Publisher's Note: There is a Blood Commentary on this article in this issue.
Key Points
A 58-gene expression-based assay aids in the molecular distinction of PMBCL and DLBCL using archival tissue biopsy specimens.
Abstract
Primary mediastinal large B-cell lymphoma (PMBCL) is recognized as a distinct entity in the World Health Organization classification. Currently, the diagnosis relies on consensus of histopathology, clinical variables, and presentation, giving rise to diagnostic inaccuracy in routine practice. Previous studies have demonstrated that PMBCL can be distinguished from subtypes of diffuse large B-cell lymphoma (DLBCL) based on gene expression signatures. However, requirement of fresh-frozen biopsy material has precluded the transfer of gene expression–based assays to the clinic. Here, we developed a robust and accurate molecular classification assay (Lymph3Cx) for the distinction of PMBCL from DLBCL subtypes based on gene expression measurements in formalin-fixed, paraffin-embedded tissue. A probabilistic model accounting for classification error, comprising 58 gene features, was trained on 68 cases of PMBCL and DLBCL. Performance of the model was subsequently evaluated in an independent validation cohort of 158 cases and showed high agreement of the Lymph3Cx molecular classification with the clinicopathological diagnosis of an expert panel (frank misclassification rate, 3.8%). Furthermore, we demonstrate reproducibility of the assay with 100% concordance of subtype assignments at 2 independent laboratories. Future studies will determine Lymph3Cx’s utility for routine diagnostic purposes and therapeutic decision making.
Visual Abstract
Introduction
Primary mediastinal large B-cell lymphoma (PMBCL) is recognized as a rare but distinct entity in the current World Health Organization classification, accounting for ∼2% to 3% of all non-Hodgkin lymphomas.1 Typically, the disease presents as a large mass in the anterior mediastinum of young adults and children with a slight female predominance. Integration of clinical features is considered critical to establish the diagnosis of PMBCL, because a reliable distinction from subtypes of diffuse large B-cell lymphoma (DLBCL), namely activated B-cell–like (ABC), germinal center B-cell–like (GCB), and unclassified DLBCL, can be difficult if solely based on morphology and immunophenotype. In addition, recent studies have identified lymphomas that share molecular and morphological features with PMBCL yet do not involve the mediastinum.2 As correct classification may impact therapeutic decision making, more robust methods to identify these cases are demanded.
Previous gene expression profiling (GEP) studies provided evidence that PMBCL can be distinguished from DLBCL on the molecular level and supported a strong relationship between PMBCL and classical Hodgkin lymphoma.3,4 Moreover, substantial similarity between classical Hodgkin lymphoma and PMBCL has been demonstrated by overlapping mutational profiles, copy-number variations,5-8 and pathway dependencies.9 However, the majority of the aforementioned studies were performed on frozen tissues, and only a limited number of approaches explored feasibility in formalin-fixed, paraffin-embedded tissues (FFPETs) to establish classification assays that can be widely used in clinical practice.10
Here, we developed a robust and accurate molecular assay for the distinction of PMBCL from DLBCL subtypes based on gene expression measurements in routinely available FFPET biopsy specimens.
Study design
All cases included in this study were retrieved from the tissue archives of participating centers. Conventional and immunohistochemically stained slides were reviewed by at least 4 members of the Lymphoma/Leukemia Molecular Profiling Project expert hematopathology panel. Information on age, biopsy location, and clinical presentation was made available whenever possible (supplemental Tables 1 and 2, available on the Blood Web site). If 3 pathologists independently agreed, then a definite diagnosis was established; all other cases were subjected to panel discussion. Although imperfect, clinicopathological diagnosis is referred to as the “gold standard” for DLBCL/PMBCL classification herein. This study was conducted with approval from institutional review boards according to the Declaration of Helsinki.
