Abstract
Key points
In mammals, the mother–offspring interaction is essential for health later in adulthood.
The impact of altered timing and quality of maternal care on the offspring's circadian system was assessed using a cross‐strain fostering approach.
Better maternal care facilitated the development of amplitudes of Bmal1 clock gene expression in the central clock, as well as the clock‐driven activity/rest rhythm, and also its entrainment to the external light/dark cycle. Worse maternal care impaired entrainment of the central clock parameters in the Wistar rat during the early developmental stages.
Better maternal care remedied the dampened amplitudes of the colonic clock, as well as cardiovascular functions.
The results provide compelling evidence that the circadian phenotype of a foster mother may affect the pathological symptoms of the offspring, even if they are genetically programmed.
Abstract
In mammals, the mother–offspring interaction is essential for health later in adulthood. Maternal care is determined by the circadian phenotype of the mother. The impact of altered timing and quality of maternal care on the circadian system was assessed using a cross‐strain fostering approach, with ‘abnormal’ (i.e. circadian misaligned) care being represented by spontaneously hypertensive rats (SHR) and ‘normal’ care by Wistar rats. The SHR mothers worsened synchrony of the central clock in the suprachiasmatic nuclei with the light/dark cycle in Wistar rat pups, although this effect disappeared after weaning. The maternal care provided by Wistar rat mothers to SHR pups facilitated the development of amplitudes of the Bmal1 expression rhythm in the suprachiasmatic nuclei of the hypothalamus, as well as the clock‐driven activity/rest rhythm and its entrainment to the external light/dark cycle. The peripheral clocks in the liver and colon responded robustly to cross‐strain fostering; the circadian phenotype of the Wistar rat foster mother remedied the dampened amplitudes of the colonic clock in SHR pups and improved their cardiovascular functions. In general, the more intensive maternal care of the Wistar rat mothers improved most of the parameters of the abnormal SHR circadian phenotype in adulthood; conversely, the less frequent maternal care of the SHR mothers worsened these parameters in the Wistar rat during the early developmental stages. Altogether, our data provide compelling evidence that the circadian phenotype of a foster mother may positively and negatively affect the regulatory mechanisms of various physiological parameters, even if the pathological symptoms are genetically programmed.
Keywords: circadian clock, maternal care, suprachiasmatic nucleus, liver, colon, heart rate, locomotor activity, development
Key points
In mammals, the mother–offspring interaction is essential for health later in adulthood.
The impact of altered timing and quality of maternal care on the offspring's circadian system was assessed using a cross‐strain fostering approach.
Better maternal care facilitated the development of amplitudes of Bmal1 clock gene expression in the central clock, as well as the clock‐driven activity/rest rhythm, and also its entrainment to the external light/dark cycle. Worse maternal care impaired entrainment of the central clock parameters in the Wistar rat during the early developmental stages.
Better maternal care remedied the dampened amplitudes of the colonic clock, as well as cardiovascular functions.
The results provide compelling evidence that the circadian phenotype of a foster mother may affect the pathological symptoms of the offspring, even if they are genetically programmed.
Introduction
In mammals, an intimate mother–offspring interaction on a daily basis is essential for health later in adulthood, as demonstrated at the level of mental and metabolic functions (Varcoe et al. 2013; Marco et al. 2016). These physiological functions are controlled by endogenous time‐keeping system that optimizes their course over the day and night. The system is composed of molecular clocks that spontaneously generate rhythmic signal at the cellular level with an ∼24 h (i.e. circadian, period) (Reppert & Weaver, 2001). The clocks temporally regulate the expression of a great array of physiologically active genes (Ripperger et al. 2000; Storch et al. 2002). During ontogenesis, the circadian system undergoes endogenously determined development and, at the same time, is affected by maternal cues (Sumova et al. 2012).
The central circadian clock, which is located in the suprachiasmatic nuclei of the hypothalamus (SCN) (Ralph et al. 1990), synchronizes the clocks present in almost every cell in the body (i.e. in the neuronal and non‐neuronal tissues) (Balsalobre et al. 1998; Yamamoto et al. 2004). The clock genes Per1,2, Cry1,2, Bmal1,2, Clock, Nr1d1 and Rora,b are basic components of the mammalian circadian clock mechanisms at the cellular level. The rhythmic regulation of their expression is based on mutually interlocked transcriptional–translational feedback loops, as has been reviewed extensively (Reppert & Weaver, 2001). Within the core regulatory feedback loop, during the night, the proteins PER1,2 and CRY1,2 inhibit the transcription of their own genes Per1,2 and Cry1,2, which were activated during the day by CLOCK and BMAL1. In the additional feedback loop, Nr1d1 and Rora gene transcription is also rhythmically activated by CLOCK and BMAL1, and their protein products feedback to regulate Bmal1 expression, thus increasing the robustness of the clock oscillations. The effective interaction of these feedback loops causes and is dependent on opposite phases of Bmal1 and Nr1d1 expression rhythms.
In rodents, the circadian system develops gradually during ontogenesis from the prenatal period until weaning (Sumova et al. 2012; Landgraf et al. 2014). The clock in the rat SCN develops to an adult‐like stage by postnatal days (P)5–10, when it begins to play the role of the central clock and generate robust rhythmic signals that are fully entrainable to external light/dark cycle. However, the peripheral clocks remain under maternal influence for a longer time and become fully independent of maternal cues after weaning, which occurs in rats at around P20–30. This course of development has been demonstrated by assessing the amplitudes and phases of clock gene expression profiles in the developing SCN (Sladek et al. 2004; Kovacikova et al. 2006) and in various peripheral tissues, such as in the liver (Sladek et al. 2007a), colon (Polidarova et al. 2014), pituitary, lung (Yoo et al. 2004), heart (Wu et al. 2015), pancreas (Muhlbauer et al. 2004), kidney (Meszaros et al. 2014) and adrenal tissue (Roa et al. 2017). The amplitudes of these rhythms reflect the degree of synchrony among individual cellular oscillators that is required for coherent rhythms at the tissue level in vivo. In the central SCN clock, this synchrony is ensured via a web of neuronal connections among the cellular oscillators (Welsh et al. 1995; Aton et al. 2005) that represents the intrinsic property of this structure. In non‐neuronal peripheral tissues, inter‐oscillator communication is lacking, and each oscillator receives rhythmic signals individually. These signals are derived from the SCN and/or from activity/rest or feeding/fasting rhythms (Stokkan et al. 2001; Guo et al. 2006). At early developmental stages after birth, the clock gene expression profiles are dependent on rhythmic maternal input (Sladek et al. 2007a; Polidarova et al. 2014).
To assess the effect of altered maternal care on the circadian clocks in pups, we used a strategy aiming to measure the impact of maternal care provided by a mother with a different circadian phenotype. We built up this approach based on our previous findings that the circadian system of spontaneously hypertensive rats (SHR) differs greatly from normotensive Wistar rats (Sladek et al. 2012; Polidarova et al. 2013), with the latter comprising a strain that SHR were originally bred from (Okamoto & Aoki, 1963). SHR spontaneously develop pathology in cardiovascular (Conrad et al. 1995) and metabolic functions (Pravenec et al. 2004) as they age. The first significant rise in blood pressure occurs at 5–6 weeks of age and this increases until adulthood when the blood pressure reaches up to 180–200 mmHg compared to 120 mmHg in Wistar–Kyoto rats (Behuliak et al. 2015). Cardiovascular disorders, such as hypertrophy of the heart and vessels, begin to develop at 40–50 weeks of age (Conrad et al. 1995). SHR are generally more responsive to stressful stimuli at 3–6 weeks of age and their hypothalamic‐pituitary‐adrenal axis is significantly more sensitive compared to Wistar–Kyoto rats (Hausler et al. 1983; Kenyon et al. 1993). Importantly, the SHR circadian system differs significantly from that of Wistar rats during ontogenesis (Olejnikova et al. 2015), as well as in adulthood (Cui et al. 2011; Sladek et al. 2012; Polidarova et al. 2013). In terms of the differences most relevant to the present study, the adult SHR have daily activity/rest cycles that are dampened and phase‐advanced relative to the light/dark cycle (Sladek et al. 2012) and they are more sensitive to changes in feeding regime (Polidarova et al. 2013). Additionally, our previous findings (Olejnikova et al. 2015), as well as those of others (Gouldsborough et al. 1998), have demonstrated that the maternal care provided by SHR mothers to pups shortly after delivery differs from that of Wistar rat mothers. In rats, maternal care consists of nest building, licking, grooming and nursing in either an arched‐back posture or when lying over the pups and/or nearby (Champagne et al. 2003). This requires physical contact with the offspring. Our infrared cameras recordings of the time that mothers spent in the nest with their pups during the day and night revealed that, compared to Wistar rat mothers, SHR mothers left the nest for a longer time, especially during the night (Olejnikova et al. 2015). Therefore, based on the differences in circadian phenotype and maternal care between SHR and Wistar rats, we designated the maternal care provided by SHR mother as ‘abnormal’ (i.e. less frequent and circadian misaligned) and the care by Wistar rats as ‘normal’. These differences in circadian and maternal behaviour between the two rat strains (SHR and Wistar rat) provide us with the unique opportunity to investigate the impact of altered maternal care on developing circadian clocks in rat pups that are reared from birth by a foster mother of a different strain. Because adverse maternal environment during gestation and lactation may have negative effect on pups and the effect may persist until adulthood (Hatanaka et al. 2017), we hypothesized that proper maternal care might affect the circadian system and possibly mitigate pathological symptoms of SHR. We assessed the effects of altered maternal care on the circadian system in pups at P10 (when pups were completely dependent on the maternal care), at P30 (when their weaning was completed) and in adulthood. To our knowledge, this is the first study to address the impact of altered maternal care on the molecular circadian clocks in pups.
Methods
Ethical approval
All experiments were approved by the Animal Care and Use Committee of the Institute of Physiology in agreement with the Animal Protection Law of the Czech Republic and the European Community Council directives 86/609/EEC. All efforts were made to minimize the suffering of the animals. The experiments conform to the principles and regulations as described in Grundy (2015).
Animals
Three‐month‐old male and female Wistar:Han and SHR/Ola rats (Institute of Physiology, Academy of Sciences of the Czech Republic) were maintained under a 12:12 h light/dark (LD) cycle (lights on 06.00 h) at 21 ± 2°C and with free access to food and water. Light was provided by overhead 40 W fluorescent tubes and illumination was ∼150 lux, depending on the cage position in the animal room.
