GTPBP10-Hz and virtually undetectable in GTPBP10-KO cells (Figure 3B) thus demonstrating a role for GTPBP10 in mitochondrial protein synthesis. The mitochondrial translation-deficient phenotype is restored by wild-type levels of expression of recombinant FLAG-tagged GTPBP10 (Figure 3C–D), thus minimizing the possibility of CRISPR-induced off-target effects. However, it is important to mention that excess of GTPBP10 may be deleterious, as recently reported (43). During the reconstitution assays, when we used a construct in which GTPBP10 expression was under the control of a standard human cytomegalovirus (CMV) intermediate early enhancer/promoter (plasmid pCMV6), GTPBP10 levels were ∼5–10-fold of WT levels (Figure 3C) but whereas the protein synthesis defect was complemented, the restoration of OXPHOS enzyme subunits was only partial (Figure 3D and E), suggesting a deleterious secondary effect by the overexpressed protein. However, when we used an attenuated CMV promoter (Δ5) (36), wild-type levels of GTPBP10 were expressed and full complementation was achieved (Figure 3D and E). Therefore, in all the complementation experiments implemented in the manuscript, we used the construct with the CMV Δ5 promoter, if not otherwise indicated.
Consistent with their protein synthesis defect, GTPBP10-KO cells presented notably decreased steady-state levels of mtDNA-encoded OXPHOS components, as analyzed by denaturing immunoblotting (Figure 3E). The OXPHOS complex deficiency resulted in decreased enzymatic activities, exemplified by the virtual absence of cytochrome c oxidase (COX) or respiratory complex IV (CIV) activity (Figure 3F).
The mitochondrial protein synthesis defect could be due to abnormalities in mitoribosome biogenesis and function. To explore whether the absence of GTPBP10 affects mitoribosome accumulation, we first tested the steady-state levels of mtLSU and mtSSU subunit markers and assembly factors (Figure 3G). Immunoblot analyses showed that most mtSSU and mtLSU proteins analyzed and several mtLSU ancillary factors (GTPBP5, MTG1, MALSU1) were markedly attenuated in GTPBP10-KO cells (Figure 3G).
We conclude that in HEK293T cells, GTPBP10 is necessary for the maintenance of mitoribosome levels, therefore for mitochondrial protein synthesis and consequently, for the biogenesis and function of mitochondrial OXPHOS complexes.
GTPBP10 is required for accumulation of mitoribosome proteins and rRNAs
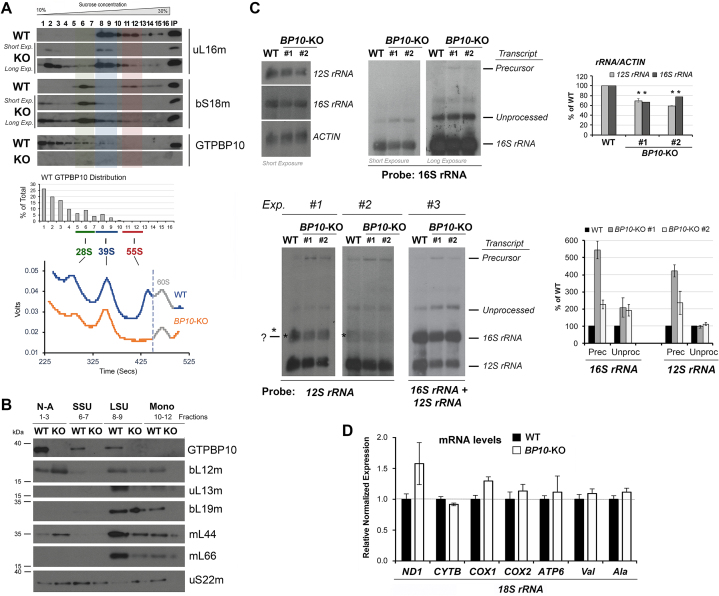
To further explore whether GTPBP10 depletion affects mitoribosome biogenesis and forces the accumulation of mitoribosome assembly intermediates, we followed the formation of mitoribonucleoprotein particles in GTPBP10-KO mitochondria by sucrose gradient sedimentation analysis of mitochondrial extracts prepared in the presence of magnesium. An apparent effect of GTPBP10-KO was the virtual absence of 55S monosomes (Figure 4A). It was also readily apparent the accumulation of unassembled or partially assembled mtSSU and mtLSU proteins, which accumulated in the top light fractions (Figure 4A). Finally, the levels of mtSSU and mtLSU sedimenting as assembled WT subunits were markedly lowered in the absence of GTPBP10.
Figure 4.
