Abstract
Constitutional loss-of-function pathogenic variants in the tumor suppressor genes BRCA1 and BRCA2 are widely associated with an elevated risk of ovarian cancer (OC). As only ~15% of OC individuals carry the BRCA1/2 pathogenic variants, the identification of other potential OC-susceptibility genes is of great clinical importance. Here, we established the prevalence and spectrum of the germline pathogenic variants in the BRCA1/2 and 23 other cancer-related genes in a large Polish population of 333 unselected OC cases. Approximately 21% of cases (71/333) carried the BRCA1/2 pathogenic or likely pathogenic variants, with c.5266dup (p.Gln1756Profs*74) and c.3700_3704del (p.Val1234Glnfs*8) being the most prevalent. Additionally, ~6% of women (20/333) were carriers of the pathogenic or likely pathogenic variants in other cancer-related genes, with NBN and CHEK2 reported as the most frequently mutated, accounting for 1.8% (6/333) and 1.2% (4/333) of cases, respectively. We also found ten pathogenic or likely pathogenic variants in other genes: 1/333 in APC, 1/333 in ATM, 2/333 in BLM, 1/333 in BRIP1, 1/333 in MRE11A, 2/333 in PALB2, 1/333 in RAD50, and 1/333 in RAD51C, accounting for 50% of all detected variants in moderate- and low-penetrant genes. Our findings confirmed the presence of the additional OC-associated genes in the Polish population that may improve the personalized risk assessment of these individuals.
Keywords: ovarian cancer, low-penetrance gene, BRCA1/2, PARP1 inhibitor, next-generation sequencing, CHEK2, NBN, BARD1, mismatch repair genes
1. Introduction
Ovarian carcinoma (OC), one of the most common gynecological malignancies, accounts for an estimated 239,000 new cases and 152,000 deaths worldwide annually, with the highest incidence rate in Central and Eastern Europe (11.4 per 100,000) [1]. Although the 5-year relative survival rate for all stages combined increased from 33.7% in 1975 to almost 50% in 2009 [2], OC is still the leading cause of death from cancer in women. Because of non-specific symptoms and the lack of effective screening tests, most individuals are diagnosed with an advanced disease (stage III or IV) with the 5-year survival rate below 30% [3].
Morphologically, the epithelial neoplasms are divided into five main subtypes (serous, endometrial, mucinous, clear cell and transitional cell tumors) that have different genetic risk factors, treatment responses, and prognosis. High-grade serous ovarian carcinoma represents approximately 70% of OC, followed by endometrial and clear cell cancer (10% each) [4]. The first-line treatment includes optimal cytoreductive surgery and platinum-based chemotherapy; however, chemotherapy sensitivity varies widely among individuals, depending mostly on the tumor’s histologic subtype with its grade, as well as the overall cancer stage [5,6].
Since the identification of tumor suppressor genes, BRCA1 and BRCA2 [7,8], of which germline pathogenic variants predispose to a significant increase in breast and ovarian cancer risk, several other cancer-related genes have been associated with their pathogenesis and progression. The application of next-generation sequencing (NGS) significantly facilitates rapid screening for new cancer susceptibility genes, and indeed, a number of pathogenic variants in the so-called moderate- and low-penetrance genes, such as ATM, BRIP1, CHEK2, PALB2 or BARD1, have been reported to be correlated with a moderate lifetime risk for breast and/or ovarian cancer [9]. However, so far for OC, these data are still limited, and only the loss-of-function pathogenic variants of the mismatch repair (MMR) genes, that is, MLH1, MSH2, MSH6, and PMS2, are clearly classified as OC-susceptibility genes [10].
In this study, we established the prevalence and spectrum of the constitutional pathogenic variants in the BRCA1/2 and 23 other cancer-related genes that may play a role in the predisposition to OC, in a large Polish population of 333 unselected OC individuals.
2. Results
2.1. Pathogenic Variants in BRCA1/2
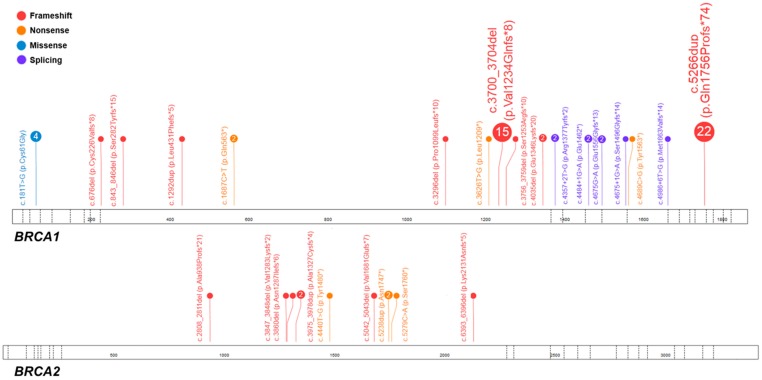
All individuals included in this study had the comprehensive BRCA1/2 molecular screening as a part of our previous study [11]. In total, pathogenic variants were found in 71 cases (n = 71/333; 21.3%), with the vast majority of variants located in BRCA1 (n = 60/71; 84.5%), and only eleven in BRCA2 (15.5%). Forty individuals (n = 40/71; 56.3%) were heterozygous for one of the most recurrent pathogenic variants observed in the Polish population, i.e., c.68_69del (p.Glu23Valfs*17), c.181T>G (p.Cys61Gly), c.3700_3704del (p.Val1234Glnfs*8), c.4035del (p.(Glu1346Lysfs*20) and c.5266dup (p.Gln1756Profs*74), all located in BRCA1 [12,13,14]. The most common pathogenic variants were c.5266dup (p.Gln1756Profs*74) and c.3700_3704del (p.Val1234Glnfs*8), accounting for 31% (n = 22/71) and 21.1% of cases (n = 15/71), respectively.