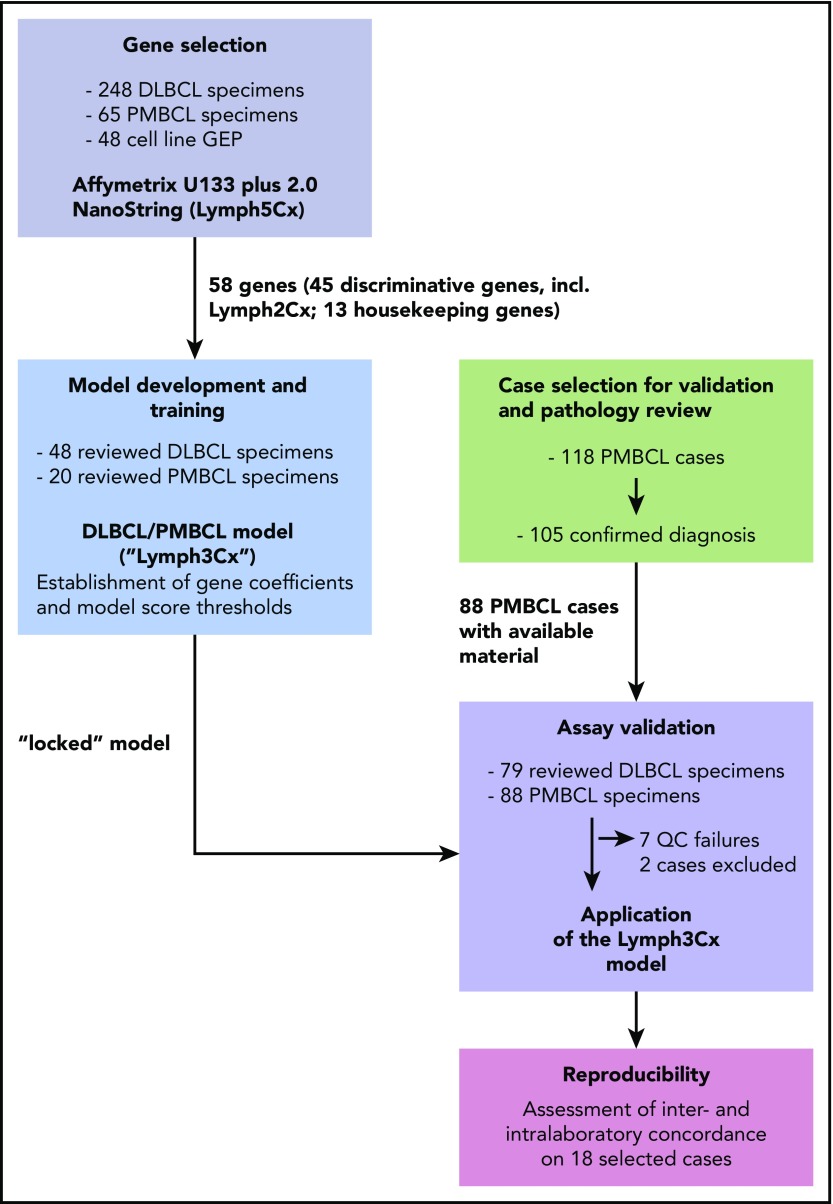
Details on study design, gene selection and model building are provided in Figure 1 and the supplemental Methods. Between the training and validation cohorts, in total, 108 PMBCL and 127 DLBCL cases were studied. Following deparaffinization, RNA was extracted from up to five 10-μm FFPET sections (tumor content ≥60% of tissue area) using the QIAGEN DNA/RNA FFPE Kit (Hilden, Germany) according to the manufacturer’s instructions. RNA was quantified using a spectrophotometer (Nanodrop; ThermoFisher, Germany). Gene expression analysis was performed on 200 ng RNA using a custom codeset on the NanoString platform (NanoString Technologies, Seattle, WA) at the “high sensitivity” setting on the Prep Station and 555 fields of view on second generation nCounter analyzers.
Figure 1.
Schematic overview of the study design. For details on clinical characteristics and pathological features of the cases refer to supplemental Tables 1 and 2. QC, quality control.