GTPBP10 is required for accumulation of mitoribosome mtLSU and mtSSU subunits and fully processed rRNAs. (A) Sucrose gradient sedimentation analyses of GTPBP10 and mitoribosome mtSSU and mtLSU markers in mitochondria prepared from HEK293T (WT) or GTPBP10 knockout (KO) cells. Two exposures of the KO immunoblots (short and long) are presented. The lower panel presents the continuous RNA profile (absorbance at 254 nm) obtained during gradient collection using a Brandel fractionation system and Brandel Peak Chart Software. Transparent green, blue and red colors mark the fractions where the 28S mtSSU, 39S mtLSU and 55S monosome sediment, respectively. (B) Immunoblot analysis of GTPBP10 and mitoribosome protein levels in fractions from the sucrose gradients presented in panel (A) where the non-assembled subunits (N-A, fractions 1–3), mtSSU (fractions 6–7), mtLSU (fractions 8–9), and monosome (fractions 10–12) peak. Equal volumes of fractions corresponding to each cell line were loaded. (C) Northern blot analyses of the steady-state levels of mitochondrial rRNAs in WT or the GTPBP10-KO clone. Multiple experimental repetitions (Exp) and X-ray film exposures are presented to display the steady-state levels of 12S-16S precursor transcript, 12S and 16S unprocessed and fully processed transcripts. The graphs on the right show the densitometry values normalized by the signal of ACTIN rRNA and expressed relative to the WT. Data represent the mean ± SD of three independent repetitions. (D) Quantitative PCR (qPCR) analyses of the steady-state levels of several mtDNA-encoded mRNAs (ND1, CYTB, COX1, COX2 and ATP6) and tRNAs (tRNA-Val and tRNA-Ala) in WT and the GTPBP10-KO clone. Data represent the mean ± SD from three independent experiments.
To better compare the accumulation of GTPBP10 and mitoribosome protein markers in WT and GTPBP10-KO mitochondrial extracts, we ran in parallel, in a single gel, samples from the relevant sucrose gradient fractions (Figure 4B). We confirmed that the levels of mtSSU and mtLSU proteins were decreased by at least 50% in the fractions where the corresponding subunit peaks (Figure 4B). The fractions from gradients presented in Figure 4A were collected with a Brandel fractionator implemented with a UV detector that allows for continuously monitoring absorbance at 254 nm throughout the gradient and therefore to analyze the rRNA distribution pattern. In GTPBP10-KO cells, levels of 12S rRNA in the 28S mtLSU fractions and of 16S rRNA in the 39S mtLSU fractions were decreased by ∼50%, and rRNAs were undetectable in the monosome fractions (Figure 4B). In agreement with these data, quantification of total 12S and 16 rRNAs by northern-blot showed that the absence of GTPBP10 lowers rRNA levels to 60-70% of WT (Figure 4C).
Importantly, when the northern-blots were overexposed, we could detect accumulation of the unprocessed precursor polycistronic transcripts that contain both the 12S and 16S rRNAs, the 12S-16S rRNA (∼2-6 fold), and unprocessed 16S rRNA transcript (∼2 fold) (Figure 4C). These data are reminiscent of the 3-4-fold increase in the levels of 23S rRNA and 16S rRNA precursors observed in the E. coli obgE mutant strain (26,30). In E. coli, rRNA processing and ribosome maturation are tightly coupled (47,48). As a consequence, mutations in LSU subunit assembly proteins can result in processing defects in both SSU and LSU rRNAs (49,50). Also in mammalian cells, mtRNA processing is rate limiting for the assembly of the mitoribosomal subunits, which has been shown to occur co-transcriptionally (51). The double-stranded circular mtDNA encodes 13 proteins, 22 tRNAs and 2 rRNAs. Genes for proteins and tRNAs are located in the two strands, which are transcribed as large polycistronic transcripts covering most of each strand. Following the original punctuation model, most genes encoding protein or rRNA are flanked by one or more tRNAs that can act as ‘punctuation’ marks for transcript processing (52). This involves cleavage at the 5′ end of tRNAs by RNase P, formed by the MRPP1-3 subunits, and cleavage of the 3′ end by the mitochondrial RNase Z known as ELAC2, although additional proteins participate (53). Notably, studies of knockout mice of the endoribonuclease component of the RNase P complex, MRPP3, have shown that mtSSU assembly cannot proceed in unprocessed 12S rRNA, whereas a mtLSU intermediate containing a subset of proteins is able to form on the unprocessed rRNA transcript (51). Therefore, our data suggest the possibility that a primary defect in mtLSU assembly in the absence of GTPBP10 could prevent efficient precursor transcript processing, ultimately impacting mtSSU maturation.
The decrease in rRNA levels does not reflect a general mtRNA instability because RT-qPCR analysis did not identify any significant changes in the steady-state levels of several tRNAs and mRNAs analyzed (Figure 4D). Consistently, no mtDNA depletion or abnormal mtDNA nucleoid morphology was observed in GTPBP10-KO cells (Supplemental Figure S2A and B).
