Abstract
Toxin-antitoxin (TA) systems play important roles in bacteria persister formation. Increasing evidence demonstrate the roles of TA systems in regulating virulence factors in pathogenic bacteria. The toxin HigB in Pseudomonas aeruginosa contributes to persister formation and regulates the expression of multiple virulence factors and biofilm formation. However, the regulatory mechanism remains elusive. In this study, we explored the HigB mediated regulatory pathways. We demonstrate that HigB decreases the intracellular level of c-di-GMP, which is responsible for the increased expression of the type III secretion system (T3SS) genes and repression of biofilm formation. By analyzing the expression levels of the known c-di-GMP metabolism genes, we find that three c-di-GMP hydrolysis genes are up regulated by HigB, namely PA2133, PA2200 and PA3825. Deletion of the three genes individually or simultaneously diminishes the HigB mediated regulation on the expression of T3SS genes and biofilm formation. Therefore, our results reveal novel functions of HigB in P. aeruginosa.
Keywords: toxin-antitoxin system, HigB/HigA, Pseudomonas aeruginosa, type III secretion system, biofilm, c-di-GMP
1. Introduction
Toxin-antitoxin (TA) systems have been found in almost all bacterial species [1]. A typical TA system is composed of a stable toxin and a labile antitoxin. Degradation of an antitoxin results in the activation of its cognate toxin, which inhibits bacterial growth by interfering with biosynthesis of macromolecules, cell division or reducing membrane potential [2]. The growth arrest facilitates bacterial survival under environmental stresses such as antibiotic treatment, nutrient deficiency and host immune clearance [3].
Based on the property of the antitoxin and the inhibitory mechanism of antitoxin on the cognate toxin, TA systems are classified into six groups (types I to VI). The type II TA systems are widely distributed in bacteria and archaea, and are composed of two proteins that are encoded in a bicistronic operon. Most type II toxins are endonucleases, the activities of which are repressed by the cognate antitoxins through direct interactions. Meanwhile, the antitoxins directly repress the expression of their own operons by binding to the promoter regions. Cross interactions between different TA systems have been reported [4,5]. Besides arresting bacterial growth, some type II TA systems have been found to influence virulence gene expression and biofilm formation [6,7,8,9,10].
P. aeruginosa is an opportunistic pathogen. It harbors various virulence factors and causes various infections in immunocompromised patients. The type III secretion system (T3SS) and biofilm formation have been demonstrated to play important roles in acute and chronic infections, respectively. In P. aeruginosa, the T3SS and biofilm formation are reciprocally regulated by a small RNA-protein (RsmY/Z-RsmA) mediated regulatory pathway and the secondary messenger molecule c-di-GMP [11].
At least 4 type II TA systems have been identified in P. aeruginosa, namely ParD/ParE, HicA/HicB, RelE/RelB and HigB/HigA [3,10,12,13,14]. Studies in Proteus vulgaris reveal that HigB cleaves mRNA with a preference in A rich sequences [15]. Wood et al found that the HigB/HigA of P. aeruginosa influences several virulence factors, including pyochelin and pyocyanin as well as swarming motility and biofilm formation [10]. We previously demonstrated that the higB/higA operon is induced by treatment with ciprofloxacin and HigB contributes to persister formation and controls the expression of T3SS genes [3]. However, the mechanism of HigB mediated regulation on the T3SS and biofilm formation remains unknown.
In this study, we found that HigB influences T3SS and biofilm formation through c-di-GMP. We further identified the HigB regulated c-di-GMP metabolism genes.
2. Results
2.1. HigB Represses Biofilm Formation
Previously, Wood et al. demonstrated reduction of biofilm formation in a higA mutant, indicating a role of HigB in the repression of biofilm formation [10]. To verify the role of HigB in biofilm formation, we constructed a ΔhigBΔhigA double mutant, which displayed normal biofilm formation (Figure 1A). In addition, overexpression of higB in wild type PA14 reduced biofilm formation (Figure 1B). These results suggest that HigB contributes to the repression of biofilm formation.
Figure 1.
HigB affects biofilm formation. Bacteria were adjusted to an OD600 of 0.025 in LB broth and inoculated in each well of a 96-well plate. The plates were incubated at 37 °C for 24 h. The biofilm was visualized and quantified by crystal violet staining. (A) Biofilm formation by wild type PA14, the higA::Tn, the higA complemented strain and the ΔhigBΔhigA mutant. ns, not significant. (B) Biofilm formation by PA14 or the ΔhigBΔhigA mutant containing the empty vector pMMB67EH or the higB overexepression plasmid pMMB67EH-higB-His. IPTG was supplemented at the indicated concentrations.