Results and discussion
To develop a classification assay, applicable to FFPET specimens, that aims at a robust discrimination between PMBCL and DLBCL as well as the DLBCL subtypes (GCB, ABC, and unclassified, respectively), we selected gene expression features from previously published datasets (supplemental Methods; supplemental Figure 1A).4,11,12 The selection process, aiming at identification of genes with the highest discriminative power and good concordance between the different analytical platforms used for GEP, yielded 58 genes for subsequent assay development (supplemental Methods; supplemental Table 3). Of those, 30 genes were employed to distinguish PMBCL from DLBCL, with 24 being overexpressed in PMBCL and 6 genes showing higher expression levels in DLBCL (supplemental Figure 1B). This approach of “balanced” gene selection was chosen to make the model less vulnerable to normalization artifacts. Additionally, in a modular assay design, the 15 genes used for DLBCL cell-of-origin (COO) assignment from the Lymph2Cx assay13 were included, and the remaining 13 genes were chosen as housekeeping genes, including all 5 from the Lymph2Cx assay. To train a linear regression model and establish model thresholds to distinguish PMBCL from DLBCL, a customized NanoString codeset including these 58 genes was then applied to a training cohort of 68 cases, of which 20 were diagnosed as PMBCL by consensus review and 48 were classified as DLBCL (Figure 1). The performance of the Lymph3Cx assay in the training cohort is shown in supplemental Results and supplemental Figures 2 and 3. The gene expression–based model, including coefficients and thresholds was “locked” and subsequently applied to the independent validation cohort comprising 167 FFPET biopsy specimens (88 PMBCL and 79 DLBCL by consensus review). None of these specimens were part of the training cohort or had been previously used to train the Lymph2Cx assay.
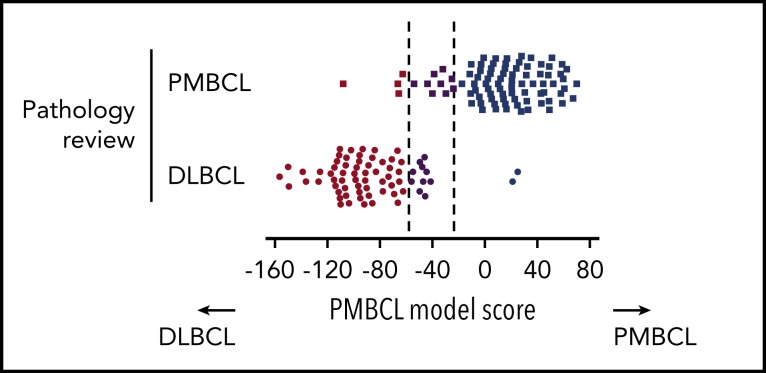
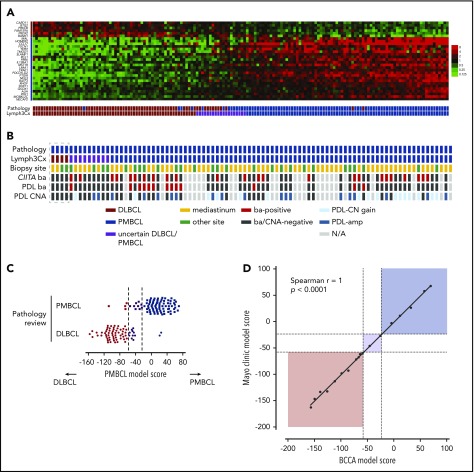
The assay yielded gene expression data of sufficient quality in 160 out of 167 cases (95.8%), leaving 88 PMBCL and 70 DLBCL cases for final analysis (2 additional DLBCL cases were excluded because of a mismatch to previously analyzed frozen biopsy specimens). Among the pathologically defined PMBCL cases, 75 (85%) were classified as such based on Lymph3Cx; 10% (9 cases) were assigned to the “uncertain” category, and 5% (4 cases) showed a molecular signature of DLBCL (Figure 2A-B). Of note, scores for most of the “misclassified” PMBCL cases (3 out of 4) were close to the cutoff (Figure 2A,C). Among the pathologically defined DLBCL cases, 58 (83%) were classified as DLBCL by the assay, 14% (10 cases) were “uncertain,” and 2 (3%) were predicted to be PMBCL (Figure 2A). A more detailed characterization of the 6 discordant cases, considering re-review of pathological and clinical variables, is included in the supplemental Results. Similar to the results obtained in the training cohort, no misclassified cases were seen with regards to DLBCL COO subclassification. Eleven cases changed between the unclassified category and ABC or GCB, respectively (supplemental Figure 4).
Figure 2.