GTPBP10 is required for correct or efficient mtLSU and mtSSU maturation during the hierarchical incorporation of MRPs
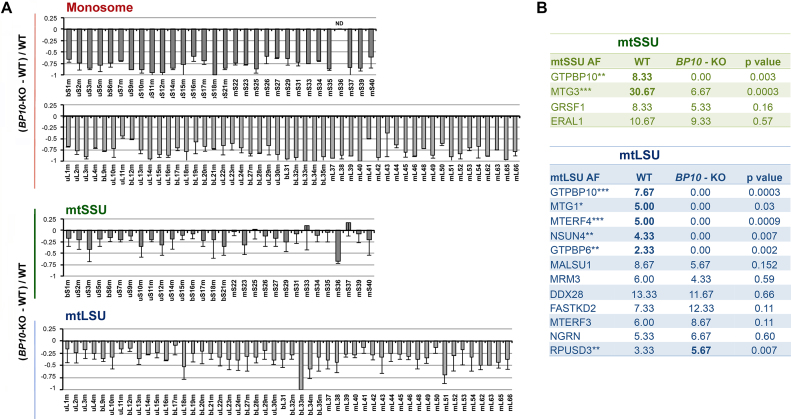
To obtain a more comprehensive picture of the differences in the composition and abundance of assembled mitoribosomal subunits, sucrose gradient fractions from Figure 4A, corresponding to the monosome, mtLSU and mtSSU were methanol/chloroform precipitated and analyzed by mass spectrometry (Figure 5A). Consistent with the data presented in Figure 4A–B, all mtLSU and mtSSU proteins from GTPBP10-KO mitochondria showed a marked decrease in the monosome fraction. They were also decreased in the mtSSU and mtLSU fractions, reflecting a 30–40% decrease of accumulated particles. Late assembly proteins such as uS3m, uS10m, bS21m and mS36 and late assembly mtLSU proteins such as bL33 or bL34 were particularly attenuated in their corresponding fractions. The data on the mtLSU is reminiscent of the observations made in E. coli obgE deletion strains, which accumulate LSU assembly intermediates containing reduced levels of L16, L33 and L34 (30). Together, our data indicate an inability to efficiently or correctly assemble the mitoribosome subunits or maintain their stability in the absence of GTPBP10.
Figure 5.
The absence of GTPBP10 alters the abundance and composition of the mtLSU and mtSSU proteomes. (A) Identity and abundance of mitoribosome proteins and assembly factors that accumulate in mitoribosome particles in mitochondrial extracts from WT and GTPBP10-KO HEK293T cells, following their accumulation in the fractions from sucrose gradient sedimentation studies presented in Figure 4A. The proteins in the fractions in which the mtSSU (fractions 6), mtLSU (fractions 8), and monosome (fractions 11) peak, were precipitated using methanol-chloroform and identified by mass spectrometry. The bar graphs represent the total unique spectral count difference between WT and GTPBP10-KO normalized by the WT count. Results represent the average ± SD of three independent repetitions. Mitoribosome proteins are identified at the bottom. (B) Accumulation of mitoribosome SSU and LSU assembly factors in the fractions corresponding to each subunit, analyzed by mass spectrometry in panel (A). Data represent the average ± SD of total unique spectral counts in WT and GTPBP10-KO samples from three independent repetitions. T-test: *P <0.05; **P <0.01, ***P <0.001.
Mass spectrometry analysis of the mitoribosome subunits from GTPBP10-KO cells allowed us to evaluate further the potential place GTPBP10 occupies within the hierarchic incorporation of assembly factors during mtSSU and mtLSU assembly (Figure 5B). Regarding the mtSSU, levels of GRSF1 and ERAL1 were similar to the WT, suggesting early incorporation and stable interaction with the mtSSU particle assembled in the absence of GTPBP10. On the contrary, levels of the GTPase MTG3 on the mtSSU were reduced in the absence of GTPBP10, indicating a labile interaction. This observation is significant from a rRNA processing perspective. MTG3 (also called NOA1 or C4orf14 in humans), is a conserved member of the YqeH family of GTPases. In yeast mitochondria, Mtg3 is required to facilitate the processing of the mtSSU rRNA precursor, jointly with the uL29 component of the mtLSU, which has been suggested that could provide a mechanism for balancing expression of the two ribosomal subunits (18). Independently of whether human MTG3 performs a similar role, its attenuated presence on the mtSSU subunit would indicate that the function or effect of GTPBP10 on the mtSSU precedes that of MTG3.
Concerning the mtLSU, levels of MALSU1, DDX28, FASTKD2, MRM3, MTERF3, NGRN and PTCD1 were unchanged and levels of RPUSD3 were increased in GTPBP10-KO mtLSU particles, which suggests that these assembly factors are recruited to the growing mtLSU particle before GTPBP10. Although levels of MALSU1 in the mtLSU particle had a tendency to decrease, they were not significantly changed in contrast with their decreased steady-state levels in the absence of GTPBP10 (Figure 3G). This suggests slow MALSU1 recycling once they have incorporated into the mtLSU intermediate upon which they act, and fast degradation of the protein fraction that is not engaged in mtLSU assembly. Several other mtLSU assembly factors, MTG1, NSUN4, MTERF4, and GTPBP6, may act after GTPBP10 since they were not detected in GTPBP10-KO mtLSU particles (Figure 5B).