Experimental groups and procedures
Female rats were mated with males and, in cases of sperm positivity in vaginal smears, were maintained individually in cages. After delivery, which was designated postnatal day 0 (P0), the dams and their pups were maintained under LD 12:12 h and divided into four groups (Fig. 1). The body weight (bw) of newborn pups (at P1) was 7.0 ± 0.5 g for Wistar rat and 5.0 ± 0.4 g for SHR. The pups of both sexes were included in the study. The pups of the control group were left undisturbed and were reared by their own mother throughout the lactation period until they were killed during the 24 h cycle at P10 or P30 (for details, see below). The other groups of pups were exposed to fostering procedures: on P1, they were transferred to a foster mother that was (i) of the different rat strain (cross‐strain foster) (i.e. the Wistar rat pups were reared by a foster SHR mother and the SHR pups were reared by a foster Wistar rat mother) or (ii) of the same rat strain (intra‐strain foster) (i.e. the Wistar rat pups were reared by a foster Wistar rat mother and the SHR pups were reared by a foster SHR mother). Pups reared by foster mothers were sampled at P10 and/or P30 as described above for the control pups. Another group of pups was exposed to a foster mother of the different strain but then returned at P10 to their own biological mother that reared them until P30 when they were sampled (cross‐strain return). Additionally, the pups of control and cross‐strain fostering groups were weaned at P30 and maintained under LD 12:12 h and their locomotor activity, blood pressure and heart rate (HR) were monitored when they were 1–2 and 6–8 months old. The rats were weighted at the age of 2 months. The bw of rats reared by their genetic mothers was 256.0 ± 12.8 g for Wistar rats and 152.5 ± 6.4 g for SHR. The bw of cross‐strain fostered Wistar rats and SHR was 233.8 ± 12.4 g and 220.4 ± 10.6 g, respectively.
Figure 1. Experimental scheme.
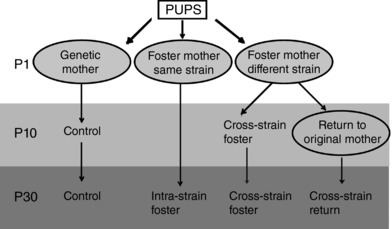
The newborn rat pups were reared either by their genetic mother (control) or exposed to a fostering procedure where on the first postnatal day (P1), the pups were transferred to a foster mother. A foster mother of the same rat strain (intra‐strain fostering) or of the other rat strain (cross‐strain fostering) reared the pups until weaning (P30) or the pups were exposed to cross‐strain fostering until P10 and then returned to their original mother for the rest of the lactation period (until P30).
Collection of tissue samples
On the day of sampling, pups of all groups and both rat strains were released into constant darkness (DD) and killed under deep anaesthesia (i.p. injections of thiopental, 50 mg kg–1) in dim red light (<1 lux) by rapid decapitation every 4 h during a 24 h cycle starting at circadian time 0 (CT0), which corresponded to the time of the previous lights on. For both rat strains, each of the above groups consisted of 35 pups (from litters of seven or eight dams) and, at each time point, 4–6 pups from the same litter were sampled to collect the brains, liver and colon.
The brains were immediately frozen on dry ice and were kept at −80°C. They were sectioned throughout the SCN into 12 μm thick slices that were used to detect the mRNA levels of clock genes by in situ hybridization. The liver samples were immediately immersed in RNAlater stabilization reagent (Qiagen, Valencia, CA, USA). Colons were dissected from the cecum to the rectum, cut longitudinally and a sample of the tissue was immersed in RNA later stabilization reagent. The samples were stored in RNAlater at –20°C until RNA isolation.
In situ hybridization
The cDNA fragments of rat rPer1 (980 bp; 581–1561; GenBank AB_002108), rPer2 (1512 bp; 369–1881; GenBank NM_031678), rNr1d1 (1109 bp; 558–1666; GenBank BC062047) and rBmal1 (841 bp; 257–1098; GenBank AB012600) were used as templates for the in vitro transcription of cRNA probes. The probes were labelled using 35S‐UTP and the in situ hybridizations were performed as described previously (Sumova et al. 2003). The brain sections were hybridized for 20 h at 60°C. Following a post‐hybridization wash, the sections were dehydrated in ethanol and dried. Finally, the slides were exposed to BIOMAX MR film (Sigma‐Aldrich, St Louis, MO, USA) for 10 days and developed using ADEFO‐MIX‐S developer and ADEFOFIX fixer (Adefo‐Chemie, Dietzenbach, Germany) in an automatic film processing machine (Protec, Oberstenfeld, Germany). Brain sections were processed simultaneously under identical conditions. Thereafter, the sections were processed for cresyl violet staining to identify the position of the SCN. Autoradiographs of the sections were analysed using an image analysis system (Image Pro; Olympus, New York, USA) to detect the relative optical density (OD) of the specific hybridization signal in the SCN (OD signal of the background measured in the close surrounding area was subtracted from the signal measured in the SCN area). For comparisons between the experimental groups, the data were normalized to the highest value of the compared daily profiles and expressed as the mean ± SD.
RNA isolation and real‐time quantitative RT‐PCR
Liver and colon samples were homogenized by ultrasound sonication and total RNA was purified using GenElute Mammalian Total RNA Miniprep Kit (Sigma‐Aldrich) in accordance with the manufacturer's instructions. RNA concentrations were determined by Nanodrop (ThermoFisher, Waltham, MA, USA) spectrophotometry at 260 nm and RNA quality was assessed by electrophoresis on 1.5% agarose gel. Moreover, the integrity of randomly selected samples of total RNA was tested using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
The real‐time quantitative RT‐PCR method, which was used to detect mRNA levels of Per1, Per2, Nr1d1 and Bmal1, has been described previously (Sladek et al. 2007a). Briefly, 1 μg of total RNA was reverse‐transcribed using a High Capacity cDNA RT Kit (ThermoFisher). Diluted cDNA was then amplified on LightCycler480 (Roche, Basel, Switzerland) using SYBR Select qPCR Master Mix (ThermoFisher) and the corresponding primers. Relative quantification was achieved using a standard curve and by subsequently normalizing the measured gene expression to the mean relative expression of β2‐microglobulin (B2M) and Gapdh. These reference genes have been used for normalization previously (Sladek et al. 2007b; Polidarova et al. 2011, 2013) and their expression was stable throughout the 24 h period. All of the primer sequences have been reported previously (Sladek et al. 2007a; Polidarova et al. 2014). In each experiment, samples from the experimental and control groups were analysed in the same quantitative RT‐PCR run.
Locomotor activity recording
The pups were weaned at P30 and housed individually to monitor locomotor activity immediately after weaning and again at the age of 6 months. The locomotor activity was monitored as described previously (Houdek et al. 2016). Briefly, rats were maintained individually in cages equipped with infrared movement detectors, which were attached above the centre of the top of the cage. A circadian activity monitoring system (Dr H. M. Cooper, INSERM, Lyon, France) was used to measure activity every minute. The data were analysed using the ClockLab toolbox (Actimetrics, Wilmette, IL, USA). Double‐plotted actograms and chi‐squared periodograms were generated to evaluate activity and calculate the period and amplitude (power of the period estimation) of its rhythm. For each group, 10–35 animals were recorded.
Direct measurements of blood pressure and HR
HR and blood pressure [mean arterial pressure (MAP), systolic blood pressure (SBP) and diastolic blood pressure (DBP)] were measured in conscious 2‐month‐old rats (for each group, 6–8 animals were measured). The measurements were performed between 14.00 h and 17.00 h, which corresponded to 1–4 h before lights off. A polyethylene catheter (PE50) was inserted into the left carotid artery under isoflurane anaesthesia (5% induction and 2–3% maintenance; Forane; AbbVie, Chicago, IL, USA). This was filled with heparinized saline, tunnelled s.c. and exteriorized in the interscapular region. One day after the surgical procedures, experiments were carried out in conscious rats kept in small transparent cages as described previously (Behuliak et al. 2015). The animals were allowed to stabilize for a period of 30 min before measurements. The arterial catheter was connected to a pressure transducer (MLT0380/D; ADInstruments Ltd, Bella Vista, NSW, Australia) that was placed at the level of the rat heart. The signal from the pressure transducer connected to bridge amplifier (QUAD Bridge; ADInstruments) was digitized with a computer‐based monitoring PowerLab system (PowerLab/8SP; ADInstruments) and recorded at a sampling rate of 400 s−1 (Hz) using LabChart software (ADInstruments). HR was derived from the arterial pressure signal as the reciprocal of the pulse interval, which was computed as the interval between two consecutive systolic peaks.
Statistical analysis
The 24 h gene expression profiles of all experimental groups for each rat strain were compared by two‐way ANOVA (effect of time, groups and interaction). If the analysis revealed significant differences between the groups or a significant interaction effect, post hoc analyses were performed with Šidák's multiple comparisons test. Additionally, the daily gene expression profiles were subjected to cosine analysis so that the data were fit to a regression model defined by: y = mesor + [amplitude*cos(2*π*(x – acrophase)/wavelength)], with a constant wavelength of 24 h. The analysis determined the timing of the peak in gene expression (acrophase) (mean ± SEM) and amplitude (mean ± SEM) of the rhythms, which were compared between the groups using two‐way ANOVA with either Šidák's (two groups) or Tukey's (four groups) multiple comparisons test. Period was analysed by a Kruskal–Wallis test with Dunn's multiple comparisons, and the other locomotor activity and cardiovascular parameters were analysed by one‐way ANOVA with Tukey's multiple comparisons test. With the exception of period, all data were assumed to have normal distribution. P < 0.05 (multiplicity‐adjusted) was considered statistically significant. All of the statistical calculations were performed with Prism, version 7 (GraphPad, La Jolla, CA, USA).
Results
The effect of altered maternal care on the circadian clocks in rat pups was tested in a series of experiments in which the pups were reared by a foster mother of the other rat strain (cross‐strain fostering) or of the same rat strain (intra‐strain fostering) for the entire lactation period (from P1 until P30), or pups were first exposed to the cross‐strain fostering and then, at P10, they were returned to their genetic mothers for the rest of the lactation period (Fig. 1).
Cross‐strain fostering significantly affects the circadian clock in the pup SCN in a strain‐specific manner
To assess the effect of altered maternal care on the SCN clock of pups, we compared daily profiles of Per2, Nr1d1 and Bmal1 expression in the SCN (Fig. 2) of 10‐day‐old and 30‐day‐old pups that were either reared by their own mothers or were exposed to cross‐strain fostering. The statistical comparisons (by two‐way ANOVA) between the experimental groups are summarized in Table 1; the significant differences in the relative expression levels at the individual time points (the results of the post‐hoc analyses) are shown in Fig. 2 A and B; and the significant differences between acrophases of the cosine fits of the expression profiles are shown in Fig. 2 C.
