Abstract
Now that induced pluripotent stem cell (iPSC)‐based transplants have been performed in humans and organizations have begun producing clinical‐grade iPSCs, it is imperative that strict quality control standards are agreed upon. This is essential as both ESCs and iPSCs have been shown to accumulate genomic aberrations during long‐term culturing. These aberrations can include copy number variations, trisomy, amplifications of chromosomal regions, deletions of chromosomal regions, loss of heterozygosity, and epigenetic abnormalities. Moreover, although the differences between iPSCs and ESCs appear largely negligible when a high enough n number is used for comparison, the reprogramming process can generate further aberrations in iPSCs, including copy number variations and deletions in tumor‐suppressor genes. If mutations or epigenetic signatures are present in parental cells, these can also be carried over into iPSCs. To maximize patient safety, we recommend a set of standards to be utilized when preparing iPSCs for clinical use. Reprogramming methods that do not involve genomic integration should be used. Cultured cells should be grown using feeder‐free and serum‐free systems to avoid animal contamination. Karyotyping, whole‐genome sequencing, gene expression analyses, and standard sterility tests should all become routine quality control tests. Analysis of mitochondrial DNA integrity, whole‐epigenome analyses, as well as single‐cell genome sequencing of large cell populations may also prove beneficial. Furthermore, clinical‐grade stem cells need to be produced under accepted regulatory good manufacturing process standards. The creation of haplobanks that provide major histocompatibility complex matching is also recommended to improve allogeneic stem cell engraftment. Stem Cells Translational Medicine 2018;7:867–875
Keywords: Cellular reprogramming, ESCs, iPSCs, Molecular cytogenetics, Quality control, Tumorigenicity
Significance Statement.
This is the first review to explicitly address the pressing need for strict quality control standards given the advent of autologous and donor induced pluripotent stem cell transplantations in two different human patients. This study focused on how to get stem cells safely from the research bench to the clinic using existing technologies. More specifically, this article reports the aberrations that commonly accumulate in stem cells during culturing, as well as the existing quality control techniques that are currently available. Recommendations are offered for quality control standards to employ when preparing embryonic and induced pluripotent stem cells for human transplantation.
Introduction
In 2006, Takahashi and Yamanaka published their seminal paper demonstrating that adult somatic cells could be restored to pluripotency through the exogenous expression of four transcription factors: Oct4, Sox2, Klf4, and c‐myc 1. These reprogrammed induced pluripotent stem cells (iPSCs) expressed classical embryonic stem cell markers, mimed ESC morphology, and could differentiate into all three germ layers 1. As iPSCs are patient‐derived, they represent an ideal stem cell source for autologous transplantation therapies. These reprogrammed cells also bridge the gap between the clinic and the laboratory with their unique ability to be used for patient‐specific disease modeling 2 and drug testing 3. Thus, iPSCs are especially appealing to the fields of precision medicine, regenerative medicine, and cell therapy. Because of the unique clinical potential of iPSCs, both the RIKEN Institute 4, 5 and the cell therapy company Lonza 6, 7 have begun producing iPSCs intended for clinical use. The RIKEN Institute, in particular, has already used iPSCs for human transplantation 4.
There are, however, serious safety concerns associated with the production of stem cells intended for clinical use. Initially, for example, it appeared that iPSCs were distinct from ESCs in a number of unflattering ways 8. Numerous studies were published indicating that iPSCs frequently harbored abnormalities compared to ESCs 8. These anomalies included epigenetic signatures reminiscent of the somatic cells they were derived from 9, premature senescence in culture 10, shortened telomeres 11, reduced telomerase activity 11, and incomplete mitochondrial remodeling 12. Such apparent iPSC‐specific defects raised serious concerns regarding the clinical utility of iPSCs, especially with regard to transplantation therapies 8. More recent data have demonstrated, however, that the differences between iPSCs and ESCs become largely negligible when a high enough n number is used for comparative studies 13. More specifically, Yamanaka demonstrated in a 2012 review that, when an n of 12 or more cell lines is used for comparison, it is difficult to consistently discern any significant differences between properly reprogrammed iPSCs and ESCs 14. Rather than iPSCs being distinct from ESCs, it appears that a large portion of the differences reported are likely due to genetic variation. An important study by Kilpinen et al. suggests that 5%–46% of all variations observed between different iPSC lines are caused by genetic differences between individuals 15. This would suggest that many of the differences reported between iPSCs and ESCs are likely due to standard variation, which bolsters the usability of iPSCs for clinical therapies.
Despite this important clarification, it remains clear that there is still significant variability between different stem cell clones derived from the same donor 8. This variation can manifest in a variety of impactful ways, such as differences in mRNA and protein expression levels of specific genes. Moreover, incomplete reprogramming or unsafe reprogramming methods may lead to both epigenetic (e.g., aberrant DNA methylation) and genetic aberrations (e.g., aneuploidy) in an iPSC line 16. Such variations raise significant clinical safety concerns with regard to the use of either ESCs or iPSCs for transplantation therapies. Corroborating these concerns, tumorigenicity is a well‐documented risk associated with pluripotent stem cell culturing and transplantation. For example, Doi et al. found that, when using ESC‐derived neural cells, remaining undifferentiated ESCs induced tumor formation when grafted into monkey brains 17. A separate group found that, when progenitors of iPSCs reprogrammed with lentiviral vectors were transplanted into immunodeficient mice, more than 90% of the recipient animals formed invasive teratocarcinoma‐like tumors 18. Conversely, tumor‐free transplantation was achieved via the combination of transgene‐free reprogramming as well as the elimination of residual stem cells 18.
Because of the concerns associated with stem cell transplantations, both the RIKEN Institute and the company Lonza have each implemented unique and strict quality control standards for the production of pluripotent stem cells intended for the clinic 4, 5, 6, 7. In humans, the RIKEN Institute used iPSCs to treat two patients with age‐related macular degeneration. The first patient in 2014 received iPSC‐retinal pigment epithelial cells derived from her own skin cells 4. The second patient in 2017 received iPSC‐retinal pigment epithelial cells derived from an anonymous donor 19. The second clinical trial was temporarily halted in 2015 after discovering a genetic abnormality in the cells used for transplantation 20.