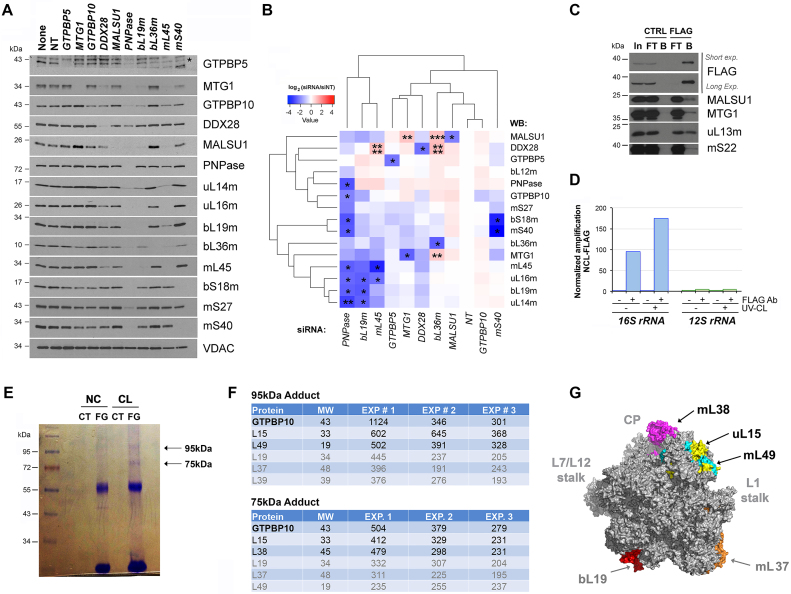
As yet another approach to further examine the role of GTPBP10, we introduced siRNA-induced silencing of selected mitoribosome proteins (Figure 6A) and assembly factors (AF) in WT cells and tested genetic interactions among them by immunoblotting (Figure 6A and B). Silencing of GTPB10 was poorly effective in four different repetitions using three different siRNA constructs but we chose to include the data to illustrate the importance of having a GTPBP10-KO cell line and have included a comparison of the effects of GTPBP10 KO and knockdown (KD) in Supplemental Figure S3. In the genetic interactions studies, we placed a major focus on mtLSU biogenesis. The selected proteins included early-assembled mtLSU proteins such as bL19 and mL45 (8). As expected, silencing of bL19 or mL45 significantly lowered the steady-state levels of most mtLSU proteins, prevented mtLSU assembly and also tended to attenuate GTPBP10 levels (Figure 6A and B), suggesting that GTPBP10 acts beyond the incorporation of early-assembled subunits. Throughout this experiment, increased or decreased GTPBP10 levels, although consistent, did not reach statistical significance, probably due to the significant fraction of the protein that is not bound to mitoribosome subunits at a given moment (as seen in sucrose gradients in Figures 2H and 4A). We observed that GTPBP10 levels also had a tendency to decrease in the absence of the early assembly factor DDX28 but to increase in the absence of the late assembly factor MTG1 (Figure 6A and B), suggesting that GTPBP10 incorporates to the mtLSU assembly line after DDX28 and before MTG1 as also suggested by the mass spectrometry analysis described earlier. Silencing of the mtLSU late-assembly protein bL36, which is incorporated following the action of MTG1 (20), resulted in increased levels of DDX28, MALSU1, GTPBP10, and MTG1. Finally, GTPBP10 levels also tend to increase in the absence of MALSU1, a result that is not fully consistent with the mass spectrometry data (see above) and may indicate that both GTPBP10 and MALSU1 could act simultaneously during mtLSU biogenesis.
Figure 6.
GTPBP10 directly interacts with the 16S rRNA and mtLSU proteins. (A and B) Knockdown (KD) of mitoribosome assembly factors and mitoribosome subunits in HEK293T cells using siRNAs for 8-9 days, verified by immunoblotting of whole cell lysates. (A) Representative image of immunoblot analysis of the steady-state levels of mitoribosome proteins after silencing of target proteins. None is a mock consisting of only transfection reagent. siRNA-NT is a non-targeting silencing control. Antibodies are listed on the right side, and VDAC was used as a loading control. (B) Following analysis in panel (A), the densitometric data obtained on the abundance of mitoribosome proteins and assembly factors accumulated after silencing of each target protein was used for cluster analysis (see Methods). The heat map, generated with the R studio software, represents a log 2 scale of the normalized average levels of ratio to control (NT) in three independent repetitions of immunoblotting analyses. 2-way ANOVA followed by a Dunnett's multiple comparisons test: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (C) Co-immunoprecipitation analysis of GTPBP10-FLAG and interacting mitoribosome proteins and assembly factors in whole cell lysates prepared in the presence of 1% NP40, by using anti-FLAG agarose beads (FLAG) or plain beads as control (CTRL). In, input. FT, flow through or unbound. B is bound. (D) qPCR analyses of reverse-transcribed control or GTPBP10-FLAG co-immunopurified RNAs after 4-thiouridine (4SU) treatment and either UV-mediated protein-RNA crosslinking (CL) or not crosslinking. (E and F) Analysis of GTPBP10 directly interacting partners by using a non-cleavable crosslinker, DSG (disuccinimidyl glutarate). Mitochondria purified from cells stably expressing GTPBP10-FLAG were incubated in the presence of DSG or DMSO as a negative control, pulled-down with anti-FLAG-agarose beads and analyzed by SDS-PAGE. After Coomassie staining, bands of ∼ 75 kDa and ∼95 kDa observed only in the crosslinked-IPed sample were cut, and its composition analyzed by mass spectrometry. The table in panel (F) presents the proteins identified in three independent experiments, sorted by their Mascot score. (G) Mitoribosome proteins listed in panel (F) mapped to the human mitoribosome structure (PDB 3J9M) (3).