2.2. HigB Controls the Intracellular c-di-GMP Level
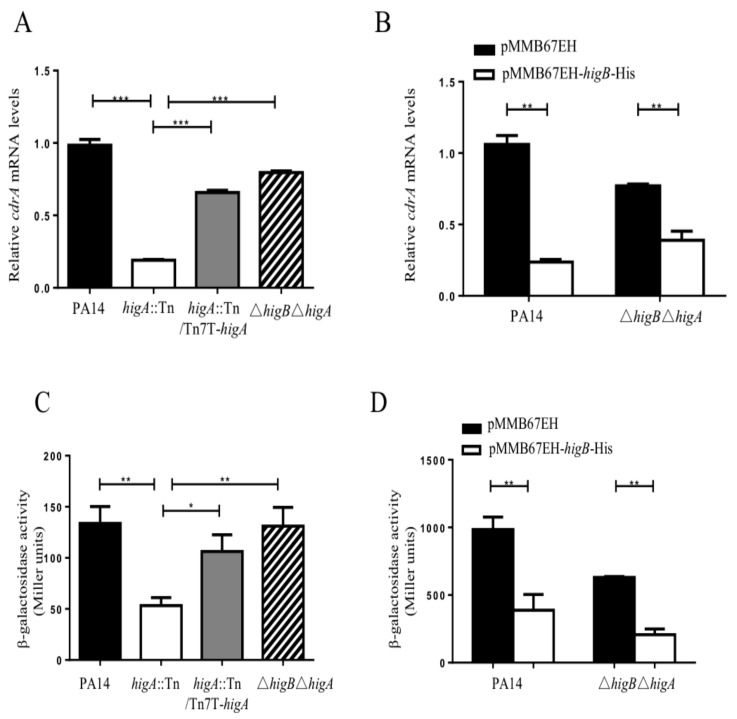
We previously found that HigB activates the expression of T3SS genes. In P. aeruginosa, the small RNAs RsmY/RsmZ and the secondary messenger molecule c-di-GMP reciprocally control T3SS and biofilm formation [16]. Therefore, we examined whether the HigB-HigA TA module influences the levels of RsmY/RsmZ and c-di-GMP. Neither mutation of higA nor overexpression of higB resulted in alteration of RsmY/RsmZ levels (Figure S1). Then, we examined c-di-GMP levels. The transcription of cdrA is controlled by the intracellular level of c-di-GMP [17]. Thus, the cdrA expression level has been used as a c-di-GMP indicator [17]. Mutation of higA or overexpression of higB reduced the mRNA level of cdrA (Figure 2A,B). To examine whether the reduction of cdrA mRNA is due to lower promoter activity, we constructed a transcriptional fusion of the cdrA promoter and a lacZ gene (PcdrA-lacZ). The expression of LacZ was lower in the higA mutant or the higB overexpression strain (Figure 2C,D), indicating a lower c-di-GMP level. Furthermore, a thiazole orange-based fluorescent assay [18] revealed lower c-di-GMP levels in the higA mutant and the strain overexpressing higB (Figure 3A,B). These results demonstrate that HigB represses the bacterial c-di-GMP level.
Figure 2.
Role of HigB in the expression of cdrA. (A) PA14, the higA::Tn mutant, the complemented strain and the ΔhigBΔhigA double mutant were grown to an OD600 of 2.0. The mRNA levels of cdrA were determined by real time PCR. (B) PA14 or the ΔhigBΔhigA mutant containing pMMB67EH or pMMB67EH-higB-His was grown in the presence of 0.1 mM IPTG. The mRNA level of cdrA were determined by real time PCR. The ribosomal protein encoding gene rpsL was used as the internal control. Error bars represent the standard errors. ** p < 0.01, *** p < 0.005 by Student’s t-test. (C,D) Expression levels of the transcriptional fusion of PcdrA-lacZ were determined in PA14, the higA:Tn mutant, the complemented strain and the ΔhigBΔhigA double mutant (C) or PA14 and the ΔhigBΔhigA mutant containing pMMB67EH or pMMB67EH-higB-His grown in the presence of 0.1 mM IPTG (D). * p < 0.05, ** p < 0.01 by Student’s t-test.