These data make it clear that, while pluripotent stem cells show invaluable therapeutic potential for the treatment of diseases, strict quality control standards need to be in place to ensure that the cells used are clinically viable. This review summarizes the aberrations that can occur in both ESCs and iPSCs. We also review the existing methods for evaluating stem cell integrity and propose new regulatory standards to streamline the progression of stem cells from the laboratory to the clinic.
Aberrations in Embryonic Stem Cells, Multipotent Stem Cells, and Progenitor Cells
Extensive expansion of pluripotent stem cells is a prerequisite to obtain the cell numbers required for human cell‐based therapies. The process of culture adaptation can, however, activate oncogenic networks and increase tumorigenicity 21. As revealed by recent cytogenetic studies, the chromosomal stability of pluripotent stem cells through extended passaging cannot be guaranteed 22. Adaptation in culture to conditions that promote cell proliferation in vitro has a lucid parallel with malignant transformation in vivo.
In murine neural stem cells procured following the neuralization from ESCs, Diaferia et al. reported that long‐term passaging was accompanied by a composite karyotype 23. Fetal‐derived neural stem cells showed especially high levels of euploidy 23. Similarly, human neural progenitor cells have been found to be susceptible to accumulation of chromosome 7 and 19 trisomy after prolonged culturing 24. As revealed by an analysis of over 400 samples of human multipotent stem cells by Ben‐David et al. 25, chromosomal abnormalities accumulate while in culture in all several different multipotent stem cell types, including pluripotent, mesenchymal, and neural stem cells.
Numerous examples specific to human embryonic stem cells (hESCs) have demonstrated how long‐term culturing of embryonic stem cells can hamper their clinical utility. The formation of a chromosomal homogeneous staining region in a single ESC line was observed by Baker et al. after extended passaging 21. Compared to early passage lines, one study found that eight of nine late‐passage hESC lines had one or more genomic alterations typically observed in cancer 26. In all, 45%, 22%, and 90% of the lines showed aberrations in copy number, mitochondrial DNA sequence, and gene promoter methylation, respectively 26. A high‐resolution DNA analysis of 17 different hESC lines identified 843 copy number variations 27. On average for the same line, 66% of the copy number variations and 24% of the loss of heterozygosity sites changed in culture between early and late passages. Many of the genes within these sites were functionally linked to cancer and showed altered expression levels 27. Other genomic anomalies, such as the occurrence of an isodicentric X chromosome during long‐term cell culturing, have also been documented 28. A gain of chromosomes 17q and 12 29, an oncogenic amplification of 20q11.21 30, a derivative chromosome 18 31, and trisomy 32 have analogously been reported in hESCs. Amplification of 20q11.21 as well as gain of chromosomes 17 and 12 both appear to be recurrent chromosomal abnormalities in ESCs 29, 30, 31. Numerous other reports exist in the literature highlighting how cell culturing of stem cells can reduce their genomic integrity and increase their tumorigenicity 22, 32, 33, 34, 35.
Select abnormalities that have been reported to occur in hESCs are summarized in Supporting Information Table S1. From an evolutionary perspective, it is easy to understand that a selection for cells with higher growth potential also results in a selection for cells with reduced clinical utility. On a more molecular mechanistic level, a large survey of pluripotent stem cell genetic variability by the International Stem Cell Initiative has identified culture‐specific genetic modifications which increase activity of the antiapoptotic gene BCL21 36. This same survey reported that prolonged culture commonly affects the genetic integrity of chromosomes 1, 12, 17, and 20 36. Given that pluripotent stem cells regulate their genomic integrity by the elimination of damaged cells via apoptosis 37, this reduction in apoptosis is likely a key factor underlying the increased growth potential and tumorigenesis observed during long‐term culturing.
Aberrations in iPSCs
Although it was initially thought that iPSCs may harbor significantly more genetic, epigenetic, and cellular abnormalities than ESCs 8, emerging data suggest that the differences between ESCs and properly reprogrammed iPSCs are minimal 14. However, there are some concerns unique to iPSCs. First, iPSCs are derived from differentiated somatic cells that may harbor existing genomic aberrations 8 (Fig. 1). These anomalies can impact the clinical quality of the transformed iPSCs 8. Exemplar of this, Yu et al. found that iPSCs reprogrammed from somatic cells with pre‐existing chromosomal mutations showed less genetic stability than iPSCs reprogrammed from cells with normal karyotypes 38. A separate study concluded that most of the genetic variation observed in iPSC clones is a consequence of iPSCs retaining their mutational history 39. This finding is further supported by the report that iPSCs harbor the same gene expression dysregulation domains as the trisomy 21 fibroblast cells they were reprogrammed from 40. Some of the remaining abnormalities in iPSCs may also be due to incomplete reprogramming or the inability of reprogramming to reverse an existing anomaly 8 (Fig. 1). Conversely, however, the reprogramming process has been reported to rectify chromosomal abnormalities present in somatic cells 41. More specifically, patient fibroblasts containing ring chromosomes underwent cell‐autonomous corrections during reprogramming into iPSCs. The resultant stem cells lost their abnormal chromosome and replaced it with a duplication of its wild‐type homologue 41. Future studies are warranted to understand what defects can and cannot be corrected by cellular reprogramming.
Figure 1.
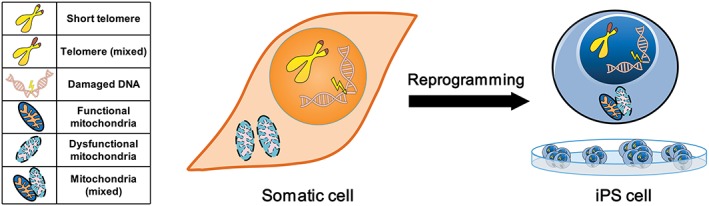
Potential anomalies can exist in induced pluripotent stem cells (iPSCs) due to pre‐existing aberrations. An aged somatic cell can accumulate aberrations such as short telomeres, damaged DNA, and dysfunctional mitochondria. During reprogramming of somatic cells into iPSCs, some of these anomalies can be fully reversed. Other anomalies, due to incomplete reprogramming or the inability of reprogramming to fully reverse an abnormality, can result in the generation of iPSCs with aberrations (e.g., shorter telomeres, dysfunctional mitochondria, and damaged DNA) uncharacteristic of pluripotent stem cells.