GTPBP10 directly interacts with the 16S rRNA and mtLSU proteins.
The E. coli homolog of GTPBP10, ObgE, possesses 16S and 23S rRNA binding activity and has been proposed to participate in rRNA refolding or stabilization of rRNA/ribosomal protein structures (30). Thus, we set out to examine whether GTPBP10 directly binds to the 12S and 16S rRNA. For this purpose, we used whole cell extracts from WT cells stably expressing functional GTPBP10-FLAG that was efficiently immunoprecipitated using anti-FLAG sepharose beads but not IgG-conjugated beads (Figure 6C). To test for potential RNA-GTPBP10 interactions, WT whole cells were subjected or not to UV-mediated protein-nucleic acid crosslinking, followed by IP, and isolation of the co-immunoprecipitated (co-IPed) RNA. Following reverse transcription and quantitative PCR analysis, the 12S rRNA was poorly detected in all the different groups (Figure 6D). In contrast, the 16S rRNA was significantly enriched in the FLAG Co-IP eluted samples compared to the control IP (Figure 6D). These data demonstrate the interaction of GTPBP10 with the 16S rRNA.
To test whether GTPBP10 could directly guide insertion of ribosomal proteins into the growing mtLSU particle as we have recently reported for MTG1 (20), we implemented several approaches using or not chemical crosslinkers to identify potential mitoriboprotein interacting partners of GTPBP10. In the assay presented in Figure 6C, whole cell lysates from HEK293T cells stably expressing FLAG-tagged GTPBP10 were lysed with 1% NP40 to disrupt most native complexes, the GTPBP10-FLAG-interacting proteins were isolated by FLAG-affinity IP and analyzed by immunoblotting. The immunoprecipitate contained only traces of the mtSSU protein mS22 but substantial amounts of mtLSU protein uL13 and assembly factors MTG1 and MALSU1 (Figure 6C), demonstrating the interaction of GTPBP10 with mtLSU proteins. These data also indicate that the effect of GTPBP10 depletion on the attenuation of assembled mtSSU levels is most probably secondary to the role of GTPBP10 on the mtLSU.
In another assay, we explored the direct interaction of GTPBP10 with mitoribosomal proteins in the presence of a non-cleavable crosslinker DSG (disuccinimidyl glutarate) followed by extraction under denaturing conditions. DSG is a homobifunctional crosslinker based on the amine-reactive N-hydroxysuccinimide (NHS) ester group, with a 7.7 Å spacer arm. Mitochondria purified from cells stably expressing GTPBP10-FLAG were incubated in the presence of DSG or DMSO as a negative control, pulled-down with anti-FLAG agarose beads and analyzed by SDS-PAGE. After Coomassie staining, two bands of ∼75 and ∼95 kDa were detected only in the crosslinked-IPed sample (Figure 6E). The bands were cut, and their composition was analyzed by mass spectrometry. We found that 34 MRPs were detected in the ∼95 kDa band and 21 in the ∼75 kDa band, many of which were overlapping (Supplementary Table S2). The table in Figure 6F presents the four top proteins with higher Mascot scores. Given the MW of these proteins, GTPBP10 could be expected to interact with one of them, although it is conceivable that more than one GTPBP10 complex of similar MW could form the ∼75 and ∼95 kDa adducts. Also, some pairs of MRPs could be crosslinked and co-migrate with the GTPBP10 adducts-MRP adduct/s. While mapping these MRPs to the structure of the mitoribosome, we noticed that the three mtLSU MRPs with the higher rank in each adduct, uL15m, mL38, and mL49 locate adjacent to each other in the mtLSU central protuberance (Figure 6G), strongly suggesting that this may be the mtLSU location to which GTPBP10 binds.
CONCLUSION
Mitoribosome maturation in vivo requires the assistance of a growing number of trans-acting factors whose functions are starting to emerge. Here, we have shown that the GTPase GTPBP10 binds to the 16S rRNA and associates with both 39S subunits and the monosome to promote their biogenesis. Secondarily, it affects the mtSSU biogenesis, which may reflect a role in coordinating the accumulation of the 28S and 39S subunits. GTPBP10 function is essential for mitoribosome maturation. It cannot be substituted for other GTPases known to act exclusively in late stages of mtLSU assembly such as MTG1 or even by the second mitochondrial member of the Obg family in human cells, GTPBP5 or MTG2 (Supplemental Figure S4A-B).
During the review of our manuscript, a characterization of GTPBP10 has been reported by Lavdovskaia et al. (43). Consistent with our data, they found that GTPBP10 interacts with the mtLSU and is involved in assisting mtLSU late-assembly steps. Although they were not able to obtain a full GTPBP10-KO cell line, they obtained a cell line expressing a partially functional GTPBP10 protein carrying a deletion of two amino acids at position 64 (R) and 65 (K) in the Obg fold domain (43), located just before the portion of the fold missing in 143B cells (Figure 2E). In contrast to what we have observed in the GTPBP10-KO line, the Gtpbp1064R65K cell line did not display any defect on mtSSU assembly or abundance. We believe that the significant GTPBP10 function retained in this cell line is the explanation for the discrepancy.