Figure 2. The effect of cross‐strain fostering on the clock gene expression in the SCN of pups.
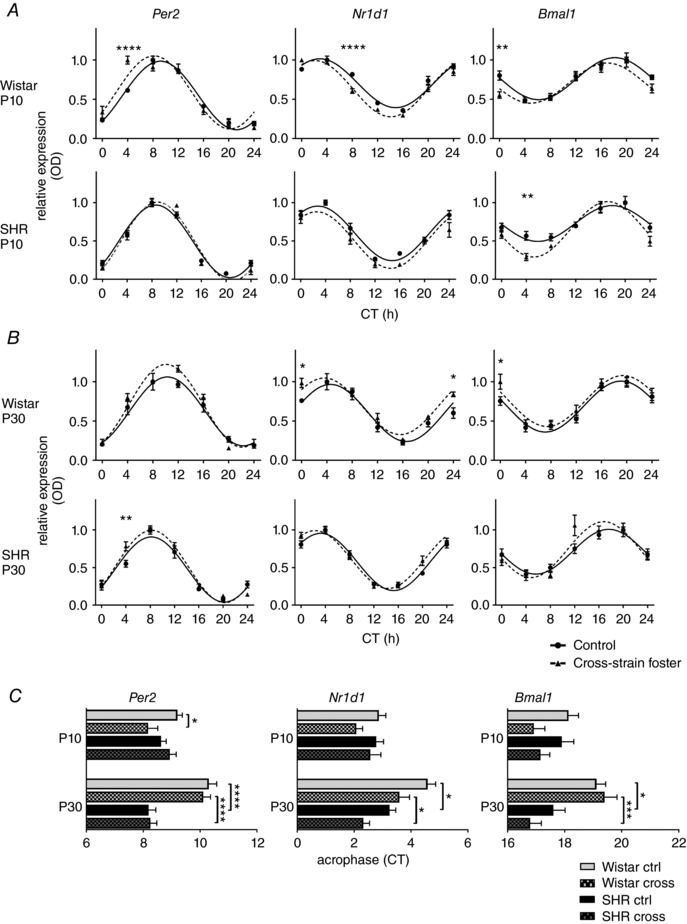
Daily Per2, Nr1d1 and Bmal1 expression profiles were analysed in 10‐day‐old (A) and 30‐day‐old (B) Wistar rat or SHR pups reared by their genetic mothers as control (full circles, full line) or exposed to cross‐strain fostering (full triangles, dashed lines). The pups were killed at 4 h intervals over 24 h. Time is expressed as circadian time (CT); CT0 represents the time of lights on in the previous LD 12:12 h cycle. The data are expressed as the mean ± SEM (n = 3–5 per time point). The results of two‐way ANOVA (Table 1) post hoc analyses (Šidák's multiple comparisons) depict differences in the expression levels between the control and cross‐strain fostered groups at individual CT. C, comparison of acrophases (mean ± SEM) of the gene expression profiles shown in (A) and (B) by two‐way ANOVA with Tukey's multiple comparisons test; the relevant significant differences are depicted. * P < 0.05; *** P < 0.001; **** P < 0.0001.
Table 1.
Results of two‐way ANOVA comparison between the daily profiles of Per2, Nr1d1 and Bmal1 expression in the pup SCN
| Per2 | Nr1d1 | Bmal1 | |||||
|---|---|---|---|---|---|---|---|
| SCN controls vs. cross‐strain | F(DFn, DFd) | P | F(DFn, DFd) | P | F(DFn, DFd) | P | |
| Wistar P10 | Interaction | F 6,52 = 6.002 | <0.0001 | F 6,51 = 5.309 | 0.0003 | F 6,51 = 2.217 | 0.0562 |
| Time | F 6,52 = 110.3 | <0.0001 | F 6,51 = 136.6 | <0.0001 | F 6,51 = 29.92 | <0.0001 | |
| Group | F 1,52 = 3.192 | 0.0798 | F 1,51 = 13.38 | 0.0006 | F 1,51 = 5.724 | 0.0205 | |
| SHR P10 | Interaction | F 6,54 = 1.468 | 0.2068 | F 6,54 = 1.105 | 0.3718 | F 6,54 = 2.231 | 0.0538 |
| Time | F 6,54 = 190.9 | <0.0001 | F 6,54 = 62.02 | <0.0001 | F 6,54 = 33.53 | <0.0001 | |
| Group | F 1,54 = 0.1834 | 0.6701 | F 1,54 = 9.511 | 0.0032 | F 1,54 = 9.711 | 0.0029 | |
| Wistar P30 | Interaction | F 6,45 = 1.821 | 0.1164 | F 6,53 = 2.128 | 0.0652 | F 6,51 = 0.9615 | 0.4605 |
| Time | F 6,45 = 100.4 | <0.0001 | F 6,53 = 55.36 | <0.0001 | F 6,51 = 32.53 | 0.0001 | |
| Group | F 1,45 = 3.114 | 0.0844 | F 1,53 = 12.31 | 0.0009 | F 1,51 = 5.194 | 0.0269 | |
| SHR P30 | Interaction | F 6,49 = 2.785 | 0.0207 | F 6,48 = 1.955 | 0.0909 | F 6,48 = 2.036 | 0.0788 |
| Time | F 6,49 = 116.1 | <0.0001 | F 6,48 = 102.7 | <0.0001 | F 6,48 = 30.21 | <0.0001 | |
| Group | F 1,49 = 2.027 | 0.1609 | F 1,48 = 3.781 | 0.0577 | F 1,48 = 0.4667 | 0.4978 | |
Daily profiles of clock gene expression in the suprachiasmatic nuclei (SCN) of 10‐day‐old (P10) and 30‐day‐old (P30) Wistar rat and SHR pups reared by their own mothers (controls) or exposed to cross‐strain fostering (cross‐strain). F test distribution ratio (F), degrees of freedom in the variance of numerator (DFn) and denominator (DFd), P value (P). Significant differences are indicated in bold.
A comparison of the clock gene expression profiles' acrophases (Fig. 2 C) between the pups of both rat strains reared by their genetic mothers showed that, at P30, the expression of all genes was significantly phase advanced in SHR compared to that in Wistar rat pups, although the advances were not significant at P10. These results demonstrate that the positive phase angle of entrainment of the locomotor activity rhythm observed in the adult SHR (Sladek et al. 2012) develops in the pups already by P30. Exposure of 10‐day‐old Wistar pups (Fig. 2 A) to the cross‐strain fostering significantly modified the clock gene expression profiles of all three tested clock genes (i.e. Per2, Nr1d1 and Bmal1) compared to the control groups (Fig. 2 A and Table 1); the acrophase (Fig. 2 C) in the cross‐strain fostered Wistar pups was advanced compared to the pups reared by their own mothers significantly for Per2 (9.2 ± 0.2 h vs. 8.2 ± 0.3 h) but not significantly for Nr1d1 (2.9 ± 0.2 vs. 2.1 ± 0.2 h) and Bmal1 (18.1 ± 0.4 vs. 16.9 ± 0.4 h) expression profiles. At P30, the cross‐strain fostering did not affect the SCN of Wistar rat pups (Fig. 2 B and C). By contrast to Wistar rat pups, the cross‐strain fostering had no effect on the phase of any of the clock gene expression profiles of SHR pups at P10 (Fig. 2 A and C) or at P30 (Fig. 2 B and C). However, at P10, there was a significant effect of the cross‐strain fostering on the amplitude of the Bmal1 expression rhythm (Fig. 2 A), which significantly increased from 0.2335 ± 0.0292 in controls to 0.3618 ± 0.0329 in cross‐strain fostered pups (P = 0.0194); this difference disappeared at P30.
These results demonstrate that, in both rat strains, the effect of cross‐strain fostering on the pup SCN clock was already significant at P10: In Wistar rat pups, SHR foster mother phase‐advanced the clock by ∼1 h, whereas, in SHR pups, Wistar rat foster mother increased the amplitude of the Bmal1 rhythm.
Cross‐strain fostering robustly affects the peripheral circadian clocks of pups in age‐ and strain‐specific manners
To assess the impact of cross‐strain fostering on pup peripheral clocks, we studied the daily profiles of Per1, Per2, Nr1d1 and Bmal1 expression in the liver (Fig. 3) and colon (Fig. 4) of the same pups that were used to detect the effects of cross‐strain fostering on the SCN clock. For the results of the statistical comparisons between the groups by two‐way ANOVA (Table 2), the results of post hoc comparisons between the groups are shown in Figs 3 and 4.
Figure 3. The effect of cross‐strain fostering on clock gene expression in the liver of pups.
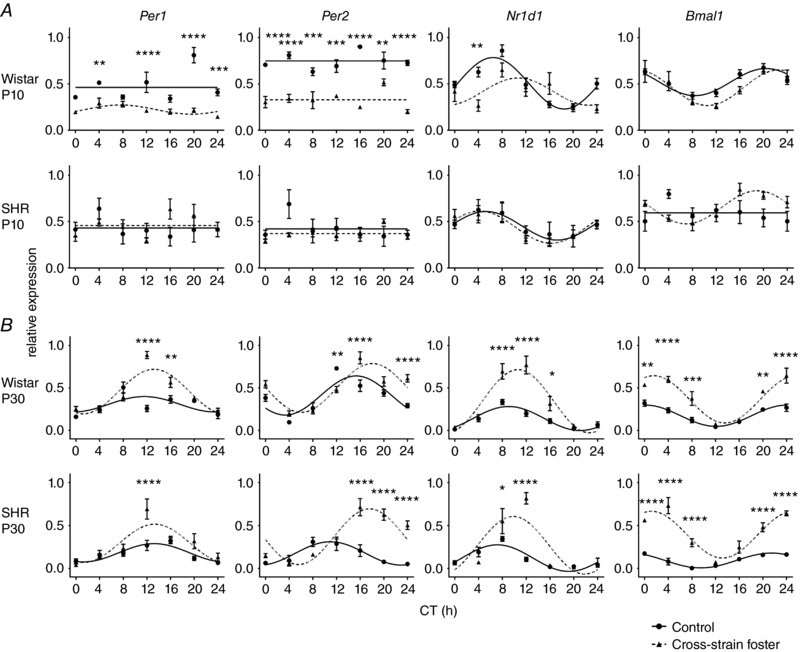
Daily Per1, Per2, Nr1d1 and Bmal1 relative expression profiles were detected in 10‐day‐old (A) and 30‐day‐old (B) Wistar rat or SHR pups reared by their genetic mothers as control (full circles, full lines) or exposed to cross‐strain fostering (full triangles, dashed lines). The pups were killed at 4 h intervals during 24 h. Time is expressed as circadian time (CT); CT0 represents the time of lights on in the previous LD 12:12 h cycle. The data are expressed as the mean ± SEM (n = 3–5 per time point). The results of two‐way ANOVA (Table 2) post hoc analyses (Šidák's multiple comparisons) of differences in the expression levels between the control and cross‐strain fostered groups at individual CT are depicted. * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
Figure 4. The effect of cross‐strain fostering on the clock gene expression in the colon of pups.