Figure 3.
HigB affects the intracellular c-di-GMP levels. The intracellular levels of c-di-GMP in PA14, the higA::Tn mutant, the complemented strain and the ΔhigBΔhigA double mutant (A) or PA14 and the ΔhigBΔhigA mutant containing pMMB67EH or pMMB67EH-higB-His grown in the presence of 0.1 mM IPTG (B) were determined by thiazole orange based fluorescence. The bacteria were grown in LB to an OD 600 of 2.0. The bacteria were lysed and the supernatant was incubated with thiazole orange to determine the c-di-GMP levels. **, p < 0.01 by Student’s t-test.
2.3. HigB Reciprocally Controls Biofilm Formation and the T3SS by Influencing c-di-GMP Levels
To examine whether the reduced c-di-GMP level is responsible for the HigB mediated repression of biofilm formation and activation of the T3SS genes, we overexpressed a diguanylate cyclases WspR, which has been shown to increase the intracellular c-di-GMP level and enhance biofilm formation [19,20,21]. In the higA mutant or the higB overexpressing strain, overexpression of wspR increased the expression level of cdrA, indicating an increase of the intracellular level of c-di-GMP (Figure 4A,B). Consistently, overexpression of wspR restored the levels of biofilm formation and down regulated the T3SS genes in the higA mutant or the higB overexpressing strain. (Figure 4C,D).
Figure 4.
Role of c-di-GMP in HigB mediated regulation on biofilm formation and T3SS gene expression. (A) The mRNA levels of cdrA in PA14 and the higA mutant carrying the empty vector pUCP20 or the higA mutant carrying pUCP20–wspR. (B) The mRNA levels of cdrA in PA14 carrying the empty vector pUCP24 or pUCP24–wspR together with pMMB67EH or pMMB67EH-higB-His. The bacteria were grown in the presence of 0.1 mM IPTG. (C) The mRNA levels of the T3SS genes, exsA, exsC and pcrV in PA14 and the higA mutant carrying pUCP20 or the higA mutant carrying pUCP20–wspR. (D) PA14 carrying the empty vector pUCP24 or pUCP24–wspR together with pMMB67EH or pMMB67EH-higB-His were grown in the presence of 0.1 mM IPTG. The mRNA levels of exsA, exsC and pcrV were determined by real time PCR. * p < 0.05, ** p < 0.01 by Student’s t-test.
2.4. Identification of c-di-GMP Metabolism Genes that Are Controlled by HigB
In P. aeruginosa, there are at least 40 genes that are involved in the metabolism of c-di-GMP [22]. Diguanylate cyclases (DGCs), containing a GGDEF motif, synthesize c-di-GMP. Phosphodiesterases (PDEs), containing a HD-GYP or EAL domain, degrade c-di-GMP [22]. To understand the mechanism of HigB mediated repression on the intracellular c-di-GMP level, we compared the mRNA levels of the 40 c-di-GMP metabolism genes between PA14 and the higA mutant. Among the diguanylate cyclases, PA3343 and PA5487 were up regulated in the higA mutant (Figure S2A), which should not lead to the reduction of c-di-GMP. PA0861, PA1727 and PA2567 were up regulated in the higA mutant (Figure S2B). However, since they contain both the GGDEF and EAL motifs, their roles in controlling the c-di-GMP level remain elusive. Of note, among the phosphodiesterases, three genes, namely PA2133, PA2200 and PA3825 were up regulated the most (≥3-fold) in the higA mutant (Figure S2C). All three genes contain an EAL domain [2]. Complementation with a higA gene reduced the expression of those genes (Figure 5A). Overexpression of HigB also increased the expression of these genes (Figure 5B). These results indicate that the three genes might be involved in the HigB mediated repression of the intracellular c-di-GMP level.
Figure 5.
HigB up-regulates the expression of PA2133, PA2200 and PA3825. (A) PA14, the higA:Tn mutant, the complemented strain and the ΔhigBΔhigA double mutant were grown to an OD600 of 2.0. The mRNA levels of PA2133, PA2200 and PA3825 were determined by real-time PCR. (B) PA14 containing pMMB67EH or pMMB67EH-higB-His was grown in the presence of 0.1 mM IPTG. The mRNA levels of PA2133, PA2200 and PA3825 were determined by real-time PCR. ** p < 0.01, *** p < 0.005 by Student’s t-test.