Second and likely the larger concern, the reprogramming process itself can generate several different anomalies due to the genomic insertion of viral vectors, global hypomethylation, the overexpression of oncogenic transcription factors, the inhibition of tumor suppressors, and the activation of oncogenes (Fig. 2) 42, 43. The formation of de novo mutations, such as copy number variations and deletions of tumor‐suppressor genes, has been reported to occur as a result of reprogramming to pluripotency 33, 44. Replicative stress associated with reprogramming underlies at least some of these de novo genetic changes 45. Lowering replication stress, either chemically or genetically, has been shown to increase the genomic stability of reprogrammed iPSCs 45. In addition to replicative stress, chromatin remodeling into an open state, which occurs during the somatic to pluripotent transition, can cause aberrations in iPSCs. (Fig. 2) 46. Epigenetic aberrations and anomalous methylation patterns caused by reprogramming have also been reported in iPSCs 37. As global hypomethylation is significantly associated with both aging and cancer development (Fig. 2) 47, any mistakes in the epigenetic remodeling process could impact the genomic stability of the resultant iPSCs.
Figure 2.
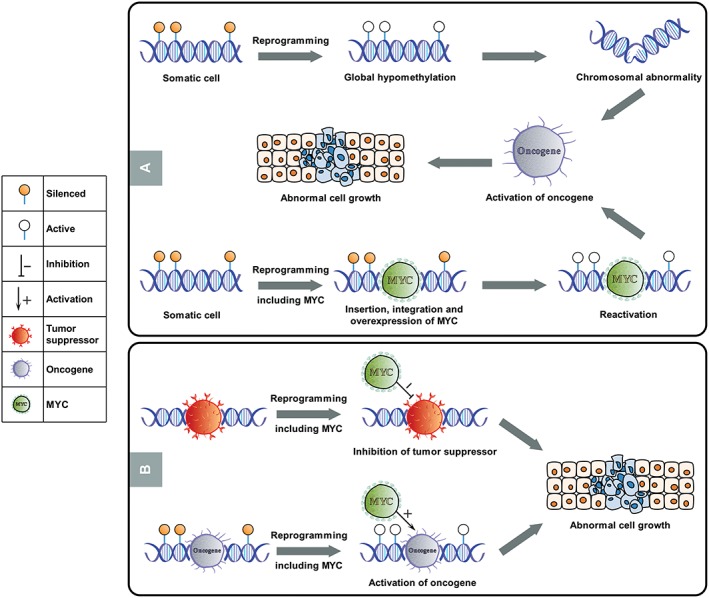
Potential risks of tumorigenicity through the induction of pluripotency. (A): The induction of somatic cells to pluripotency using transcription factors causes significant cellular remodeling and reprogramming, including the resetting of epigenetic states. This resetting includes global hypomethylation, which removes methylated, silenced signatures in parental cells. This global hypomethylation could potentially lead to the generation of chromosomal abnormalities, the activation of previously silenced oncogenes, and abnormal cell growth. The insertion, integration, overexpression, and reactivation of oncogenic transcription factors such as C‐Myc, especially via viral integration methods, may similarly result in the reactivation of oncogenic networks and abnormal cell growth following reprogramming. (B): The insertion and integration of C‐Myc may also inhibit tumor suppressors or reactivate previously silenced oncogenes, thereby triggering abnormal cell growth.
To avoid direct genomic integration during reprogramming, multiple nonintegrating gene delivery systems have been generated that avoid the use of lentiviral or retroviral vectors 48. These include minicircle vectors, nonintegrating episomes, purified protein, and mRNA 8, 49. Each of these strategies has a different reprogramming efficiency rate and produces different degrees of aneuploidy 50, highlighting the importance of the reprogramming method used. The RNA reprogramming method, for example, produces a low aneuploidy rate compared to methods using viral vectors 50. The downside of this method is that it requires an increased laboratory workload and that it can be more challenging to reprogram cells with RNA versus other methods 50. The tumorigenic transcription factor C‐Myc (Fig. 2) has also been found to be dispensable for the reprogramming process 51, though the overall efficiency is decreased without it 52. Other alternatives, such as L‐Myc 53 and small molecules, have been identified 52 that help to boost efficiency in lieu of C‐Myc. It is therefore possible that many of the genomic risks which can occur through reprogramming can be minimized via safer and less invasive reprogramming methods. However, it is also possible that some of these alternative strategies may in turn have off‐target effects capable of promoting oncogenesis.
Just like ESCs, iPSCs also become more tumorigenic and accumulate genomic abnormalities during long‐term culturing. Recurrent copy number variations due to culturing have been documented in iPSCs 54. Chromosomal copy number variations as well as moderate numbers of deletions have additionally been reported in expanded iPSCs 33. In addition to summarizing common abnormalities seen in ESCs during prolonged culture conditions, Supporting Information Table S1 also highlights typical aberrations reported in cultured iPSCs.
Common Methods for Evaluating the Genetic Integrity of Stem Cells
Given the prevalence of genetic abnormalities that can arise in stem cells, particularly those that are cultured, the ability to effectively analyze the genomic stability of a pluripotent stem cell line is essential. Several different types of cytogenetic analyses are currently available to perform studies on cellular genetic integrity. The most common techniques used to study the cytogenetics of ESCs and iPSCs as well as their merits and shortcomings are summarized in Supporting Information Table S2.
A conventional approach is provided via conventional karyotyping by GTG banding, which is also known as G banding or Giemsa banding 55. This method provides a quick snapshot of the entire genome and can reveal both numerical and structural aberrations that exist in a single cell's chromosomes. Additionally, large abnormalities and mosaicism can be quickly detected by GTG banding. Uniquely, this is the only major method that can detect structural abnormalities such as balanced translocations and inversions. GTG banding is more cost effective than other cytogenetic techniques, such as CGH and SNP arrays. Its main limitation is that it has a limited resolution and can only detect chromosomal aberrations >5 Mb (Supporting Information Table S2).