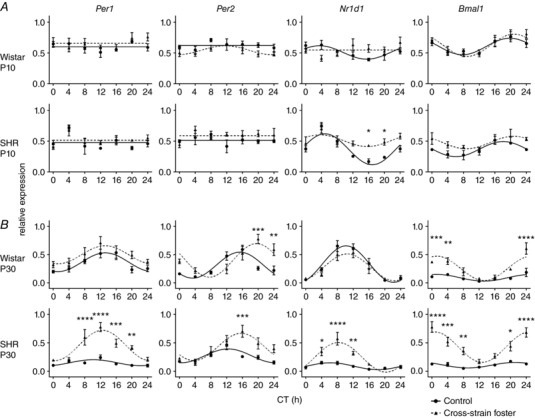
Daily Per1, Per2, Nr1d1 and Bmal1 expression profiles were detected in 10‐day‐old (A) and 30‐day‐old (B) Wistar rat or SHR pups reared by their genetic mothers as control (full circles, full lines) or exposed to cross‐strain fostering (full triangles, dashed lines). The pups were sacrificed in 4‐h intervals over 24 h. Time is expressed as circadian time (CT); CT0 represents the time of lights on in the previous LD12:12 cycle. The data are expressed as the mean ± SEM, n = 3−5/time point. For statistical comparisons, see Table 3.
Table 2.
Results of two‐way ANOVA comparison between the daily profiles of Per1, Per2, Nr1d1 and Bmal1 expression in the pup liver
| Per1 | Per2 | Nr1d1 | Bmal1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Liver | F(DFn. DFd) | P | F(DFn. DFd) | P | F(DFn. DFd) | P | F(DFn. DFd) | P | ||
| P10 | Wistar controls vs. cross‐strain | Interaction | F 6,55 = 7.083 | <0.0001 | F 6,54 = 4.636 | 0.0007 | F 6,54 = 4.012 | 0.0022 | F 6,55 = 1.115 | 0.3655 |
| Time | F 6,55 = 7.846 | <0.0001 | F 6,54 = 3.350 | 0.0070 | F 6,54 = 9.820 | <0.0001 | F 6,55 = 10.43 | <0.0001 | ||
| Group | F 1,55 = 115.9 | <0.0001 | F 1,54 = 274.8 | <0.0001 | F 1,54 = 7.621 | 0.0079 | F 1,55 = 2.501 | 0.1195 | ||
| SHR controls vs. cross‐strain | Interaction | F 6,49 = 1.702 | 0.1405 | F 6,49 = 2.246 | 0.0541 | F 6,49 = 0.3672 | 0.8962 | F 6,47 = 3.731 | 0.0041 | |
| Time | F 6,49 = 1.659 | 0.1513 | F 6,49 = 1.912 | 0.0975 | F 6,49 = 4.793 | 0.0006 | F 6,47 = 1.432 | 0.2227 | ||
| Group | F 1,49 = 0.4769 | 0.4931 | F 1,49 = 1.791 | 0.1869 | F 1,49 = 0.0999 | 0.7533 | F 1,47 = 3.272 | 0.0769 | ||
| P30 | Wistar controls vs. cross‐strain | Interaction | F 6,54 = 16.68 | <0.0001 | F 6,54 = 9.325 | 0.0001 | F 6,54 = 12.07 | 0.0001 | F 6,54 = 6.609 | <0.0001 |
| Time | F 6,54 = 25.20 | <0.0001 | F 6,54 = 34.65 | 0.0001 | F 6,54 = 42.71 | <0.0001 | F 6,54 = 33.10 | <0.0001 | ||
| Group | F 1,54 = 35.95 | <0.0001 | F 1,54 = 20.09 | <0.0001 | F 1,54 = 54.16 | <0.0001 | F 1,54 = 101.7 | <0.0001 | ||
| Wistar controls vs. cross‐strain return | Interaction | F 6,56 = 22.80 | <0.0001 | F 6,56 = 4.001 | 0.0021 | F 6,56 = 13.04 | <0.0001 | F 6,56 = 3.657 | 0.0039 | |
| Time | F 6,56 = 25.65 | <0.0001 | F 6,56 = 39.05 | 0.0001 | F 6,56 = 105.0 | <0.0001 | F 6,56 = 31.00 | <0.0001 | ||
| Group | F 1,56 = 11.06 | 0.0016 | F 1,56 = 0.0161 | 0.8994 | F 1,56 = 9.823 | 0.0027 | F 1,56 = 20.00 | <0.0001 | ||
| Wistar controls vs. intra‐strain | Interaction | F 6,53 = 7.124 | <0.0001 | F 6,55 = 4.832 | 0.0005 | F 6,54 = 14.26 | <0.0001 | F 6,55 = 1.739 | 0.1292 | |
| Time | F 6,53 = 8.803 | <0.0001 | F 6,55 = 34.64 | 0.0001 | F 6,54 = 82.71 | <0.0001 | F 6,55 = 17.11 | <0.0001 | ||
| Group | F 1,53 = 0.9994 | 0.3220 | F 1,55 = 1.586 | 0.2132 | F 1,54 = 39.29 | <0.0001 | F 1,55 = 6.884 | 0.0112 | ||
| SHR controls vs. cross‐strain | Interaction | F 6,48 = 5.162 | 0.0004 | F 6,48 = 13.91 | <0.0001 | F 6,48 = 13.02 | <0.0001 | F 6,48 = 11.87 | 0.0001 | |
| Time | F 6,48 = 16.24 | <0.0001 | F 6,48 = 11.81 | <0.0001 | F 6,48 = 26.93 | <0.0001 | F 6,48 = 23.09 | 0.0001 | ||
| Group | F 1,48 = 14.13 | 0.0005 | F 1,48 = 49.75 | <0.0001 | F 1,48 = 16.61 | 0.0002 | F 1,48 = 224.9 | 0.0001 | ||
| SHR controls vs. cross‐strain return | Interaction | F 6,51 = 3.892 | 0.0028 | F 6,51 = 10.43 | <0.0001 | F 6,51 = 0.8747 | 0.5200 | F 6,51 = 1.152 | 0.3465 | |
| Time | F 6,51 = 18.00 | <0.0001 | F 6,51 = 36.87 | <0.0001 | F 6,51 = 15.89 | <0.0001 | F 6,51 = 20.56 | <0.0001 | ||
| Group | F 1,51 = 0.4959 | 0.4845 | F 1,51 = 3.921 | 0.0531 | F 1,51 = 0.5494 | 0.4620 | F 1,51 = 0.0804 | 0.7780 | ||
| SHR controls vs. intra‐strain | Interaction | F 6,50 = 4.443 | 0.0011 | F 6,47 = 7.096 | <0.0001 | F 6,49 = 4.847 | 0.0006 | F 6,48 = 6.770 | <0.0001 | |
| Time | F 6,50 = 22.19 | 0.0001 | F 6,47 = 26.66 | <0.0001 | F 6,49 = 33.60 | 0.0001 | F 6,48 = 31.03 | <0.0001 | ||
| Group | F 1,50 = 13.74 | 0.0005 | F 1,47 = 13.22 | 0.0007 | F 1,49 = 0.0017 | 0.9675 | F 1,48 = 17.18 | 0.0001 | ||
Daily profiles of clock gene expression in the liver of 10‐day‐old (P10) and 30‐day‐old (P30) Wistar rat and SHR pups reared by their own mothers (controls), the foster mother of the different strain (cross‐strain), the foster mother of the same strain (intra‐strain) or they were reared by the foster mother of the different strain from P1 until P10 and then returned to their own mother (cross‐strain return). F test distribution ratio (F), degrees of freedom in the variance of numerator (DFn) and denominator (DFd), P value (P). Significant differences are indicated in bold.
Table 3.