Performance of the Lymph3Cx assay in the validation cohort and inter-laboratory concordance. (A) Heatmap of gene expression in the independent validation cohort of 88 pathologically defined PMBCL cases and 70 DLBCL cases. Each column represents an individual case, and the discriminating gene features included in the Lymph3Cx assay are ordered in rows. The top 6 genes are overexpressed in DLBCL, and the other 24 show higher expression levels in PMBCL. Housekeeping genes and the features used for DLBCL COO assignment are not displayed for visualization purposes. Cases are ordered according to their respective model score, with cases that obtained the highest score on the right. (B) Displayed are only the pathologically defined PMBCL cases with their respective assignment by the molecular gene expression–based assay (Lymph3Cx). No bias was observed with regards to mediastinal/nonmediastinal location of biopsy site, providing evidence that the assay is also applicable to PMBCL(-like) cases arising outside the mediastinum or nonmediastinal biopsy specimens, respectively. Interestingly, the 4 cases with a molecular signature of DLBCL (box) did not harbor chromosomal rearrangements of CIITA and/or the programmed death ligand (PDL) loci, or PDL copy-number (CN) alterations. (C) Distribution of model scores in the validation cohort. (D) Comparison of Lymph3Cx scores for selected cases of the validation cohort from 2 independent laboratories (BC Cancer Agency [BCCA] and Mayo Clinic). Dotted lines represent the defined thresholds to discriminate PMBCL (dark blue) from DLBCL (brown) using the Lymph3Cx assay (uncertain category displayed in purple). Of note, no case changed subtype assignment between the different laboratories. amp, amplification; ba, break-apart; CNA, copy-number alteration; N/A, not assessable.
For 66 cases with available COO predictions and model scores from the Lymph2Cx assay,13,14 we compared the results to Lymph3Cx and revealed a high correlation coefficient between both assays (Spearman r = 0.9937). We did not observe any classification changes, demonstrating the robustness of DLBCL COO assignment across these assays (supplemental Figure 5). We next performed experiments to determine intralaboratory reproducibility and inter-laboratory concordance of the Lymph3Cx assay. Eighteen biopsy specimens were selected on the basis that their model scores were equally distributed across the population and thus are representative of the validation cohort. For interlaboratory comparison, separate tissue scrolls or unstained sections were distributed to an independent laboratory (Mayo Clinic, Scottsdale, AZ) where RNA was extracted and run on the Lymph3Cx assay. The concordance was excellent (Spearman r2 = 0.996; Figure 2D), demonstrating the robustness of the assay. For details on intralaboratory comparison, see supplemental Results.
Our study establishes a feasible and reproducible molecular biomarker assay, Lymph3Cx, for routine diagnostic purposes in clinical pathology laboratories and as a tool to support clinical trials. Development of our assay leveraged the NanoString platform used for subtyping of DLBCL13 and breast cancer15 and can be considered a modular upgrade to the Lymph2Cx assay, thus creating synergy for diagnostic workflows. While avoiding the uncertainty of current consensus diagnostic procedures, the clinical utility of a molecular PMBCL subtyping test remains to be demonstrated. With the emergence of novel therapeutic approaches, such as dose-adjusted EPOCH-R16 or PD1 inhibition with nivolumab17 or pembrolizumab18 that might have preferential benefit in PMBCL, as compared with DLBCL not-otherwise-specified subtypes, accurate molecular subtyping is projected to become more important.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This project was funded by a grant from the National Cancer Institute, National Institutes of Health (“Molecular Diagnosis and Prognosis in Aggressive Lymphoma [SPECS II] grant 1U01CA157581-01). A.M. received support through postdoctoral fellowship awards from the Mildred-Scheel-Cancer-Foundation (Deutsche Krebshilfe) and the Michael Smith Foundation for Health Research and Lymphoma Canada and is currently supported by the Clinician Scientist Program of the Medical Faculty at the University of Ulm. E.C. received funding from the Ministerio de Economia y Competitividad (grant SAF2015-64885-R). C.S. is supported by research grants from the Canadian Institutes of Health Research (2202) and a Terry Fox Research Institute team grant (1061). C.S. is the recipient of a Michael Smith Foundation for Health Research Career Investigator award.