Fluorescent in situ hybridization (FISH) and spectral karyotyping (SKY) are two additional cytogenetic techniques that can be used to assess the genomic health of stem cells. These techniques, which use fluorescent DNA probes that can bind to specific chromosomes and/or chromosomal regions, are capable of identifying structural aberrations such as translocations or duplications 56. Both methods work by hybridizing chemically labeled DNA probes with metaphase/interphase cells and then visualizing them with fluorescent signals. FISH and SKY are well‐suited for identifying additional attributable chromosomal material, such as marker chromosomes. SKY specifically enables the determination of balanced/unbalanced translocations and complex rearrangements. The resolution limit of FISH and SKY is approximately >1–2 Mb 56. Despite this resolution improvement over GTG banding, both FISH and SKY are disadvantaged in only being able to detect larger abnormalities. Other techniques are better suited for the detection of smaller aberrations (Supporting Information Table S2).
More recent molecular cytogenetic techniques such as CGH and SNP arrays as well as whole genome sequencing are extremely sensitive. CGH and SNP arrays are capable of detecting chromosomal regions as small as 25 kilobases as well as mosaicism 57, 58. While both CGH and SNP arrays can detect copy number variations, SNP arrays are advantageous in their ability to detect uniparental disomy and loss of heterozygosity 59, 60. Whole genome sequencing is even more sensitive with its ability to both detect single base changes and identify mosaicism 61. Another molecular cytogenetic technique is global gene expression meta‐analysis that can detect the function of genes affected by chromosomal changes 34. However, this technique it is not as sensitive as CGH or SNP arrays and can only detect large abnormalities >10 megabases. This approach also cannot detect mosaicism (Supporting Information Table S2).
Taken together, each of the discussed techniques has its own advantages and limitations (Supporting Information Table S2). Thus, it is frequently valuable to use a combination of conventional karyotyping and molecular cytogenetic techniques as complementary tools to evaluate the genomic stability and clinical potential of both ESCs and iPSCs.
Quality Control Standards for the Safe Clinical Application of Pluripotent Stem Cells
Generation of clinical‐grade ESC and iPSC lines for use in patients should be done in compliance with good manufacturing practice (GMP). Put differently, it is imperative that strict standards and quality controls be adhered to prior to using pluripotent stem cells for clinical applications. The United States Food and Drug Administration (USFDA), the European Medicines Agency (EMA), the Japanese Pharmaceuticals and Medical Devices Agency (PMDA), and other regulatory agencies currently provide GMPs to promote the safe use of therapies for patients. With regard to stem cells, the aim of a GMP is to consider all of the potential risks involved and set guidelines for manufacturing, testing, and application of the final product. Given the requirement for large cell numbers, any stem cell procedure must also be amenable to large‐scale production 62.
The cell production company Lonza has already generated clinical‐grade iPSCs manufactured under a GMP‐compliant process 6. These iPSCs are available for clinical use 6. After establishing a tissue acquisition program, Lonza's manufacturing process began with the isolation of CD34+ cells from fresh cord blood units. Because of the safety concerns associated with the use of integrating viral vectors for reprogramming, Lonza utilized a nonintegrative episomal‐based technology developed by Chou et al. 63. Reprogrammed iPSCs were expanded in a feeder‐free serum‐free cell culture medium 6. Eliminating the use of feeders and serum served to abrogate the risk of transmitting animal pathogens to human subjects. The resultant iPSCs were then subjected to a series of safety evaluations, namely endotoxin, sterility, mycoplasma, short tandem repeat, plasmid clearance, and karyotype tests. Whole genome expression analyses was then performed on several different lines and compared to previously characterized iPSCs. A panel of approximately 250 markers was additionally analyzed to assess markers of ectoderm, mesoderm, endoderm, gender, imprint, and pluripotency 6.
RIKEN, which is the largest publicly funded research organization in Japan, has also produced clinical‐grade iPSCs which have received approval from the Japanese health ministry for human application. As mentioned heretofore 19, the RIKEN Institute has pioneered the world's first iPSC‐based transplants in two different patients. The first patient received autologous iPSC‐based cells 4 and the second patient received iPSC‐based cells from an anonymous donor that were banked 19. Reprogrammed iPSCs were generated using nonintegrating episomal vectors and then differentiated into retinal pigment epithelial cell sheets 5. In the preliminary work justifying the use in humans, these cells were shown to exhibit gene expression patterns similar to those of native retinal pigment epithelium. The differentiated cells also showed classical retinal pigment epithelial characteristics and caused no tumor or rejection issues when transplanted into nonhuman primates 5. Prior to use in the first patient recipient, autologous iPSC retinal pigment epithelial cell sheets underwent whole‐genome sequencing, expression analyses, and whole‐genome methylation profiling 4. Single‐cell quantitative polymerase chain reaction was performed to check that expression levels of retinal pigment epithelial‐specific genes were consistent with those seen in native tissue. Mandai et al. further used immunodeficient mice to assess the tumorigenic potential of the patient‐derived cells 4. No tumors were observed in the transplanted mice and extensive testing revealed that there were no de novo insertions, deletions, or DNA copy number alterations in protein‐coding regions. The plasmid DNA used for reprogramming had also not been integrated into the genomic DNA 4.