Results of two‐way ANOVA comparison between the daily profiles of Per1, Per2, Nr1d1 and Bmal1 expression in the pup colon
| Per1 | Per2 | Nr1d1 | Bmal1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Colon | F(DFn. DFd) | P | F(DFn. DFd) | P | F(DFn. DFd) | P | F(DFn. DFd) | P | ||
| P10 | Wistar controls vs. cross‐strain | Interaction | F 6,54 = 0.5541 | 0.7646 | F 6,54 = 0.4797 | 0.8205 | F 6,55 = 3,268 | 0.0081 | F 6,55 = 0.5623 | 0.7584 |
| Time | F 6,54 = 1,335 | 0.2580 | F 6,54 = 1,850 | 0.1067 | F 6,55 = 0.9945 | 0.4383 | F 6,55 = 7,519 | <0.0001 | ||
| Group | F 1,54 = 2,010 | 0.1620 | F 1,54 = 8,594 | 0.0049 | F 1,55 = 0.8589 | 0.3581 | F 1,55 = 0.6903 | 0.4097 | ||
| SHR controls vs. cross‐strain | Interaction | F 6,56 = 0.7186 | 0.6362 | F 6,56 = 1,102 | 0.3731 | F 6,56 = 2,497 | 0.0327 | F 6,56 = 0.5723 | 0.7506 | |
| Time | F 6,56 = 4,969 | 0.0004 | F 6,56 = 1,096 | 0.3765 | F 6,56 = 12,40 | <0.0001 | F 6,56 = 3,502 | 0.0052 | ||
| Group | F 1,56 = 1,520 | 0.2228 | F 1,56 = 4,745 | 0.0336 | F 1,56 = 20.76 | 0.0001 | F 1,56 = 13,35 | 0.0006 | ||
| P30 | Wistar controls vs. cross‐strain | Interaction | F 6,48 = 0.4782 | 0.8212 | F 6,48 = 6,481 | <0.0001 | F 6,48 = 0.6245 | 0.7097 | F 6,48 = 4,806 | 0.0007 |
| Time | F 6,48 = 6,314 | <0.0001 | F 6,48 = 8,902 | <0.0001 | F 6,48 = 25,45 | <0.0001 | F 6,48 = 14,99 | <0.0001 | ||
| Group | F 1,48 = 11,17 | 0.0016 | F 1,48 = 11,81 | 0.0012 | F 1,48 = 1,810 | 0.1848 | F 1,48 = 59,94 | <0.0001 | ||
| Wistar controls vs. cross‐strain return | Interaction | F 6,50 = 4,866 | 0.0006 | F 6,50 = 5,697 | 0.0001 | F 6,51 = 4,123 | 0.0019 | F 6,50 = 0.7635 | 0.6020 | |
| Time | F 6,50 = 8,968 | <0.0001 | F 6,50 = 8,416 | <0.0001 | F 6,51 = 16,49 | <0.0001 | F 6,50 = 12,21 | <0.0001 | ||
| Group | F 1,50 = 4,989 | 0.0300 | F 1,50 = 14,00 | 0.0005 | F 1,51 = 18,23 | <0.0001 | F 1,50 = 0.4523 | 0.5043 | ||
| SHR controls vs. cross‐strain | Interaction | F 6,48 = 6,239 | <0.0001 | F 6,48 = 2,923 | 0.0164 | F 6,48 = 7,680 | <0.0001 | F 6,48 = 6,294 | <0.0001 | |
| Time | F 6,48 = 11,09 | <0.0001 | F 6,48 = 7,394 | <0.0001 | F 6,48 = 15,89 | <0.0001 | F 6,48 = 9,065 | <0.0001 | ||
| Group | F 1,48 = 80.35 | <0.0001 | F 1,48 = 19,00 | <0.0001 | F 1,48 = 33,66 | <0.0001 | F 1,48 = 131,6 | <0.0001 | ||
| SHR controls vs. cross‐strain return | Interaction | F 6,49 = 3,026 | 0.0135 | F 6,50 = 1,294 | 0.2772 | F 6,50 = 2,008 | 0.0820 | F 6,50 = 0.3778 | 0.8897 | |
| Time | F 6,49 = 13,30 | <0.0001 | F 6,50 = 22,05 | <0.0001 | F 6,50 = 13,34 | <0.0001 | F 6,50 = 8,787 | <0.0001 | ||
| Group | F 1,49 = 1,121 | 0.2948 | F 1,50 = 48,84 | <0.0001 | F 1,50 = 5,921 | 0.0186 | F 1,50 = 2,825 | 0.0990 | ||
Daily profiles of clock gene expression in the liver of 10‐day‐old (P10) and 30‐day‐old (P30) Wistar rat and SHR pups reared by their own mothers (controls), the foster mother of the different strain (cross‐strain), or they were reared by the foster mother of the different strain from P1 until P10 and then returned to their own mother (cross‐strain return). F test distribution ratio (F), degrees of freedom in the variance of numerator (DFn) and denominator (DFd), P value (P). Significant differences are indicated in bold.
In the liver of 10‐day‐old Wistar pups (Fig. 3 A), cross‐strain fostering significantly down‐regulated overall Per1 and Per2 expression levels and decreased the amplitude of Nr1d1 expression rhythm, although the rhythm of Bmal1 expression was not affected. By marked contrast, cross‐strain fostering had no significant effect on the expression profile of any of the studied genes in the liver of 10‐day‐old SHR pups (Fig. 3 A). In 30‐day‐old pups (Fig. 3 B), cross‐strain fostering affected the clock gene expression profiles in a similar manner in both strains, in that the amplitudes of the expression rhythms of all of the genes studied (Fig. 5 A; cross‐strain) and/or their expression levels (Fig. 3 A) were increased.
Figure 5. Amplitudes of the cosine fits of the Per1, Per2, Nr1d1 and Bmal1 expression profiles.
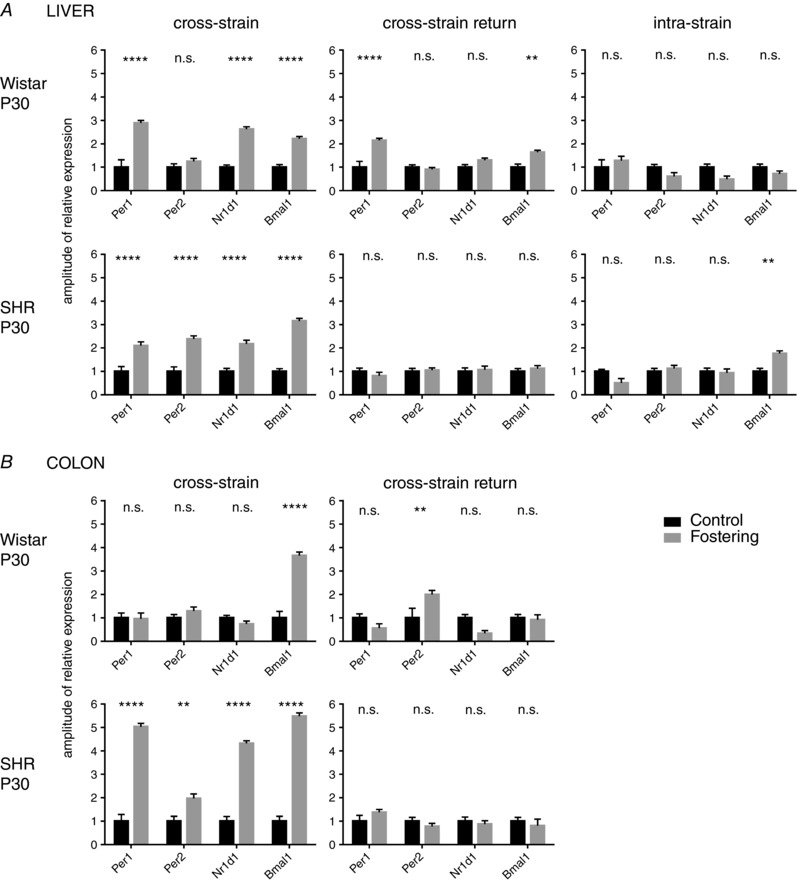
Amplitudes of the cosine fits of the Per1, Per2, Nr1d1 and Bmal1 expression profiles in the liver (A) and colon (B) of 30‐day‐old (P30) Wistar rat and SHR pups. For each clock gene, the amplitudes from pups reared by their own mother (control) were normalized and compared with profiles from pups reared by a foster mother of the other (cross‐strain fostering) or same (intra‐strain fostering) strain or with pups who were first exposed to cross‐strain fostering and at 10 days of age returned back to their genetic mother (cross‐strain return). The data are expressed as the mean ± SEM. The results of comparisons between groups by two‐way ANOVA with Šidák's multiple comparisons test are shown. ** P < 0.01; *** P < 0.001; **** P < 0.0001.
Unlike the hepatic clock, the clock in the colon of 10‐day‐old Wistar pups was not sensitive to cross‐strain fostering (Fig. 4 A) because none of the studied gene expression profiles was significantly affected. In 10‐day‐old SHR pups, cross‐strain fostering selectively dampened the amplitude of the Nr1d1 expression profile at the same time as leaving the expression of the other clock genes unaffected (Fig. 4 A). By contrast, at P30, the effect of cross‐strain fostering was more pronounced and was similar to the effect seen in the liver (i.e. cross‐strain fostering increased the amplitudes of expression profiles of all studied clock genes in SHR pups) (Figs 4 B and 5 B; cross‐strain) and had a less dramatic effect in Wistar pups, where it significantly increased only the amplitude of Bmal1 expression rhythm (Fig. 5 B; cross‐strain) and also modified the profile of Per2 (Fig. 4 B). Because our previous results demonstrated that, in adult SHR, the amplitudes of clock gene expression in the colon were significantly lower compared to Wistar rats (Sladek et al. 2012), we performed an additional comparison of the amplitudes in the colon of 30‐day‐old control and cross‐strain fostered pups of both strains. The results of this comparison (Fig. 6) revealed that, already at P30, the rhythm amplitudes of Per1 and Nr1d1 in control SHR were significantly lower than in Wistar rats, as previously reported in adults (Sládek et al. 2012), and the cross‐strain fostering significantly increased the amplitudes for all genes (with the exception of Per1 for which the amplitude was increased non‐significantly). For Wistar rat pups, the comparison confirmed the significant effect of cross‐strain fostering only on amplitude of the Bmal1 expression rhythm (Fig. 5 B; cross‐foster).
Figure 6. Comparisons of the amplitudes of the cosine fits of the Per1, Per2, Nr1d1 and Bmal1 expression profiles.
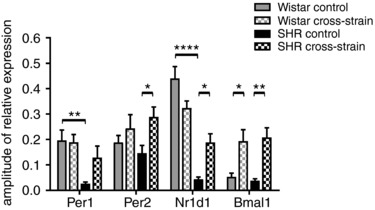
Comparisons of the amplitudes of the cosine fits of the Per1, Per2, Nr1d1 and Bmal1 expression profiles in the colon between 30‐day‐old Wistar rat and SHR pups that were reared by their own mothers (control) or by a foster mother of a different strain (cross‐strain). The data are expressed as the mean ± SEM. The results of comparisons between groups by two‐way ANOVA with Tukey's multiple comparisons test are shown. * P < 0.05; ** P < 0.01; **** P < 0.0001.
Altogether, these results demonstrate that the impact of cross‐strain fostering was dependent on age, peripheral tissue and the clock gene studied. At P10, the procedure significantly suppressed the amplitude of the hepatic clock of Wistar pups but not SHR pups and it had almost no effect on the colonic clock of both strains. Notably, at P30, this procedure had a significant impact on the amplitudes of the clock in both of the studied peripheral tissues of pups from both strains, although the effect was more pronounced in SHR. Overall, the amplitudes of the clock gene expression profiles of cross‐strain fostered 30‐day‐old pups were elevated compared to controls of the same strain and, in the colon, a Wistar rat foster mother increased the dampened amplitudes of SHR pups to the same (or higher) levels seen in Wistar rat pups.
The effect of cross‐strain fostering on the amplitudes of peripheral circadian clocks is due to the different circadian phenotype of the mother
The robust impact of cross‐strain fostering on the peripheral clocks in pups at P30 led us to examine whether the effect was a consequence of maternal care provided by a mother of a different strain or a result of the presence of a foster mother per se. Accordingly, we performed two additional experiments (see scheme in Fig. 1). In the first experiment, the pups were exposed to cross‐strain fostering at P1, as in the previous experiment, although they were returned to their own genetic mother at P10 and remained with her until P30 (cross‐strain return). In the second experiment, the pups were exposed to the fostering procedure at P1, although the foster mother was of the same strain (intra‐strain fostering) and the pups remained with the foster mother until they were sampled at P30.