2.5. PA2133, PA2200 and PA3825 Contribute to the HigB Mediated Decrease of c-di-GMP
To explore the roles of PA2133, PA2200 and PA3825 in the reduction of c-di-GMP, each individual gene was knocked out in wild type PA14. Compared to wild type PA14, mutation of each of the genes increased the biofilm formation and the expression of cdrA as well as partially reduced the expression of T3SS genes upon overexpression of higB (Figure 6). Then we constructed a triple mutant by deleting all the three genes, resulting in ΔPA2133ΔPA2200ΔPA3825. Upon overexpressing higB, the triple mutant displayed similar levels of biofilm formation, expression of cdrA and the T3SS genes as those in wild type PA14 (Figure 7). These results suggest that the up regulation of PA2133, PA2200 and PA3825 are involved in the HigB mediated reduction of intracellular level of c-di-GMP and the reciprocal regulation of the T3SS genes and biofilm formation.
Figure 6.
Roles of PA2133, PA2200 and PA3825 in HigB mediated regulation on biofilm formation and the expression of cdrA and T3SS genes. Biofilm formation, mRNA levels of cdrA and the T3SS genes were examined in PA14 and the indicated mutant containing the empty vector pMMB67EH or the higB overexpression plasmid pMMB67EH-higB-His. Biofilm formation was observed and quantified by staining with crystal violet. The mRNA levels of indicated genes were examined by real-time PCR. Comparison between PA14 and the ΔPA2133 (A–C), the ΔPA2200 (D–F) and the ΔPA3825 mutants (G–I). Error bars represent the standard errors. * p < 0.05, ** p < 0.01, *** p < 0.005 by Student’s t-test.
Figure 7.
Effects of simultaneous deletion of PA2133, PA2200 and PA3825 on HigB mediated regulation on biofilm formation, cdrA and T3SS gene expression. (A) Biofilm formation by PA14 and the ΔPA2133ΔPA2200ΔPA3825 mutant containing the empty vector pMMB67EH or pMMB67EH-higB-His. IPTG was supplemented at 0.1 mM. (B) PA14 or the ΔPA2133ΔPA2200ΔPA3825 mutant containing pMMB67EH or pMB67EH-higB-His was grown to an OD600 of 2.0 in the presence of 0.1 mM IPTG. The mRNA level of cdrA (C) and exsA, exsC and pcrV were determined by real time PCR. Error bars represent the standard errors. ** p < 0.01, *** p < 0.005 by Student’s t-test.
3. Discussion
In this study, we found that HigB controls the intracellular c-di-GMP level in P. aeruginosa. Studies on P. vulgaris HigB demonstrate that the toxin cleaves mRNA in a ribosome dependent manner with a preference of A rich sequences [15]. Overexpression of the Mycobacterium tuberculosis higB gene resulted in the down regulation of IdeR and Zur regulated genes. In addition, the M. tuberculosis HigB cleaves tmRNA, which is involved in ribosome rescue [23,24]. The antitoxin HigA of M. tuberculosis has been confirmed to bind to the promoter of the higB-higA operon, however, no other binding site of HigA was identified on the M. tuberculosis chromosome [25]. These results suggest that HigB might specifically regulate a subset of genes in M. tuberculosis. Studies in Caulobacter crescentus revealed that HigB controls the bacterial tolerance to nalidixic acid and cell cycle by targeting the mRNAs of acrB2 and ctrA, respectivel and that the preferred cleavage site of HigB was found to be UCG [26]. Considering the ribosome (translation) dependent cleavage by HigB and the possible random distribution of the A rich coding sequences or UCG, ribosome stalling and the sequences adjacent to HigB preferred regions might be involved in the cleavage specificity and efficiency of HigB.
In P. aeruginosa, HigB controls a subset of virulence factors, including pyochelin, pyocyanin, T3SS, swarming motility and biofilm formation [3,10]. The amino acid residues G17, I66, W72 and R73 of HigB are conserved in various species and might be the active site. Point mutation of each of the residues reduced the growth inhibitory effect of HigB (Figure S3). We further found that HigB represses the intracellular c-di-GMP level though up regulation of the expression of three c-di-GMP degradation genes, namely PA2133, PA2200 and PA3825. Based on the endonuclease function of HigB, we suspect that HigB might repress the expression of the negative regulator(s) of those three genes. Further studies are needed to elucidate this aspect of the regulatory mechanism.