Highlighting the importance of several quality control methods, the company Lonza and the RIKEN Institute had considerable overlap in their quality control approach. Both utilized nonintegrating episomal vectors to reprogram somatic cells into iPSCs and both avoided the use of serum or feeders 4, 6. Similarly, both organizations assessed gene expression of the resultant cells. There were also differences in their approach. For example, the RIKEN Institute performed whole‐genome sequencing and whole‐genome methylation profiling while Lonza performed karyotyping and whole‐genome expression analysis 4, 6. A comparison of their quality control methods highlights the need for a cohesive, international set of standards to guide the optimization of pluripotent stem cells geared towards patient application. Only nonintegrating methods should be used for reprogramming and standard tests to ensure that the cells are sterile and free of mycoplasma should be accepted practice. With regard to quality control of stem cells, we would minimally recommend karyotyping and whole‐genome sequencing to assess genomic stability. Gene expression analyses should become routine as well to ensure that the gene expression profiles are within the normal range, which is something that also needs to be clearly defined. We would additionally recommend whole‐genome methylation profiling. Neither organization assessed mitochondrial DNA integrity or performed single‐cell genome sequencing of cellular populations. While adding to the expense and time of the quality control process, these may be valuable techniques to further maximize patient safety. More research is required to determine how valuable it would be to additionally incorporate these quality control techniques.
In 2015, the International Stem Cell Banking Initiative published a collaborative paper suggesting detailed guidelines for the development of human pluripotent stem cell seed stocks 64. In this article, the authors outline two main categories of concerns—microbiological hazards and stem cell genetic integrity. For the former, they suggest performing virological, sterility, and mycoplasma testing. They minimally recommend GTG karyotyping and also mention that other cytogenetic techniques like FISH, SKY, CGH arrays, and whole‐genome sequencing would be useful to identify information that GTG karyotyping cannot acquire. The authors also recommend some form of standardized tumorigenicity testing 64. In 2017, the International Stem Cell Banking Initiative published a follow‐up manuscript where they summarized contemporary banking efforts and cytogenetic screening protocols utilized. The latter include a 300,000 SNP genome array as well as whole‐exome sequencing 65. The methods implemented by RIKEN and Lonza largely agree with and exceed these criteria 4, 5, 6, 7.
Once cells are prepared for transplantation, autologous transplantations are clearly more desirable than allogeneic transplantations from an immunogenic perspective. For example, a direct comparison between the two in cynomolgus monkeys previously found that autologous iPSC‐derived neurons elicited a very minimal immune reaction while allografts induced an acquired immune response that resulted in the activation of microglia and the infiltration of lymphocytes 66. Additionally, a higher number of transplanted dopaminergic neurons survived in the autologous transplantation compared to the allogeneic transplantation 66. Unfortunately, the ideal autologous transplantation model is impractical for a number of reasons 67. Generating a robust patient‐derived iPSC line, characterizing various clones of that line, and then selecting an appropriately suitable clone are all processes that can take months to complete. Extra time equates to substantial extra cost in any large‐scale manufacturing process 67. Just like what occurred in the first human iPSC‐based transplant, where the trial was halted due to the discovery of a genetic abnormality in the transplanted cells used 20, new lines carry a risk of harboring a risk‐associated genetic anomaly. Correcting these abnormalities prior to differentiation and transplantation further adds to the cost and time of this production process 67. If immunogenic reactions were a nonissue, allogeneic transplantations would be much more practical due to their ability to be flexibly utilized for a wide variety of patients.
A recent 2017 study from the laboratories of Yamanaka and Takahashi has shown that major histocompatibility complex (MHC) matching improves the engraftment of iPSC‐derived cells in macaque monkeys 68. More specifically, iPSCs obtained from a MHC homozygous cynomolgus macaque were differentiated into neurons for neural grafting experiments. In response to a MHC mismatch, neuroinflammation was revealed via positron emission tomography. In contrast, MHC matching reduced the inflammatory response by suppressing the graft accumulation of both lymphocytes and microglia 68. As MHC matching enhanced engraftment of iPSC‐derived neurons in the brain and MHC matching is known to improve graft survival rates post‐organ transplantation 69, this study makes a strong case for MHC matching in humans. While MHC matching is an intelligent strategy for minimizing immune concerns, it is important to note that other immune responses may still hinder the utility of allopathic transplantations compared to autologous transplantations 70.
With the allopathic transplantation strategy in mind, Yamanaka is currently establishing an iPSC bank with the goal of being able to MHC‐match donors to recipients 19. The near‐term goal is to generate enough cell lines to be able to match more prospective recipients. Probabilistic modeling by Gourraud et al. indicates that a bank comprising iPSC lines representing the 20 most frequent human leukocyte antigen haplotypes would match more than 50% of European Americans and 22% of African Americans 71. A haplobank comprised 100 iPSC lines with the most frequent human leukocyte antigen in each ethnic population would match 45%, 52%, 63%, and 78% of African Americans, Hispanics, Asians, and European Americans, respectively 71. Other estimates suggest that as few as 50 cell lines would be sufficient to match 90% of the Japanese population and that a bank from 150 HLA‐typed volunteers could match 93% of the U.K. population 67. The development of a global iPSC haplobank that is human leukocyte antigen‐compatible for 50%–90% of respective human populations will likely improve the therapeutic potential of iPSCs. It is important to note that, even with perfect HLA matching, autologous transplantations may still be ideal for transplantations that are thought to be especially immunogenic, such as the lung, gut, liver, and spleen transplantation therapies 67.
In order to authorize iPSCs from these haplobanks for clinical use, international regulators will need to confirm that one GMP‐compliant iPSC cell line generated at one site is sufficiently similar to the one derived from another site. The establishment of an international advisory group as well as extensive international collaborations would be required to bring this idea to fruition. Both the International Stem Cell Initiative and the International Stem Cell Banking Initiative are good examples of such collaborative efforts working to establish cohesive guidelines.