The return of pups exposed to cross‐strain fostering to their genetic mothers ameliorated or abolished the effect of cross‐strain fostering on the increase in amplitudes of the peripheral clocks in a strain‐dependent manner (Fig. 5; cross‐strain return). In Wistar rat pups, the effect was blocked partially in the liver (only for Nr1d1) (Fig. 5 A) but almost completely in the colon (Fig. 5 B). However, in SHR pups, the return to their genetic mother abolished the effect of cross‐strain fostering completely both in the liver (Fig. 5 A) and colon (Fig. 5 B).
The fostering procedure per se (i.e. the exchange of a genetic mother with a foster mother of the same rat strain) (Fig. 5 A; intra‐strain) did not increase the amplitudes of the clock gene expression profiles in the liver of Wistar rats as observed because of cross‐strain fostering. In the liver of SHR, the intra‐strain fostering procedure caused only a small increase in the amplitude of Bmal1. The effect on intra‐strain fostering on the clock in the colon was not examined.
Cross‐strain fostering has a long‐lasting impact on circadian behaviour
The effect of the cross‐strain fostering procedure on the SCN‐driven behavioural activity rhythms was assessed in the rats monitored shortly after weaning: after P35 (1 month old) and again in adulthood (6 months old) (Fig. 7). The results of the statistical comparisons between the groups are shown in Fig. 7. The total activity was not affected by the cross‐strain fostering conditions, nor did it change with age under LD 12:12 h and DD (Fig. 7 A). The amplitude of the locomotor activity rhythm was measured as an activity/rest ratio (Fig. 7 B). Under LD 12:12 h, the amplitude increased significantly with age in control Wistar rats but not in control SHR. Consequently, the amplitude was not different between 1‐month‐old control Wistar rats and SHR, whereas, in 6‐month‐old rats, it was significantly higher in control Wistar rats than in SHR. The cross‐strain fostering had no effect on the amplitudes of the activity rhythms of Wistar rats at either age or in the 1‐month‐old SHR; however, it significantly increased the amplitude in 6‐month‐old SHR compared to age‐matched controls, reaching approximately the same level seen in Wistar rats. In rats maintained under DD conditions, the amplitude in control SHR was significantly lower than in Wistar rats (similar to that seen in LD12:12) but an effect of cross‐strain fostering was not present. These results suggest that the improvement in the activity/rest ratio as a result of cross‐strain fostering was related to a change in the phase angle of the rhythm relative to the LD cycle. Therefore, the phase angle of the entrainment of the locomotor activity rhythm was measured as the level of activity during the interval in the 3 h before lights off under LD 12:12 h (Fig. 7 C). This pre‐lights‐off activity was significantly increased in SHR compared to Wistar rats at 1 month of age, and even more so at 6 months of age. The cross‐strain fostering significantly decreased the pre‐lights‐off activity in SHR at 6 months of age, when activity attained the same level as that seen in Wistar rats. In 1‐month‐old SHR, an effect was suggested, although this was not significant. Therefore, the data demonstrate that a small positive phase angle of entrainment is already present in 1‐month‐old SHR and progressively develops later in adulthood. The cross‐strain fostering procedure completely blocked the development of the positive phase angle of entrainment in adulthood and thus improved the entrainment of locomotor activity rhythm to LD 12:12 h in SHR, which attained the same level of entrainment as Wistar rats. The period of free‐running locomotor activity rhythm measured in 6‐month‐old rats maintained under DD (Fig. 7 D) was significantly shorter in SHR compared to Wistar rats and was not affected by cross‐strain fostering. Therefore, in contrast to the phase, the free‐running period is a genetic trait that was not changed by maternal care in this experimental model.
Figure 7. Analyses of locomotor activity parameters of 1‐ and 6‐month‐old Wistar rats and SHR maintained under a LD regime or DD.
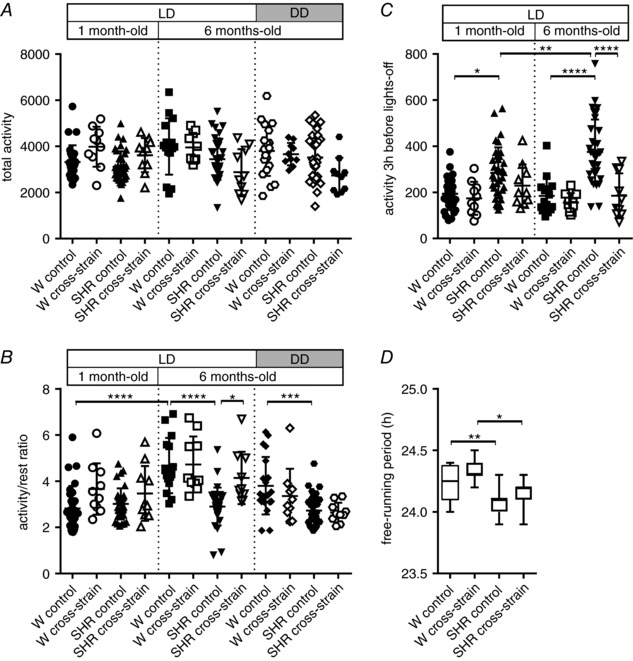
Total activity (A), activity/rest ratio (B), activity in the 3‐h interval before lights‐off (C) and free‐running period (D) were measured in rats that were reared until weaning by their own mothers (control) and in those that were exposed to cross‐strain fostering (cross‐strain). The data are expressed as the mean ± SD (n = 10–35 per group); box plots in (D) show median (line) with 25th to 75th percentile (box) and minimal to maximal value (whiskers). The results of the statistical comparisons between groups are depicted. * P < 0.05; *** P < 0.001; **** P < 0.0001.
Cross‐strain fostering affects cardiovascular function
The effects of cross‐strain fostering on HR, MAP, SBP and DBP were studied in 2‐month‐old animals. The results of the statistical comparisons are shown in Fig. 8. A two‐way ANOVA revealed that control SHR had significantly higher values in all four parameters compared to control Wistar rats. These results confirm that cardiovascular pathology developed with age in our SHR, as reported previously (Conrad et al. 1995). The maternal care of Wistar rat foster mothers significantly decreased the elevated HR of SHR, whereas the maternal care of SHR foster mothers had no effect on the parameters in Wistar rats.
Figure 8. Analyses of cardiovascular function parameters of 2‐month‐old Wistar rats and SHR maintained under a LD regime.
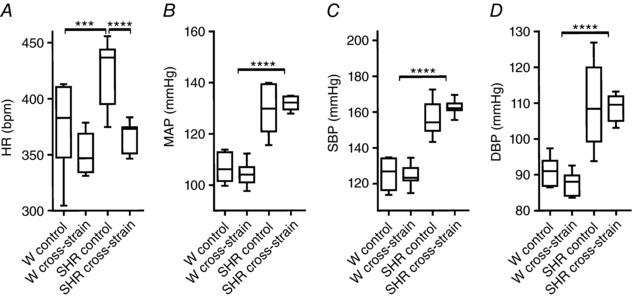
The HR (A), MAP (B), SBP (C) and DBP (D) were measured in rats that were reared until weaning by their own mothers (control) and those that were exposed to cross‐strain fostering (cross‐strain). The data are expressed as median with 25th to 75th percentile (box) and minimal to maximal value (whiskers) (n = 6–8 per group). HR is expressed as beats min–1 and blood pressure is expressed as mmHg. The results of the statistical comparisons between groups are depicted. * P < 0.05; *** P < 0.001; **** P < 0.0001.
Discussion
The results of the present study provide strong evidence for the unexpectedly robust impact of altered maternal care provided by a foster mother on developing circadian clocks and on behaviour and cardiovascular function in adulthood. Notably, altered maternal care significantly affected the developing circadian clocks despite the pups being maintained under an LD regime that is considered the dominant cue for entraining the circadian clocks, thus demonstrating the relevance of these findings under natural conditions. We found that altered maternal care affected the SCN and peripheral clocks of pups at the molecular level already during the early part of ontogenesis. Moreover, the effect of altered maternal care was exhibited in adulthood at the level of circadian behaviour and HR. In general, the more intensive maternal care of Wistar rat mothers improved most of the parameters seen with the abnormal SHR circadian phenotype and, during early developmental stages, the less frequent maternal care of SHR mothers worsened these parameters in Wistar rats.
In the present study, we found that, in Wistar rat pups, the maternal care provided by SHR foster mothers disrupted the proper entrainment of their central SCN clock to the external light/dark cycle because it significantly phase‐advanced the clock. The effect imposed on the pup's clock corresponded to the circadian phenotype of the SHR mother. In adult SHR, the phase of the SCN clock is abnormally advanced, which causes a positive phase angle of entrainment of the SCN‐driven locomotor activity rhythm relative to the external light/dark cycle. Thus, the adult SHR have already begun to be active before lights off, whereas the Wistar rats become active only at the time of lights off (Sladek et al. 2012). The magnitude of the SCN clock phase advance in 10‐day‐old Wistar pups reared by SHR foster mothers was ∼1 h (1.0 h for Per2, 0.8 h for Nr1d1 and 1.2 h for Bmal1), which is comparable to that previously observed in adult SHR (1.4 h for Per2, 0.8 h for Nr1d1 and 1.7 h for Bmal1). Therefore, the abnormal behavioural phenotype of SHR mothers was completely imprinted on the phase of the SCN clock of 10‐day‐old pups although the pups were kept in the light/dark cycle that entrains their SCN clock at this age (Mateju et al. 2009). Nevertheless, this detrimental maternal effect on the phase of the SCN clock of pups was not detectable at P30 or after weaning.
In SHR pups reared by their own mothers, the clock gene expression rhythms in the SCN spontaneously phase‐advanced between P10 and P30 and, in adulthood, their SCN drove locomotor activity rhythm that was phase‐advanced relative to the light/dark cycle. The maternal care provided to SHR pups by the Wistar rat foster mothers significantly affected this process because it completely abolished the development of the positive phase‐angle of the entrainment of the locomotor activity rhythm in adult SHR. Additionally, the maternal care of Wistar rat foster mothers facilitated the development of the amplitude of Bmal1 expression rhythm in the SCN of SHR, which has been found to be delayed in SHR compared to Wistar rats (Olejnikova et al. 2015). The cross‐strain fostering procedure also completely reversed the lower amplitude of locomotor activity rhythm found in adult SHR (Sladek et al. 2012). Therefore, the presumably better (more intensive and properly aligned) maternal care provided by Wistar rat mothers to SHR pups increased the robustness of the SCN clock‐driven rhythm and improved its entrainment to the external light/dark cycle in adulthood.