Recently, PA2133 has been demonstrated to control biofilm formation through degrading c-di-GMP [27]. However, PA2133 represses the expression of T3SS genes under a T3SS inducing condition independent of its phosphodiesterase activity [27]. In our study, we found that mutation of PA2133 in PA14 did not affect the expression of the T3SS genes under a T3SS non-inducing condition (grown in LB), but reduced HigB mediated up regulation of the T3SS genes. Therefore, PA2133 might possess dual functions in controlling c-di-GMP and the T3SS.
Overall, we found that HigB represses biofilm formation through reducing the intracellular c-di-GMP level. Previous studies demonstrate enhanced persister cell formation in biofilm [28,29]. In P. aeruginosa, HigB promoted persister cells might be defective in biofilm formation. We suspect that the cells with activated HigB might be embedded in the extracellular matrix generated by other cells, thus forming a heterogeneous community that is highly tolerant to antibiotics and host immune attack.
4. Materials and Methods
4.1. Strains, Plasmids and Growth Conditions
The bacterial strains, plasmids and primers used in this study are listed in Table S1. The Escherichia coli and P. aeruginosa strains were cultured in the Luria–Bertani (LB) broth (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, pH 7.4) at 37 °C. When needed, antibiotics were used at the following concentrations (μg/mL): For E.coli, tetracycline 100 ampicillin 100; for P. aeruginosa, gentamicin 100, carbenicillin 150 (BBI life sciences). Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at indicated concentrations. Chromosomal gene mutation was constructed as described previously [30].
4.2. RNA Preparation and qRT-PCR Analysis
RNA was purified with a RNAprep Kit (Tiangen Biotech, Beijing, China). Briefly, bacteria were grown to the stationary phase (OD600 = 2). 200 μL bacterial culture was collected, followed by RNA purification. cDNA was synthesized from each RNA sample by using a Prime-Script Reverse Transcriptase (TaKaRa, Dalian, China). Real-time PCR was performed with the SYBR Premix ExTaq II (TaKaRa) in a CFX Real-Time system (Bio-Rad, Hercules, CA, USA). rpsL (the 30s ribosomal protein coding gene) was used as the internal control.
4.3. Biofilm Assays
Biofilm formation was examined as previously described [31]. Briefly, bacteria were grown to an OD600 of 0.8. Then the bacteria were diluted to an OD600 of 0.025 in LB broth and 200 μL of the bacterial suspension was inoculated in each well of a 96-well plate. The plates were incubated at 37 °C for 24 h. Then the wells were washed with 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, pH 7.4) 3 times and dried in an incubator for 20 min. The wells were stained with 0.05% crystal violet for 20 min. Afterwards, the wells were washed twice with PBS. 200 μL methanol was added to each well and incubated at room temperature for 15 min. The solubilized crystal violet and the cell density were quantified by measuring its optical density (OD) at 590 nm and 600 nm with the microplate reader Varioskan Flash (Thermo Scientific, Billerica, MA, USA).
4.4. Quantification of c-di-GMP by Thiazole Orange
The concentration of c-di-GMP was measured as described previously [18]. Bacteria at the OD600 of 2.0 were harvested by centrifugation at 13,000× g for 3 min. The pellet was resuspended in 1 mL 10 mM Tris-HCl (pH 8.0) with 100 mM NaCl. The cells were lysed through sonication. The protein concentrations were measured with a Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA, USA). 12% perchloric acid was then added to the mixture and incubated for 10 min on ice. The solution was neutralized by aqueous alkali (0.4 M Tris, 3 M KOH and 2 M KCl) [32]. The solution was subjected to centrifugation at 12,000× g for 5 min at 4 °C. The supernatant was then filtered through a 0.2 μm filter and a 3 kD exclusion column. The supernatant was heated at 95 °C for 5 min and then cooled to room temperature for 15 min, followed by addition of thiazole orange (TO) at a final concentration of 30 μM. The mixture was incubated for 12 h at 4 °C. The signal was detected with a Varioskan Flash microplate reader (Thermo Scientific, Billerica, MA, USA) at 10 °C with the following settings: excitation 508 nm, emission 533 nm [18,32].