Future Research and Directions for Quality Control
While a considerable amount of work has gone into identifying genomic alterations in ESCs and iPSCs for cell therapy approaches, considerably less work has investigated the integrity of mitochondrial DNA in pluripotent stem cells. This is significant as we know that mitochondrial DNA mutations occur at a high rate and that there are several human diseases due to mutations in mitochondrial DNA, such as Kearns–Sayre syndrome and Pearson syndrome 72. Work by Prigione et al. has revealed that, upon the induction of pluripotency, iPSCs can harbor novel single‐base mitochondrial DNA mutations that were not present in the parental cells they were derived from 73. However, these iPSCs showed a proper reprogramming of mitochondrial energy metabolism reflecting mitochondrial remodeling and a metabolic switch toward glycolysis 73. A more recent 2017 study found that mitochondrial copy number fluctuated throughout long‐term cultivation in iPSCs reprogrammed from a patient with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke‐like episodes 74. A third study reported that the frequency of mitochondrial DNA defects in iPSCs increased with age of the donor and that many of these defects resided in or were nonsynonymous with RNA coding genes 75. These data indicate that the integrity of mitochondrial DNA as well as genomic DNA should be assessed in pluripotent stem cells prior to clinical application. This as well as other data indicate that age of the donor may impact the genomic integrity of reprogrammed iPSCs 8. Aging may also impact the functionality of other clinically relevant cells, such as progenitor cells 76. In fact, the age of the donor has been reported to impact the migration efficiency of transplanted mesenchymal stem cells, indicating that using stem cells derived from old donors may be less than ideal 77. Future research should aim to better understand the extent to which ESCs and iPSCs are sensitive to aging as well as prone to aberrations in mitochondrial DNA.
At the moment, many powerful cytogenetic techniques are available for evaluating the genomic integrity of stem cells (Supporting Information Table S2). However, there are more innovative, recent methods that are not yet being heavily utilized. Single‐cell genome sequencing, for example, can be used to sequence large populations of single cells to understand their genetic heterogeneity. In human iPSCs differentiated into active neurons, single‐cell RNA‐seq was used along with patch clamping to define a continuum of low to high electrophysiological states 78. This technique has been applied to detect clonal evolution within solid tumors and reveal somatic copy number variations in single cells 79. The costs associated with single‐cell genome sequencing are steadily decreasing, though technical hurdles such as data mining and increasing sensitivity remain to be addressed 80. The discussion of future quality control screening also brings up the topic of epigenetic screening for iPSCs and ESCs. It is currently unclear how important it is to do more thorough epigenetic screens for pluripotent stem cells prior to clinical application. Reports that genomic imprinting in iPSCs is variably lost between different lines 81, 82 would indicate that it would be beneficial to standardize epigenetic quality control tests. Future research should aim to better understand the cost‐benefit of making such screens routine. It would also be interesting to explore the possibility of using single‐cell RNA‐seq to identify unique subpopulations that are especially suited for a patient‐specific therapy.
Conclusion
Since Yamanaka's and Takahashi's Nobel Prize‐winning discovery in 2006 1, the field of regenerative medicine has made significant research progress. In a little over a decade, this progress has culminated in historic experiments involving clinical transplantation of both autologous and donor iPSCs in human patients 19. Substantial work remains to be done, however, before ESCs and iPSCs can fulfill their therapeutic dream in the clinic. Given that stem cells can accumulate genomic aberrations in culture and unsafe or incomplete reprogramming can create further anomalies in iPSCs, the global community needs to agree upon strict quality control standards that permit the generation of clinical‐grade stem cells. Novel, emerging techniques such a single‐cell genome sequencing of large cell populations will likely teach us more about genomic integrity within pluripotent stem cell lines. Regardless, it is essential that the field progress safely and take the requisite time needed to optimize quality control before foraying too much further into human therapies.
Author Contributions
L.R.: collected and assembled data, helped provide the concept for the manuscript, and helped write the manuscript. A.A.J.: collected and assembled data, helped write the manuscript, and helped shape the concept of the manuscript. P.N.: helped with the design and construction of the figures. D.E.R.: performed editing. H.U.: performed editing. H.H.: collected and assembled data, helped provide the concept for the manuscript, and performed editing.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Supporting Information Table S1. Summary of genomic aberrations that can occur during prolonged culturing for both human embryonic stem cells and induced pluripotent stem cells.
Supporting Information Table S2. Comparison of methods for evaluating the genomic integrity of pluripotent stem cells.
Acknowledgments
The work presented in this article was made possible by funding from the Canadian Institutes of Health Research, São Paulo Research Foundation FAPESP, and the National Council for Scientific and Technological Development (CNPq) in Brazil.
References
- 1. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 2. Marmorstein AD, Johnson AA, Bachman LA et al. Mutant best1 expression and impaired phagocytosis in an iPSC model of autosomal recessive bestrophinopathy. Sci Rep 2018;8:4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Inoue H, Nagata N, Kurokawa H et al. iPS cells: a game changer for future medicine. EMBO J 2014;33:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mandai M, Watanabe A, Kurimoto Y et al. Autologous induced stem‐cell‐derived retinal cells for macular degeneration. N Engl J Med 2017;376:1038–1046. [DOI] [PubMed] [Google Scholar]
- 5. Kamao H, Mandai M, Okamoto S et al. Characterization of human induced pluripotent stem cell‐derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports 2014;2:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baghbaderani BA, Tian X, Neo BH et al. cGMP‐Manufactured human induced pluripotent stem cells are available for pre‐clinical and clinical applications. Stem Cell Reports 2015;5:647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baghbaderani BA, Syama A, Sivapatham R et al. Detailed characterization of human induced pluripotent stem cells manufactured for therapeutic applications. Stem Cell Rev 2016;12:394–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rohani L, Johnson AA, Arnold A et al. The aging signature: a hallmark of induced pluripotent stem cells? Aging Cell 2014;13:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim K, Doi A, Wen B et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010;467:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng Q, Lu SJ, Klimanskaya I et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 2010;28:704–712. [DOI] [PubMed] [Google Scholar]