Apart from the altered circadian phenotype and maternal care, nursing regime also differs between Wistar rat and SHR dams. Not only is the presence in the nest more fragmented especially during the first few days after birth (Olejnikova et al. 2015), but also the milk production is lower in SHR dams (Rose & McCarty, 1994). Because breast milk of SHR differs in the content of electrolytes, proteins and fatty acids to Wistar rats (Mills et al. 1990; Azar et al. 1991; McCarty & Tong, 1995), the poor dietary quality of SHR breast milk might contribute to this effect of cross‐strain fostering on the hepatic circadian clock of 10‐day‐old Wistar rat pups. Perinatal protein restriction was found to alter expression of genes regulating cellular processes, including the cluster of genes that link temporal and nutritional cues to metabolism through their tight interaction with the circadian clock (Orozco‐Solis et al. 2011). Changes in maternal feeding regime have been shown to have a significant effect on the developing peripheral clocks located in the liver (Sladek et al. 2007a) and colon (Polidarova et al. 2013). Consistent with these findings, we found a significant negative impact of the abnormal maternal care of the SHR mother on the circadian clocks in the liver (but not on the colon) of Wistar rat pups at P10; cross‐strain fostering robustly down‐regulated the expression of Per1, Per2 and Nr1d1 genes. However, the more intensive maternal care provided by the foster Wistar rat mothers to SHR pups had much smaller effect on their clocks in the liver and almost no effect on the clock in the colon at P10. Unexpectedly, in 30‐day‐old pups, cross‐strain fostering had enormous opposite impact on their peripheral clocks in both strains. The expression of most studied clock genes was robustly upregulated during the day and night and/or the amplitudes of the rhythmic expression profiles were significantly increased. The general pattern of the effect was very similar in both of the peripheral tissues tested but was more pronounced in the SHR than in the Wistar rat pups. These results led us to investigate whether the effect was caused by the presence of a foster mother of a different circadian phenotype providing altered maternal care or by the fostering procedure per se. We demonstrated that the effect was indeed specific to presence of a foster mother with a different circadian phenotype because, when the pups were reared by a foster mother of the same strain, the amplitudes of the clock gene expression profiles in the liver and colon did not change at P30, with the exception of a small increase in the amplitude of Bmal1 of SHR pups. Moreover, returning the pups of both strains exposed to cross‐strain fostering to their genetic mothers at P10 prevented the increase in the amplitudes of the expression rhythms in the liver and colon of these pups (in Wistar rat pups, the prevention was partial, whereas in SHR pups, it was complete). These results revealed that, in contrast to the effect on the SCN clock, the peripheral clocks responded more robustly to cross‐strain fostering, with the response being stronger in SHR than Wistar rats. The interval between P10 and P30 represents a developmental period during which pups gain sight and, together with a gradual decline in breast‐milk intake, start to consume solid food at night, which affects the phases of their peripheral clocks (Sladek et al. 2007a; Polidarova et al. 2014). Importantly, dietary factors were probably not involved in the effects of cross‐strain fostering on peripheral clocks observed after weaning because cross‐strain fostering induced (and return to genetic mother abolished) these effects in pups of both rat strains. It appears that, despite a gradual weaning of maternal breast feeding, the presence of a mother whose circadian system is not aligned with the genetically programmed pups' circadian system during this interval still has a significant impact on the pups' peripheral molecular clocks. The greater response of the peripheral clocks of SHR to cross‐strain fostering is in agreement with previous studies reporting that this strain exhibits a significantly higher sensitivity to environmental cues (Hausler et al. 1983), as well as to changes in feeding regime not only in adulthood (Polidarova et al. 2013), but also during ontogenesis (Olejnikova et al. 2015). The generally higher sensitivity of the autonomous nervous system in SHR (Hausler et al. 1983) may play a role in the higher sensitivity of their peripheral clocks because the activity of the nervous system is considered causal in their cardiovascular pathology. It innervates these peripheral tissues and, furthermore, the clocks in the liver (Terazono et al. 2003) and colon (Malloy et al. 2012) are directly responsive to changes in adrenergic tonus. Apart from sensitivity to feeding regime, SHR also differ in sleep patterns (Carley et al. 1996). The altered sleep pattern of a foster mother may also affect the sleep patterns of her pups, which could account for the observed effects on the circadian clocks. Importantly, the circadian phenotype of the Wistar rat foster mother remedied the dampened amplitudes of the colonic clock in SHR pups. The beneficial effect of the care provided by the Wistar rat foster mothers for SHR pups was also confirmed by cardiovascular function measurements. Our data show that cross‐strain fostering normalized the increased HR in 2‐month‐old SHR to the Wistar rat control levels, although it did not affect blood pressure (MAP, SBP and DBP). The cross‐strain fostering had no effect on any of these parameters in normotensive control pups. The low birth weight accompanied by catch‐up growth during childhood has been associated with the development of hypertension in adulthood (Eriksson et al. 2000). It is possible that the SHR pups, who are born with a lower body weight, experienced a catch‐up growth as a result of cross‐strain fostering. The absence of the effect of cross‐strain fostering on blood pressure contrasts with previous studies demonstrating a reduction in blood pressure as a result of cross‐strain fostering (McCarty et al. 1992; Gouldsborough & Ashton, 1998). Siew‐Keah et al. (2014) reported that, in SHR, cross‐strain fostering significantly postponed the increase in SBP with age. In the present study, such effect was not observed. Importantly, it should be noted that the effect of cross‐strain fostering on SHR blood pressure is contradictory because, in accordance with our results, the effect was also not observed in the original study by McMurtry et al. (1981). The discordance may result from a different methodology of blood pressure measurement (i.e. indirect tail‐cuff method) (Siew‐Keah et al. 2014) vs. our direct blood pressure measurement using catheters inserted into the carotid arteries of conscious rats. Additionally, we cannot completely exclude the possibility that the different outcome was a result of variability in SHR colonies and/or the use of a different strain of normotensive controls, or even that the presumed delay in blood pressure rise occurred earlier and was thus missed in our cross‐strain fostered SHR. Nevertheless, our finding that cross‐strain fostering protected the HR rise in SHR confirms its positive effect of on SHR cardiovascular functions.
In conclusion, although the importance of the maternal circadian system in the entrainment of the rat SCN has been suggested previously (Reppert & Schwartz, 1986; Reppert et al. 1988; Ohta et al. 2002; Varcoe et al. 2013), in the present study, we demonstrate that the maternal care of a mother with misaligned circadian clocks may be a strong cue positively or negatively affecting the circadian clocks in the SCN and the periphery of pups and may also have long lasting effects on the circadian regulation of behaviour later in adulthood. It is tempting to speculate that the effects of altered maternal behaviour on the circadian system, as well as other physiological parameters, may be mediated via epigenetic programming because maternal care (grooming and licking behaviour) is known to have an impact on the DNA methylation of rat pups (Weaver et al. 2004). Additionally, the rat model of nurturing was previously suggested to support the hypothesis of a developmental origin of health and disease (DOHaD hypothesis) based on empirical observations of effects of nutrition and stress during perinatal period on outcomes in childhood and adulthood. Potential epigenetic mechanisms that may underlie these observations and theory have been suggested (Wadhwa et al. 2009). Altogether, our data provide compelling evidence that the maternal care of a foster mother of a different circadian phenotype may have a significant impact on the regulatory mechanisms of various physiological parameters, even if the pathological symptoms are genetically programmed.
Additional information
Competing interests
The authors declare that they have no competing interests.
Author contributions
L.O. performed experiments, analyzed data and prepared figures; L.P. performed experiments; M.B. performed blood pressure measurements; M.S. analyzed locomotor activity data, prepared figures, performed statistical analyses; A.S conceived and designed the study and wrote the manuscript; L.O.; L.P., M.B., M.S., A.S. approved final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
The study was supported by the Czech Science Foundation grant No. 16‐03932S (to AS) and Research Project RV0: 67985823.
Acknowledgements
The authors thank to Eva Suchanova and Lucie Heppnerova for their excellent technical help.