4.5. Transcriptional Reporter Assay
To construct the cdrA promoter and lacZ transcriptional fusion (PcdrA-lacZ), a DNA fragment containing the cdrA promoter was amplified by PCR, and cloned upstream of the promoter-less lacZ in the plasmid pDN19lacZΩ. The empty vector pDN19lacZΩ was used as a negative control. To measure the LacZ levels, overnight bacterial cultures were diluted at 1:100 in fresh LB and grown to an OD600 of 2.0. The β-galactosidase activity was determined with ortho-nitrophenyl-galactopyranoside (ONPG) (BBI life sciences) as described previously [33,34].
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/10/11/424/s1, Figure S1: Role of HigB in the expression of rsmY and rsmZ, Figure S2: Expression levels of c-di-GMP metabolism genes in PA14 and the higA mutant, Figure S3: Effects of point mutations on the growth inhibitory function of HigB, Table S1: Bacterial strains, plasmids and primers used in this study.
Author Contributions
Conceived and designed the experiments: W.W., S.J., Y.Z. Performed the experiments: Y.Z., B.X., M.L., J.S., Y.L. Analyzed the data: Y.Z., Y.J., F.B., Z.C., S.J., W.W. Wrote the paper: Y.Z., W.W., S.J.
Funding
This work was supported by National Science Foundation of China (31670130, 41831287, and 31600110); Science and Technology Program of Sichuan Province (2018JZ0069); Science and Technology Committee of Tianjin (15JCYBJC53900 and 15JCZDJC33000) and the State Key Laboratory of Medicinal Chemical Biology.
Conflicts of Interest
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Key Contribution
Identification of a novel regulatory pathway through which the toxin HigB influences the level of c-di-GMP, which subsequently controls the type III secretion system and biofilm formation in Pseudomonas aeruginosa.
References
- 1.Pandey D.P., Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schureck M.A., Repack A., Miles S.J., Marquez J., Dunham C.M. Mechanism of endonuclease cleavage by the HigB toxin. Nucleic Acids Res. 2016;44:7944–7953. doi: 10.1093/nar/gkw598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M., Long Y., Liu Y., Liu Y., Chen R., Shi J., Zhang L., Jin Y., Yang L., Bai F., et al. HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced. Front. Cell. Infect. Microbiol. 2016;6:125. doi: 10.3389/fcimb.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goeders N., Melderen L.V. Toxin-antitoxin systems as multilevel interaction systems. Toxins. 2014;6:304–324. doi: 10.3390/toxins6010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocker A., Meinhart A. Type II toxin: Antitoxin systems. More than small selfish entities? Curr. Genet. 2016;62:287–290. doi: 10.1007/s00294-015-0541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (mqsr, b3022) J. Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y., Wang X., Ma Q., Zhang X.-S., Wood T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through yjgk (taba) and fimbriae. J. Bacteriol. 2009;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Wood T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y., Wang X., Zhang X.-S., Grigoriu S., Page R., Peti W., Wood T.K. Escherichia coli toxin/antitoxin pair mqsr/mqsa regulate toxin cspd. Environ. Microbiol. 2010;12:1105–1121. doi: 10.1111/j.1462-2920.2009.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood T.L., Wood T.K. The higb/higa toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. MicrobiologyOpen. 2016;5:499–511. doi: 10.1002/mbo3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moscoso J.A., Mikkelsen H., Heeb S., Williams P., Filloux A. The Pseudomonas aeruginosa sensor rets switches type iii and type vi secretion via c-di-gmp signalling. Environ. Microbiol. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 12.Li G., Shen M., Lu S., Le S., Tan Y., Wang J., Zhao X., Shen W., Guo K., Yang Y., et al. Identification and characterization of the hicab toxin-antitoxin system in the opportunistic pathogen Pseudomonas aeruginosa. Toxins. 2016;8:113. doi: 10.3390/toxins8040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen S.B., Ghout M., Griffin A.S., Petersen B., Johansen H.K., Molin S. Diversity, prevalence, and longitudinal occurrence of type II toxin-antitoxin systems of Pseudomonas aeruginosa infecting cystic fibrosis lungs. Front. Microbiol. 2017;8:1180. doi: 10.3389/fmicb.2017.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams J.J., Halvorsen E.M., Dwyer E.M., DiFazio R.M., Hergenrother P.J. Toxin-antitoxin (ta) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant staphylococcus aureus. FEMS Microbiol. Lett. 2011;322:41–50. doi: 10.1111/j.1574-6968.2011.