The biogenesis of the mitoribosome small and large subunits was initially thought to proceed largely independently, since mutations affecting mtSSU proteins or assembly factors do not usually affect the biogenesis of the mtLSU, and vice versa (12,14,20). However, recent studies on human mitoribosome assembly kinetics have shown that mitoribosome proteins are imported in large excess and that unassembled copies are degraded relatively rapidly (8) probably to avoid excessive accumulation or proteins that could compromise mitochondrial proteostasis. Therefore, the attenuated steady-state levels of mtSSU proteins observed in the GTPBP10-KO cell line could be partially explained by a failure in the assembly of the mtLSU. However, our data clearly indicate that not only proteins but also levels of matured mtSSU particles are partially depleted in the absence of GTPBP10.
The question of how a mtLSU assembly defect can affect the biogenesis or steady-state levels or the mtSSU is intriguing. In an attempt to answer this question, we have followed two hypotheses regarding the coordination of mtSSU and mtLSU assembly.
Following a first hypothesis, proposed by the group of A. Spinazzola (54), the mtLSU assembly needs to reach an undefined intermediate stage to facilitate the release of an early mtSSU intermediate from the mtDNA nucleoids in order to proceed with its maturation within the mitochondrial RNA granule compartment. If this does not occur, the mtDNA nucleoids aggregate and are enriched in mtSSU proteins. Specifically, gene silencing of the mtLSU assembly factor MPV17L2, which co-sediments on sucrose gradients with the mtLSU and the monosome, results in a marked decrease in these structures and also in the mtSSU (54). Furthermore, although MPV17L2 depletion does not affect mtDNA levels, it also results in aggregation of the mitochondrial nucleoids where mtSSU proteins were shown to accumulate (54). It was proposed that if mtSSU assembly starts at the mtDNA nucleoids, MPV17L2 may coordinate mtLSU biogenesis with the release from the nucleoids of mtSSU assembly intermediates or fully assembled particles (54). In our case, as for MPV17L2 silencing, the absence of GTPBP10 did not lead to mtDNA depletion. On the contrary, mtDNA nucleoids in GTPBP10-KO cells have a morphology and distribution similar to wild-type cells, thus suggesting different mechanisms underlying the mtSSU biogenesis defects in cells depleted of MPV17L2 or GTPBP10.
A second hypothesis is suggested by work by Rackham, O., et al. (51), who showed that mitoribosome assembly proceeds co-transcriptionally and starts on a 12S-16S rRNA precursor transcript that may be only processed once mtLSU maturation has reached a certain intermediate stage. This is concluded from the observation that an unprocessed 16S rRNA transcript was shown to be sufficient to stimulate the incorporation of a large subset of mtLSU ribosomal proteins (at least 27 were reported) but not the assembly of the mature mtLSU subunits (51). On the contrary, mtSSU assembly (incorporation of proteins to the 12S rRNA) does not occur on the precursor and requires processing (51). In vivo, as explained earlier, canonical 5′ tRNA cleavage by the RNAse P complex has been shown to precede 3′ tRNA processing by ELAC2, and this is required for the correct biogenesis of the mitochondrial ribosomal subunits (51). With this information in mind, we have shown that indeed a 12S-16S rRNA precursor transcript accumulates several folds in GTPBP10-KO mitochondria (Figure 4C). We propose that the mtLSU assembly step catalyzed by GTPBP10 is required to stimulate efficient 12S-16S rRNA precursor transcript processing by the nucleases and therefore delays or impairs mtSSU assembly. This model provides a rationale for a direct role of GTPBP10 in mtLSU biogenesis secondarily affecting mtSSU maturation.
Focusing on the role of GTPBP10 on mtLSU biogenesis, Lavdovskaia et al. (43) found that GTPBP10 co-precipitated in native conditions with mtLSU particles containing several known late-stage assembly factors, such as MALSU1 and SMCR7L (55), but also several additional proteins involved in 16S rRNA chaperoning, stability or modification such as DDX28, NGRN, RPUSD4, TRUB2, and FASTKD2 (43). Although valuable to link GTPBP10 to the biogenesis of the mtLSU, these observations do not necessarily define the substrate upon which GTPBP10 acts. Here, we believe that the interaction of GTPBP10 with the 16SrRNA and GTPBP10 cross-linking with specific mtLSU proteins that we have described, as well as our data on the hierarchical incorporation of assembly factors to the mtLSU assembly line, provides further insight into the direct and precise effect of GTPBP10 in mtLSU maturation. We propose that GTPBP10 could catalyze 16S rRNA conformation remodeling and interactions with proteins in the central protuberance to facilitate incorporation of late assembly subunits, similar to what we have recently described for MTG1 (20). Our data suggest that GTPBP10 would act in an intermediate that contains DDX28, FASTKD2, MRM3, MTERF3, NGRN, and RPUSD3, perhaps concurrently with MALSU1, and before the action of MTG1, NSUN4, MTERF4, and GTPBP6. As reported for MTG1 (20) and MALSU1 (55), GTPBP10 remains bound to the mtLSU until mtSSU joints to form the functional monosome. Therefore, these mitoribosome assembly factors may serve redundant roles in preventing premature subunit joining and translation initiation.