Footnotes
Presented in part at the 14th International Conference on Malignant Lymphoma, Lugano, Switzerland, 15 June 2017.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.M., G.W., D.W.S., L.M.S., C.S., and L.M.R. designed the study and interpreted data; A.M., G.W., C.R., and B.J.G.-G. performed research; A.M., A.R., G.O., E.C., R.M.B., J.D., D.D.W., J.Y.S., W.C.C., J.R.C., K.F., T.G., J.M.C., K.J.S., E.S., H.H., R.D.G., E.S.J., and L.M.R. provided clinical data and/or performed pathology review; A.M., G.W., C.S., and L.M.R. wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: A.M., G.W., C.S., L.M.S., D.W.S., and L.M.R. are named inventors on a patent filed by the National Cancer Institute (“Methods for determining lymphoma type”). D.W.S., G.W., A.R., G.O., E.C., R.M.B., J.D., D.D.W., W.C.C., J.R.C., T.G., E.S., H.H., R.D.G., L.M.S., E.S.J., J.M.C., and L.M.R. are inventors on a patent filed by the National Cancer Institute (“Methods for selecting and treating lymphoma types”) licensed to NanoString Technologies. D.W.S. is consulting for Celgene and Janssen. K.J.S. is consulting and has an advisory role for Bristol-Meyers Squibb. K.J.S. received honoraria from Bristol-Meyers Squibb and Merck. C.S. is consulting for Seattle Genetics, Inc. and Hoffmann-La Roche. The remaining authors declare no competing financial interests.
Correspondence: Christian Steidl, Centre for Lymphoid Cancer, BC Cancer, 675 West 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: csteidl@bccancer.bc.ca.
REFERENCES
- 1.Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. [Google Scholar]
- 2.Yuan J, Wright G, Rosenwald A, et al. ; Lymphoma Leukemia Molecular Profiling Project (LLMPP). Identification of primary mediastinal large B-cell lymphoma at nonmediastinal sites by gene expression profiling. Am J Surg Pathol. 2015;39(10):1322-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage KJ, Monti S, Kutok JL, et al. . The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102(12):3871-3879. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Leroy K, et al. . Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198(6):851-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wessendorf S, Barth TF, Viardot A, et al. . Further delineation of chromosomal consensus regions in primary mediastinal B-cell lymphomas: an analysis of 37 tumor samples using high-resolution genomic profiling (array-CGH). Leukemia. 2007;21(12):2463-2469. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz R, Hansmann ML, Bohle V, et al. . TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206(5):981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steidl C, Shah SP, Woolcock BW, et al. . MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471(7338):377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunawardana J, Chan FC, Telenius A, et al. . Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat Genet. 2014;46(4):329-335. [DOI] [PubMed] [Google Scholar]
- 9.Dunleavy K, Steidl C. Emerging biological insights and novel treatment strategies in primary mediastinal large B-cell lymphoma. Semin Hematol. 2015;52(2):119-125. [DOI] [PubMed] [Google Scholar]
- 10.Bobée V, Ruminy P, Marchand V, et al. . Determination of molecular subtypes of diffuse large B-cell lymphoma using a reverse transcriptase multiplex ligation-dependent probe amplification classifier: a CALYM study. J Mol Diagn. 2017;19(6):892-904. [DOI] [PubMed] [Google Scholar]
- 11.Lenz G, Wright G, Dave SS, et al. ; Lymphoma/Leukemia Molecular Profiling Project. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alizadeh AA, Eisen MB, Davis RE, et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. [DOI] [PubMed] [Google Scholar]
- 13.Scott DW, Wright GW, Williams PM, et al. . Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123(8):1214-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott DW, Mottok A, Ennishi D, et al. . Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33(26):2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen T, Wallden B, Schaper C, et al. . Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014;14(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunleavy K, Pittaluga S, Maeda LS, et al. . Dose-adjusted EPOCH-rituximab therapy in primary mediastinal B-cell lymphoma. N Engl J Med. 2013;368(15):1408-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu-Monette ZY, Zhou J, Young KH. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood. 2018;131(1):68-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zinzani PL, Ribrag V, Moskowitz CH, et al. . Safety and tolerability of pembrolizumab in patients with relapsed/refractory primary mediastinal large B-cell lymphoma. Blood. 2017;130(3):267-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.