- 11. Vaziri H, Chapman KB, Guigova A et al. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regen Med 2010;5:345–363. [DOI] [PubMed] [Google Scholar]
- 12. Varum S, Rodrigues AS, Moura MB et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 2011;6:e20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson AA, Andrews‐Pfannkoch C, Nelson TJ et al. Disease modeling studies using induced pluripotent stem cells: Are we using enough controls? Regen Med 2017;12:899–903. [DOI] [PubMed] [Google Scholar]
- 14. Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 2012;10:678–684. [DOI] [PubMed] [Google Scholar]
- 15. Kilpinen H, Goncalves A, Leha A et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 2017;546:370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell 2013;13:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doi D, Morizane A, Kikuchi T et al. Prolonged maturation culture favors a reduction in the tumorigenicity and the dopaminergic function of human ESC‐derived neural cells in a primate model of Parkinson's disease. Stem Cells 2012;30:935–945. [DOI] [PubMed] [Google Scholar]
- 18. El Khatib MM, Ohmine S, Jacobus EJ et al. Tumor‐free transplantation of patient‐derived induced pluripotent stem cell progeny for customized islet regeneration. Stem Cells Transl Med 2016;5:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cyranoski D. Japanese man is first to receive 'reprogrammed' stem cells from another person. Nature News 2017. doi:10.1038/nature.2017.21730 [Google Scholar]
- 20. Garber K. RIKEN suspends first clinical trial involving induced pluripotent stem cells. Nat Biotechnol 2015;33:890–891. [DOI] [PubMed] [Google Scholar]
- 21. Baker DE, Harrison NJ, Maltby E et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 2007;25:207–215. [DOI] [PubMed] [Google Scholar]
- 22. Lund RJ, Narva E, Lahesmaa R. Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet 2012;13:732–744. [DOI] [PubMed] [Google Scholar]
- 23. Diaferia GR, Conti L, Redaelli S et al. Systematic chromosomal analysis of cultured mouse neural stem cell lines. Stem Cells Dev 2011;20:1411–1423. [DOI] [PubMed] [Google Scholar]
- 24. Sareen D, McMillan E, Ebert AD et al. Chromosome 7 and 19 trisomy in cultured human neural progenitor cells. PLoS One 2009;(4):e7630 10.1371/journal.pone.0007630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ben‐David U, Mayshar Y, Benvenisty N. Large‐scale analysis reveals acquisition of lineage‐specific chromosomal aberrations in human adult stem cells. Cell Stem Cell 2011;9:97–102. [DOI] [PubMed] [Google Scholar]
- 26. Maitra A, Arking DE, Shivapurkar N et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet 2005;37:1099–1103. [DOI] [PubMed] [Google Scholar]
- 27. Narva E, Autio R, Rahkonen N et al. High‐resolution DNA analysis of human embryonic stem cell lines reveals culture‐induced copy number changes and loss of heterozygosity. Nat Biotechnol 2010;28:371–377. [DOI] [PubMed] [Google Scholar]
- 28. Inzunza J, Sahlen S, Holmberg K et al. Comparative genomic hybridization and karyotyping of human embryonic stem cells reveals the occurrence of an isodicentric X chromosome after long‐term cultivation. Mol Hum Reprod 2004;10:461–466. [DOI] [PubMed] [Google Scholar]
- 29. Draper JS, Smith K, Gokhale P et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol 2004;22:53–54. [DOI] [PubMed] [Google Scholar]
- 30. Lefort N, Feyeux M, Bas C et al. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol 2008;26:1364–1366. [DOI] [PubMed] [Google Scholar]
- 31. Spits C, Mateizel I, Geens M et al. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat Biotechnol 2008;26:1361–1363. [DOI] [PubMed] [Google Scholar]
- 32. Taapken SM, Nisler BS, Newton MA et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol 2011;29:313–314. [DOI] [PubMed] [Google Scholar]
- 33. Laurent LC, Ulitsky I, Slavin I et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 2011;8:106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayshar Y, Ben‐David U, Lavon N et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 2010;7:521–531. [DOI] [PubMed] [Google Scholar]
- 35. Abeyta MJ, Clark AT, Rodriguez RT et al. Unique gene expression signatures of independently‐derived human embryonic stem cell lines. Hum Mol Genet 2004;13:601–608. [DOI] [PubMed] [Google Scholar]
- 36. Amps K, Andrews PW, Anyfantis G et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 2011;29:1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Desmarais JA, Hoffmann MJ, Bingham G et al. Human embryonic stem cells fail to activate CHK1 and commit to apoptosis in response to DNA replication stress. Stem Cells 2012;30:1385–1393. [DOI] [PubMed] [Google Scholar]
- 38. Yu Y, Chang L, Zhao H et al. Chromosome microduplication in somatic cells decreases the genetic stability of human reprogrammed somatic cells and results in pluripotent stem cells. Sci Rep 2015;5:10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Young MA, Larson DE, Sun CW et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell 2012;10:570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Letourneau A, Santoni FA, Bonilla X et al. Domains of genome‐wide gene expression dysregulation in Down's syndrome. Nature 2014;508:345–350. [DOI] [PubMed] [Google Scholar]
- 41. Bershteyn M, Hayashi Y, Desachy G et al. Cell‐autonomous correction of ring chromosomes in human induced pluripotent stem cells. Nature 2014;507:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bayart E, Cohen‐Haguenauer O. Technological overview of iPS induction from human adult somatic cells. Curr Gene Ther 2013;13:73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ronen D, Benvenisty N. Genomic stability in reprogramming. Curr Opin Genet Dev 2012;22:444–449. [DOI] [PubMed] [Google Scholar]
- 44. Hussein SM, Batada NN, Vuoristo S et al. Copy number variation and selection during reprogramming to pluripotency. Nature 2011;471:58–62. [DOI] [PubMed] [Google Scholar]
- 45. Ruiz S, Lopez‐Contreras AJ, Gabut M et al. Limiting replication stress during somatic cell reprogramming reduces genomic instability in induced pluripotent stem cells. Nat Commun 2015;6:8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liang G, Zhang Y. Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective. Cell Res 2013;23:49–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson AA, Akman K, Calimport SR et al. The role of DNA methylation in aging, rejuvenation, and age‐related disease. Rejuvenation Res 2012;15:483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gonzalez F, Boue S, Izpisua Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nat Rev Genet 2011;12:231–242. [DOI] [PubMed] [Google Scholar]