Collectively, our results suggest that GTPBP10 might act as a quality control checkpoint molecule in the final stages of 39S subunit assembly. It binds to the 16S rRNA possibly to promote or stabilize rRNA folding and proper assembly of rRNA with mtLSU proteins. Our data have also uncovered potential links between 39S mtLSU maturation, 12S-16S precursor transcript processing, and biogenesis of the 28S mtSSU, which could provide a mechanism for balancing or coordinating the maturation of the two ribosomal subunits in mammalian mitochondria.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: P.M. generated all the presented data on the GTPBP10-KO studies, protein localization, PAR-CLIP assays and siRNA silencing. H-J.K. generated data on siRNA silencing and GTPBP10 protein crosslinking. Y.T. produced data on the DDX28 interactome. P.M. and A.B. designed the study. P.M., H.J.K. and A.B. analyzed results and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Medical Sciences Maximizing Investigator's Award (NIGMS-MIRA) [R35GM118141 to A.B.]; Muscular Dystrophy Association Research Grant [MDA-381828 to A.B.]; American Heart Association predoctoral fellowship (to H.J.K.). Funding for open access charge: National Institutes of Health Medical Sciences Maximizing Investigator’s Award (NIGMS-MIRA) [R35GM118141 to A.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., Ban N.. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015; 348:303–308. [DOI] [PubMed] [Google Scholar]
- 2. Greber B.J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N., Aebersold R., Ban N.. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014; 515:283–286. [DOI] [PubMed] [Google Scholar]
- 3. Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V.. The structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown A., Amunts A., Bai X.C., Sugimoto Y., Edwards P.C., Murshudov G., Scheres S.H., Ramakrishnan V.. Structure of the large ribosomal subunit from human mitochondria. Science. 2014; 46:718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim H.J., Maiti P., Barrientos A.. Mitochondrial ribosomes in cancer. Semin. Cancer Biol. 2017; 47:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Silva D., Tu Y.T., Amunts A., Fontanesi F., Barrientos A.. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015; 14:2226–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeng R., Smith E., Barrientos A.. Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab. 2018; 27:645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bogenhagen D.F., Ostermeyer-Fay A.G., Haley J.D., Garcia-Diaz M.. Kinetics and mechanism of mammalian mitochondrial ribosome assembly. Cell Rep. 2018; 22:1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bogenhagen D.F., Martin D.W., Koller A.. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 2014; 19:618–629. [DOI] [PubMed] [Google Scholar]
- 10. Iborra F.J., Kimura H., Cook P.R.. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004; 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Antonicka H., Sasarman F., Nishimura T., Paupe V., Shoubridge E.A.. The mitochondrial RNA-binding protein GRSF1 Localizes to RNA granules and Is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013; 17:386–398. [DOI] [PubMed] [Google Scholar]
- 12. Antonicka H., Shoubridge E.A.. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 13. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tu Y.T., Barrientos A.. The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 2015; 10:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrientos A. Mitochondriolus: assembling mitoribosomes. Oncotarget. 2015; 6:16800–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrientos A., Korr D., Barwell K.J., Sjulsen C., Gajewski C.D., Manfredi G., Ackerman S., Tzagoloff A.. MTG1 codes for a conserved protein required for mitochondrial translation. Mol. Biol. Cell. 2003; 14:2292–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Datta K., Fuentes J.L., Maddock J.R.. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell. 2005; 16:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paul M.F., Alushin G.M., Barros M.H., Rak M., Tzagoloff A.. The putative GTPase encoded by MTG3 functions in a novel pathway for regulating assembly of the small subunit of yeast mitochondrial ribosomes. J. Biol. Chem. 2012; 287:24346–24355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kotani T., Akabane S., Takeyasu K., Ueda T., Takeuchi N.. Human G-proteins, ObgH1 and Mtg1, associate with the large mitochondrial ribosome subunit and are involved in translation and assembly of respiratory complexes. Nucleic Acids Res. 2013; 41:3713–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim H.-J., Barrientos A.. MTG1 couples mitoribosome large subunit assembly and intersubunit bridge formation. Nucleic Acid Res. 2018; 46:8435–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirano Y., Ohniwa R.L., Wada C., Yoshimura S.H., Takeyasu K.. Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes Cells. 2006; 11:1295–1304. [DOI] [PubMed] [Google Scholar]
- 22. He J., Cooper H.M., Reyes A., Di Re M., Kazak L., Wood S.R., Mao C.C., Fearnley I.M., Walker J.E., Holt I.J.. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012; 40:6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolanczyk M., Pech M., Zemojtel T., Yamamoto H., Mikula I., Calvaruso M.A., van den Brand M., Richter R., Fischer B., Ritz A. et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell. 2011; 22:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang S., Haldenwang W.G.. Guanine nucleotides stabilize the binding of Bacillus subtilis Obg to ribosomes. Biochem. Biophys. Res. Commun. 2004; 322:565–569. [DOI] [PubMed] [Google Scholar]