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurley J.M., Woychik N.A. Bacterial toxin higb associates with ribosomes and mediates translation-dependent mrna cleavage at a-rich sites. J. Biol. Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabi N., Zheng Y., Opoku-Temeng C., Du Y., Bonsu E., Sintim H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- 17.Rybtke M., Berthelsen J., Yang L., Høiby N., Givskov M., Tolker-Nielsen T. The lapg protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the cdra adhesin on the cell surface. MicrobiologyOpen. 2015;4:917–930. doi: 10.1002/mbo3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama S., Kelsey I., Wang J., Roelofs K., Stefane B., Luo Y., Lee V.T., Sintim H.O. Thiazole orange-induced c-di-gmp quadruplex formation facilitates a simple fluorescent detection of this ubiquitous biofilm regulating molecule. J. Am. Chem. Soc. 2011;133:4856–4864. doi: 10.1021/ja1091062. [DOI] [PubMed] [Google Scholar]
- 19.Valentini M., Filloux A. Biofilms and cyclic di-gmp (c-di-gmp) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016;291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Güvener Z.T., Harwood C.S. Subcellular location characteristics of the Pseudomonas aeruginosa ggdef protein, wspr, indicate that it produces cyclic-di-gmp in response to growth on surfaces. Mol. Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman J.W., Tifrea D.F., Harwood C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha D.-G., Richman M.E., O’Toole G.A. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa pa14. Appl. Environ. Microbiol. 2014;80:3384–3393. doi: 10.1128/aem.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuessler D.L., Cortes T., Fivian-Hughes A.S., Lougheed K.E.A., Harvey E., Buxton R.S., Davis E.O., Young D.B. Induced ectopic expression of higb toxin in mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mrnas and cleavage of tmrna. Mol. Microbiol. 2013;90:195–207. doi: 10.1111/mmi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y., Jin S., Wu W. Regulation of bacterial gene expression by ribosome stalling and rescuing. Curr. Genet. 2016;62:309–312. doi: 10.1007/s00294-015-0545-3. [DOI] [PubMed] [Google Scholar]
- 25.Fivian-Hughes A.S., Davis E.O. Analyzing the regulatory role of the higa antitoxin within mycobacterium tuberculosis. J. Bacteriol. 2010;192:4348–4356. doi: 10.1128/JB.00454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkpatrick C.L., Martins D., Redder P., Frandi A., Mignolet J., Chapalay J.B., Chambon M., Turcatti G., Villier P.H. Growth control switch by a DNA-damage-inducible toxin-antitoxin system in Caulobacter crescentus. Nat. Microbiol. 2016;1:16008. doi: 10.1038/nmicrobiol.2016.8. [DOI] [PubMed] [Google Scholar]
- 27.Rossello J., Lima A., Gil M., Duarte J.R., Correa A., Carvalho P.C., Kierbel A., Durán R. The EAL-domain protein FcsR regulates flagella, chemotaxis and type III secretion system in Pseudomonas aeruginosa by a phosphodiesterase independent mechanism. Sci. Rep. 2017;7:10281. doi: 10.1038/s41598-017-09926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flemming H.C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 30.Hoang T.T., Karkhoff-Schweizer R.R., Kutchma A.J., Schweizer H.P. A broad-host-range flp-frt recombination system for site-specific excision of chromosomally-located dna sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 31.Li K., Xu C., Jin Y., Sun Z., Liu C., Shi J., Chen G., Chen R., Jin S., Wu W. Suhb is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio. 2013;4:e00419–13. doi: 10.1128/mBio.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G., Zhao Q., Zhu F., Chen R., Jin Y., Liu C., Pan X., Jin S., Wu W., Cheng Z. Oligoribonuclease is required for the type iii secretion system and pathogenesis of Pseudomonas aeruginosa. Microbiol. Res. 2016;188–189:90–96. doi: 10.1016/j.micres.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Ha U.-H., Kim J., Badrane H., Jia J., Baker H.V., Wu D., Jin S. An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-exsa to suppress the type iii secretion system. Mol. Microbiol. 2004;54:307–320. doi: 10.1111/j.1365-2958.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- 34.Deng X., Li M., Pan X., Zheng R., Liu C., Chen F., Liu X., Cheng Z., Jin S., Wu W. Fis regulates type iii secretion system by influencing the transcription of exsa in Pseudomonas aeruginosa strain PA14. Front. Microbiol. 2017;8:669. doi: 10.3389/fmicb.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.