- 49. Rohani L, Fabian C, Holland H et al. Generation of human induced pluripotent stem cells using non‐synthetic mRNA. Stem Cell Res 2016;16:662–672. [DOI] [PubMed] [Google Scholar]
- 50. Schlaeger TM, Daheron L, Brickler TR et al. A comparison of non‐integrating reprogramming methods. Nat Biotechnol 2015;33:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wernig M, Meissner A, Cassady JP et al. c‐Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008;2:10–12. [DOI] [PubMed] [Google Scholar]
- 52. Desponts C, Ding S. Using small molecules to improve generation of induced pluripotent stem cells from somatic cells. Methods Mol Biol 2010;636:207–218. [DOI] [PubMed] [Google Scholar]
- 53. Nakagawa M, Takizawa N, Narita M et al. Promotion of direct reprogramming by transformation‐deficient Myc. Proc Natl Acad Sci U S A 2010;107:14152–14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martins‐Taylor K, Nisler BS, Taapken SM et al. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol 2011;29:488–491. [DOI] [PubMed] [Google Scholar]
- 55. Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet 2005;6:782–792. [DOI] [PubMed] [Google Scholar]
- 56. Imataka G, Arisaka O. Chromosome analysis using spectral karyotyping (SKY). Cell Biochem Biophys 2012;62:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cross J, Peters G, Wu Z et al. Resolution of trisomic mosaicism in prenatal diagnosis: estimated performance of a 50K SNP microarray. Prenat Diagn 2007;27:1197–1204. [DOI] [PubMed] [Google Scholar]
- 58. Ballif BC, Rorem EA, Sundin K et al. Detection of low‐level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A 2006;140:2757–2767. [DOI] [PubMed] [Google Scholar]
- 59. Yu Y, Chang L, Zhao H et al. Chromosome microduplication in somatic cells decreases the genetic stability of human reprogrammed somatic cells and results in pluripotent stem cells. Sci Rep 2015;5:10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Keren B. The advantages of SNP arrays over CGH arrays. Mol Cytogenet 2014;7:I31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. King DA, Sifrim A, Fitzgerald TW et al. Detection of structural mosaicism from targeted and whole‐genome sequencing data. Genome Res 2017;27:1704–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ausubel LJ, Lopez PM, Couture LA. GMP scale‐up and banking of pluripotent stem cells for cellular therapy applications. Methods Mol Biol 2011;767:147–159. [DOI] [PubMed] [Google Scholar]
- 63. Chou BK, Mali P, Huang X et al. Efficient human iPS cell derivation by a non‐integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res 2011;21:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andrews PW, Baker D, Benvinisty N et al. Points to consider in the development of seed stocks of pluripotent stem cells for clinical applications: International Stem Cell Banking Initiative (ISCBI). Regen Med 2015;10:1–44. [DOI] [PubMed] [Google Scholar]
- 65. Kim JH, Kurtz A, Yuan BZ et al. Report of the International Stem Cell Banking Initiative workshop activity: current hurdles and progress in seed‐stock banking of human pluripotent stem cells. Stem Cells Transl Med 2017;6:1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morizane A, Doi D, Kikuchi T et al. Direct comparison of autologous and allogeneic transplantation of iPSC‐derived neural cells in the brain of a non‐human primate. Stem Cell Reports 2013;1:283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Solomon S, Pitossi F, Rao MS. Banking on iPSC‐‐is it doable and is it worthwhile. Stem Cell Rev 2015;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morizane A, Kikuchi T, Hayashi T et al. MHC matching improves engraftment of iPSC‐derived neurons in non‐human primates. Nat Commun 2017;8:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Opelz G, Dohler B. Effect of human leukocyte antigen compatibility on kidney graft survival: comparative analysis of two decades. Transplantation 2007;84:137–143. [DOI] [PubMed] [Google Scholar]
- 70. Bravery CA. Do human leukocyte antigen‐typed cellular therapeutics based on induced pluripotent stem cells make commercial sense? Stem Cells Dev 2015;24:1–10. [DOI] [PubMed] [Google Scholar]
- 71. Gourraud PA, Gilson L, Girard M et al. The role of human leukocyte antigen matching in the development of multiethnic "haplobank" of induced pluripotent stem cell lines. Stem Cells 2012;30:180–186. [DOI] [PubMed] [Google Scholar]
- 72. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet 2005;6:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Prigione A, Lichtner B, Kuhl H et al. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell‐like metabolic reprogramming. Stem Cells 2011;29:1338–1348. [DOI] [PubMed] [Google Scholar]
- 74. Yahata N, Matsumoto Y, Omi M et al. TALEN‐mediated shift of mitochondrial DNA heteroplasmy in MELAS‐iPSCs with m.13513G>A mutation. Sci Rep 2017;(7):15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kang E, Wang X, Tippner‐Hedges R et al. Age‐related accumulation of somatic mitochondrial DNA mutations in adult‐derived human iPSCs. Cell Stem Cell 2016;18:625–636. [DOI] [PubMed] [Google Scholar]
- 76. Scutt N, Johnson AA, Scutt A et al. Tissue‐specific ageing of rat tendon‐derived progenitor cells. J Stem Cell Res Ther 2015;5:309 10.4172/2157-7633.1000309. [DOI] [Google Scholar]
- 77. Fabian C, Naaldijk Y, Leovsky C et al. Distribution pattern following systemic mesenchymal stem cell injection depends on the age of the recipient and neuronal health. Stem Cell Res Ther 2017;8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bardy C, van den Hurk M, Kakaradov B et al. Predicting the functional states of human iPSC‐derived neurons with single‐cell RNA‐seq and electrophysiology. Mol Psychiatry 2016;21:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Navin NE. The first five years of single‐cell cancer genomics and beyond. Genome Res 2015;25:1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Linnarsson S, Teichmann SA. Single‐cell genomics: coming of age. Genome Biol 2016;17:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Takikawa S, Ray C, Wang X et al. Genomic imprinting is variably lost during reprogramming of mouse iPS cells. Stem Cell Res 2013;11:861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hiura H, Toyoda M, Okae H et al. Stability of genomic imprinting in human induced pluripotent stem cells. BMC Genet 2013;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table S1. Summary of genomic aberrations that can occur during prolonged culturing for both human embryonic stem cells and induced pluripotent stem cells.
Supporting Information Table S2. Comparison of methods for evaluating the genomic integrity of pluripotent stem cells.