- 25. Scott J.M., Ju J., Mitchell T., Haldenwang W.G.. The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 2000; 182:2771–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato A., Kobayashi G., Hayashi H., Yoshida H., Wada A., Maeda M., Hiraga S., Takeyasu K., Wada C.. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005; 10:393–408. [DOI] [PubMed] [Google Scholar]
- 27. Wout P., Pu K., Sullivan S.M., Reese V., Zhou S., Lin B., Maddock J.R.. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 2004; 186:5249–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feng B., Mandava C.S., Guo Q., Wang J., Cao W., Li N., Zhang Y., Zhang Y., Wang Z., Wu J. et al. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014; 12:e1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kint C., Verstraeten N., Hofkens J., Fauvart M., Michiels J.. Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit. Rev. Microbiol. 2014; 40:207–224. [DOI] [PubMed] [Google Scholar]
- 30. Jiang M., Datta K., Walker A., Strahler J., Bagamasbad P., Andrews P.C., Maddock J.R.. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006; 188:6757–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tan J., Jakob U., Bardwell J.C.. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 2002; 184:2692–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kallstrom G., Hedges J., Johnson A.. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003; 23:4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M.. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003; 23:4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calvo S.E., Clauser K.R., Mootha V.K.. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016; 44:D1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. King M.P., Attadi G.. Mitochondria-mediated transformation of human rho (0) cells. Methods Enzymol. 1996; 264:304–313. [DOI] [PubMed] [Google Scholar]
- 36. Morita E., Arii J., Christensen D., Votteler J., Sundquist W.I.. Attenuated protein expression vectors for use in siRNA rescue experiments. Biotechniques. 2012; 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leary S.C., Sasarman F.. Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol. Biol. 2009; 554:143–162. [DOI] [PubMed] [Google Scholar]
- 38. Bourens M., Barrientos A.. A CMC1-Knockout reveals translation-independent control of human mitochondrial Complex IV biogenesis. EMBO Rep. 2017; 18:477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Enriquez J.A., Attardi G.. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996; 264:183–196. [DOI] [PubMed] [Google Scholar]
- 40. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bogenhagen D.F., Rousseau D., Burke S.. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008; 283:3665–3675. [DOI] [PubMed] [Google Scholar]
- 42. Borowski L.S., Dziembowski A., Hejnowicz M.S., Stepien P.P., Szczesny R.J.. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013; 41:1223–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lavdovskaia E., Kolander E., Steube E., Mai M.M., Urlaub H., Richter-Dennerlein R.. The human Obg protein GTPBP10 is involved in mitoribosomal biogenesis. Nucleic Acids Res. 2018; 46:8471–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buglino J., Shen V., Hakimian P., Lima C.D.. Structural and biochemical analysis of the Obg GTP binding protein. Structure. 2002; 10:1581–1592. [DOI] [PubMed] [Google Scholar]
- 45. Kok J., Trach K.A., Hoch J.A.. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J. Bacteriol. 1994; 176:7155–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kobayashi G., Moriya S., Wada C.. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 2001; 41:1037–1051. [DOI] [PubMed] [Google Scholar]
- 47. Srivastava A.K., Schlessinger D.. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:7144–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith B.A., Gupta N., Denny K., Culver G.M.. Characterization of 16S rRNA processing with pre-30S subunit assembly intermediates from E. coli. J. Mol. Biol. 2018; 430:1745–1759. [DOI] [PubMed] [Google Scholar]
- 49. Gutgsell N.S., Deutscher M.P., Ofengand J.. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA. 2005; 11:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Charollais J., Pflieger D., Vinh J., Dreyfus M., Iost I.. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003; 48:1253–1265. [DOI] [PubMed] [Google Scholar]
- 51. Rackham O., Busch J.D., Matic S., Siira S.J., Kuznetsova I., Atanassov I., Ermer J.A., Shearwood A.M., Richman T.R., Stewart J.B. et al. Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 2016; 16:1874–1890. [DOI] [PubMed] [Google Scholar]
- 52. Ojala D., Montoya J., Attardi G.. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981; 290:470–474. [DOI] [PubMed] [Google Scholar]
- 53. Sanchez M.I., Mercer T.R., Davies S.M., Shearwood A.M., Nygard K.K., Richman T.R., Mattick J.S., Rackham O., Filipovska A.. RNA processing in human mitochondria. Cell Cycle. 2011; 10:2904–2916. [DOI] [PubMed] [Google Scholar]
- 54. Dalla Rosa I., Durigon R., Pearce S.F., Rorbach J., Hirst E.M., Vidoni S., Reyes A., Brea-Calvo G., Minczuk M., Woellhaf M.W. et al. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 2014; 42:8500–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brown A., Rathore S., Kimanius D., Aibara S., Bai X.C., Rorbach J., Amunts A., Ramakrishnan V.. Structures of the human mitochondrial ribosome in native states of assembly. Nat. Struct. Mol. Biol. 2017; 24:866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heinicke S., Livstone M.S., Lu C., Oughtred R., Kang F., Angiuoli S.V., White O., Botstein D., Dolinski K.. The Princeton Protein Orthology Database (P-POD): a comparative genomics analysis tool for biologists. PLoS One. 2007; 2:e766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.