Edited by: Laura Bennet & Dino Giussani
References
- Aton SJ, Colwell CS, Harmar AJ, Waschek J & Herzog ED (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azar SH, Hensleigh HC, Matthys E & Azar MM (1991). Oviductal‐uterine and nursing environment alter blood pressure development in spontaneously hypertensive and normotensive rats. J Hypertens Suppl 9, S294–295. [PubMed] [Google Scholar]
- Balsalobre A, Damiola F & Schibler U (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. [DOI] [PubMed] [Google Scholar]
- Behuliak M, Vavrinova A, Bencze M, Polgarova K, Ergang P, Kunes J, Vaneckova I & Zicha J (2015). Ontogenetic changes in contribution of calcium sensitization and calcium entry to blood pressure maintenance of Wistar‐Kyoto and spontaneously hypertensive rats. J Hypertens 33, 2443–2454. [DOI] [PubMed] [Google Scholar]
- Carley DW, Trbovic S & Radulovacki M (1996). Sleep apnea in normal and REM sleep‐deprived normotensive Wistar‐Kyoto and spontaneously hypertensive (SHR) rats. Physiol Behav 59, 827–831. [DOI] [PubMed] [Google Scholar]
- Conrad CH, Brooks WW, Hayes JA, Sen S, Robinson KG & Bing OH (1995). Myocardial fibrosis and stiffness with hypertrophy and heart failure in the spontaneously hypertensive rat. Circulation 91, 161–170. [DOI] [PubMed] [Google Scholar]
- Cui H, Kohsaka A, Waki H, Bhuiyan ME, Gouraud SS & Maeda M (2011). Metabolic cycles are linked to the cardiovascular diurnal rhythm in rats with essential hypertension. PLoS ONE 6, e17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Forsen T, Tuomilehto J, Osmond C & Barker D (2000). Fetal and childhood growth and hypertension in adult life. Hypertension 36, 790–794. [DOI] [PubMed] [Google Scholar]
- Gouldsborough I & Ashton N (1998). Effect of cross‐fostering on neonatal sodium balance and adult blood pressure in the spontaneously hypertensive rat. Clin Exp Pharmacol Physiol 25, 1024–1031. [DOI] [PubMed] [Google Scholar]
- Gouldsborough I, Black V, Johnson IT & Ashton N (1998). Maternal nursing behaviour and the delivery of milk to the neonatal spontaneously hypertensive rat. Acta Physiol Scand 162, 107–114. [DOI] [PubMed] [Google Scholar]
- Grundy D (2015). Principles and standards for reporting animal experiments in The Journal of Physiology and Experimental Physiology. J Physiol 593, 2547–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Lehman MN & Bittman EL (2006). Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker. J Neurosci 26, 6406–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Kabuta T & Wada K (2017). Disturbance in maternal environment leads to abnormal synaptic instability during neuronal circuitry development. Front Neurosci 11, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler A, Girard J, Baumann JB, Ruch W & Otten UH (1983). Stress‐induced secretion of ACTH and corticosterone during development of spontaneous hypertension in rats. Clin Exp Hypertens A 5, 11–19. [DOI] [PubMed] [Google Scholar]
- Houdek P, Novakova M, Polidarova L, Sladek M & Sumova A (2016). Melatonin is a redundant entraining signal in the rat circadian system. Horm Behav 83, 1–5. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Francis DD, Mar A & Meaney MJ (2003). Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiol Behav 79, 359–371. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ, Panarelli M, Holloway CD, Dunlop D, Morton JJ, Connell JM & Fraser R (1993). The role of glucocorticoid activity in the inheritance of hypertension: studies in the rat. J Steroid Biochem Mol Biol 45, 7–11. [DOI] [PubMed] [Google Scholar]
- Kovacikova Z, Sladek M, Bendova Z, Illnerova H & Sumova A (2006). Expression of clock and clock‐driven genes in the rat suprachiasmatic nucleus during late fetal and early postnatal development. J Biol Rhythms 21, 140–148. [DOI] [PubMed] [Google Scholar]
- Landgraf D, Achten C, Dallmann F & Oster H (2014). Embryonic development and maternal regulation of murine circadian clock function. Chronobiol Int, 1–12. [DOI] [PubMed] [Google Scholar]
- Malloy JN, Paulose JK, Li Y & Cassone VM (2012). Circadian rhythms of gastrointestinal function are regulated by both central and peripheral oscillators. Am J Physiol Gastrointest Liver Physiol 303, G461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco EM, Velarde E, Llorente R & Laviola G (2016). Disrupted circadian rhythm as a common player in developmental models of neuropsychiatric disorders. Curr Top Behav Neurosci 29, 155–181. [DOI] [PubMed] [Google Scholar]
- Mateju K, Bendova Z, El‐Hennamy R, Sladek M, Sosniyenko S & Sumova A (2009). Development of the light sensitivity of the clock genes Period1 and Period2, and immediate‐early gene c‐fos within the rat suprachiasmatic nucleus. Eur J Neurosci 29, 490–501. [DOI] [PubMed] [Google Scholar]
- McCarty R, Cierpial MA, Murphy CA, Lee JH & Fields‐Okotcha C (1992). Maternal involvement in the development of cardiovascular phenotype. Experientia 48, 315–322. [DOI] [PubMed] [Google Scholar]
- McCarty R & Tong H (1995). Development of hypertension in spontaneously hypertensive rats: role of milk electrolytes. Clin Exp Pharmacol Physiol Suppl 22, S215–217. [DOI] [PubMed] [Google Scholar]
- McMurtry JP, Wright GL & Wexler BC (1981). Spontaneous hypertension ion cross‐suckled rats. Science 211, 1173–1175. [DOI] [PubMed] [Google Scholar]
- Meszaros K, Pruess L, Szabo AJ, Gondan M, Ritz E & Schaefer F (2014). Development of the circadian clockwork in the kidney. Kidney Int 86, 915–922. [DOI] [PubMed] [Google Scholar]
- Mills DE, Ward RP & Huang YS (1990). Fatty acid composition of milk from genetically normotensive and hypertensive rats. J Nutr 120, 431–435. [DOI] [PubMed] [Google Scholar]
- Muhlbauer E, Wolgast S, Finckh U, Peschke D & Peschke E (2004). Indication of circadian oscillations in the rat pancreas. FEBS Lett 564, 91–96. [DOI] [PubMed] [Google Scholar]
- Ohta H, Honma S, Abe H & Honma K (2002). Effects of nursing mothers on rPer1 and rPer2 circadian expressions in the neonatal rat suprachiasmatic nuclei vary with developmental stage. Eur J Neurosci 15, 1953–1960. [DOI] [PubMed] [Google Scholar]
- Okamoto K & Aoki K (1963). Development of a strain of spontaneously hypertensive rats. Jpn Circ J 27, 282–293. [DOI] [PubMed] [Google Scholar]
- Olejnikova L, Polidarova L, Pauslyova L, Sladek M & Sumova A (2015). Diverse development and higher sensitivity of the circadian clocks to changes in maternal‐feeding regime in a rat model of cardio‐metabolic disease. Chronobiol Int 32, 531–547. [DOI] [PubMed] [Google Scholar]
- Orozco‐Solis R, Matos RJ, Lopes de Souza S, Grit I, Kaeffer B, Manhaes de Castro R & Bolanos‐Jimenez F (2011). Perinatal nutrient restriction induces long‐lasting alterations in the circadian expression pattern of genes regulating food intake and energy metabolism. Int J Obes (Lond) 35, 990–1000. [DOI] [PubMed] [Google Scholar]
- Polidarova L, Olejnikova L, Pauslyova L, Sladek M, Sotak M, Pacha J & Sumova A (2014). Development and entrainment of the colonic circadian clock during ontogenesis. Am J Physiol Gastrointest Liver Physiol 306, G346–356. [DOI] [PubMed] [Google Scholar]
- Polidarova L, Sladek M, Novakova M, Parkanova D & Sumova A (2013). Increased sensitivity of the circadian system to temporal changes in the feeding regime of spontaneously hypertensive rats ‐ a potential role for Bmal2 in the liver. PLoS ONE 8, e75690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidarova L, Sladek M, Sotak M, Pacha J & Sumova A (2011). Hepatic, duodenal, and colonic circadian clocks differ in their persistence under conditions of constant light and in their entrainment by restricted feeding. Chronobiol Int 28, 204–215. [DOI] [PubMed] [Google Scholar]
- Pravenec M, Zidek V, Landa V, Simakova M, Mlejnek P, Kazdova L, Bila V, Krenova D & Kren V (2004). Genetic analysis of “metabolic syndrome” in the spontaneously hypertensive rat. Physiol Res 53(Suppl 1), S15–22. [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC & Menaker M (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978. [DOI] [PubMed] [Google Scholar]
- Reppert SM & Schwartz WJ (1986). Maternal suprachiasmatic nuclei are necessary for maternal coordination of the developing circadian system. J Neurosci 6, 2724–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM & Weaver DR (2001). Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63, 647–676. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR & Rivkees SA (1988). Maternal communication of circadian phase to the developing mammal. Psychoneuroendocrinology 13, 63–78. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM & Schibler U (2000). CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev 14, 679–689. [PMC free article] [PubMed] [Google Scholar]
- Roa SLR, Martinez EZ, Martins CS, Antonini SR, de Castro M & Moreira AC (2017). Postnatal ontogeny of the circadian expression of the adrenal clock genes and corticosterone rhythm in male rats. Endocrinology 158, 1339–1346. [DOI] [PubMed] [Google Scholar]
- Rose JL & McCarty R (1994). Maternal influences on milk intake in SHR and WKY pups. Physiol Behav 56, 901–906. [DOI] [PubMed] [Google Scholar]
- Siew‐Keah L, Sundaram A, Sirajudeen KN, Zakaria R & Singh HJ (2014). Effect of melatonin supplementation and cross‐fostering on renal glutathione system and development of hypertension in spontaneously hypertensive rats. J Physiol Biochem 70, 73–79. [DOI] [PubMed] [Google Scholar]
- Sladek M, Jindrakova Z, Bendova Z & Sumova A (2007a). Postnatal ontogenesis of the circadian clock within the rat liver. Am J Physiol Regul Integr Comp Physiol 292, R1224–R1229. [DOI] [PubMed] [Google Scholar]
- Sladek M, Polidarova L, Novakova M, Parkanova D & Sumova A (2012). Early chronotype and tissue‐specific alterations of circadian clock function in spontaneously hypertensive rats. PLoS ONE 7, e46951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek M, Rybova M, Jindrakova Z, Zemanova Z, Polidarova L, Mrnka L, O'Neill J, Pacha J & Sumova A (2007b). Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology 133, 1240–1249. [DOI] [PubMed] [Google Scholar]
- Sladek M, Sumova A, Kovacikova Z, Bendova Z, Laurinova K & Illnerova H (2004). Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci USA 101, 6231–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y & Menaker M (2001). Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH & Weitz CJ (2002). Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83. [DOI] [PubMed] [Google Scholar]
- Sumova A, Jac M, Sladek M, Sauman I & Illnerova H (2003). Clock gene daily profiles and their phase relationship in the rat suprachiasmatic nucleus are affected by photoperiod. J Biol Rhythms 18, 134–144. [DOI] [PubMed] [Google Scholar]
- Sumova A, Sladek M, Polidarova L, Novakova M & Houdek P (2012). Circadian system from conception till adulthood. Prog Brain Res 199, 83–103. [DOI] [PubMed] [Google Scholar]
- Terazono H, Mutoh T, Yamaguchi S, Kobayashi M, Akiyama M, Udo R, Ohdo S, Okamura H & Shibata S (2003). Adrenergic regulation of clock gene expression in mouse liver. Proc Natl Acad Sci USA 100, 6795–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varcoe TJ, Boden MJ, Voultsios A, Salkeld MD, Rattanatray L & Kennaway DJ (2013). Characterisation of the maternal response to chronic phase shifts during gestation in the rat: implications for fetal metabolic programming. PLoS ONE 8, e53800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa PD, Buss C, Entringer S & Swanson JM (2009). Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med 27, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M & Meaney MJ (2004). Epigenetic programming by maternal behavior. Nat Neurosci 7, 847–854. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M & Reppert SM (1995). Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706. [DOI] [PubMed] [Google Scholar]
- Wu T, Tao Y, Tsang F, Abe K, Xu L, Jiang Q, Xu L, Fu H & Fu Z (2015). The effect of L‐carnosine on the circadian resetting of clock genes in the heart of rats. Mol Biol Rep 42, 87–94. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T & Takumi T (2004). Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M & Takahashi JS (2004). PERIOD2::LUCIFERASE real‐time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101, 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]