Among BRCA1/2 positive cases, the most common histological subtype was serous OC (n = 59/71; 83.1%), followed by low differentiated (n = 5/71; 7%) and endometrial (n = 3/71; 4.2%) tumors. Clear cell and mesonephroid tumors each were represented in less than 3% (n = 2/71; 2.8%) of positive cases.
The average age of onset in BRCA1/2 carriers was 58.2 years (range: 27–87), with ten years of difference in diagnosis between BRCA1 and BRCA2 individuals (49.3 versus 59 years). All pathogenic variants identified in the BRCA1 and BRCA2 genes are displayed in Table 1 and Figure 1.
Table 1.
Clinical, histopathological and molecular data of the ovarian cancer individuals heterozygous for the BRCA1/2 pathogenic variants.
| Case No. | Exon/Intron | Variant in Corresponding cDNA | Predicted Amino Acid Sequence | Variant Type | dbSNP ID 1 |
ACMG Classification 2 | Age, Years | FIGO Stage | Histology | Family History |
|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 (NM_007294.3; LRG_292t1) | ||||||||||
| M115 | 5 | c.181T>G | p.(Cys61Gly) | M | rs28897672 | Pathogenic (PS3 + PS4 + PM2 + PP1+ PP5) | 36 | IIIC | serous | - |
| M296 | 54 | IIIC | serous | + | ||||||
| K199 | 46 | IIIC | serous | - | ||||||
| K221 | 51 | IIIC | serous | - | ||||||
| D166 | 11 | c.676del | p.(Cys226Valfs*8) | F | rs80357941 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 43 | ND | serous | - |
| K187 | 11 | c.843_846del | p.(Ser282Tyrfs*15) | F | rs80357919 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 46 | IIIC | serous | - |
| D18 | 11 | c.1292dup | p.(Leu431Phefs*5) | F | rs80357528 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 42 | IIIC | serous | + |
| D138 | 11 | c.1687C>T | p.(Gln563*) | N | rs80356898 | Pathogenic (PVS1+PS4+PM1+PM2 + PP1 + PP5) | 49 | ND | serous | ND |
| K100 | 45 | IIB | serous | + | ||||||
| M397 | 11 | c.3296del | p.(Pro1099Leufs*10) | F | rs80357815 | Pathogenic (PVS1 + PM2 + PM4 + PP5) | 59 | IIIC | serous | ND |
| D53 | 11 | c.3626T>G | p.(Leu1209*) | N | rs786203884 | Pathogenic (PVS1 + PS4 + PM2 + PM4 + PP1 + PP5) | 59 | ND | low differentiated | ND |
| M22 | 11 | c.3700_3704del | p.(Val1234Glnfs*8) | F | rs80357609 | Pathogenic (PVS1 + PM1 + PM2 + PP5) | 61 | IIIB | serous | ND |
| M38 | 43 | IIIC | serous | - | ||||||
| M66 | 52 | ND | serous | ND | ||||||
| D3 | 45 | IIIC | serous | + | ||||||
| D23 | 47 | IV | serous | - | ||||||
| D63 | 55 | IIIC | serous | + | ||||||
| D70 | 48 | ND | serous | ND | ||||||
| D71 | 47 | ND | serous | - | ||||||
| D104 | 56 | ND | low differentiated | - | ||||||
| D136 | 43 | IIIB | serous | - | ||||||
| D156 | 47 | ND | serous | - | ||||||
| K65 | 64 | IIIC | serous | + | ||||||
| K125 | 50 | IIIC | serous | - | ||||||
| K152 | 66 | IV | serous | + | ||||||
| K189 | 63 | IIIC | low differentiated | + | ||||||
| K74 | 11 | c.3756_3759del | p.(Ser1253Argfs*10) | F | rs80357868 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 45 | IV | serous | + |
| D15 | 11 | c.4035del | p.(Glu1346Lysfs*20) | F | rs80357711 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 54 | IIB | serous | - |
| K222 | ND | IIB | low differentiated | + | ||||||
| M92 | 13 | c.4357+2T>G; r.[4186_4357del];[=] |
p.(Arg1377Tyrfs*2) | S | rs80358152 | Pathogenic (PVS1 + PS2 + PM2 + PP1 + PP3 + PP5) | 48 | IIIC | serous | + |
| M95 | 13 | 45 | IIIC | serous | - | |||||
| M395 | 14 | c.4484+1G>A; r.[4358_4484del];[=] | p.(Glu1462*) | S | rs80358063 | Pathogenic (PVS1 + PS3 + PP1 + PP3 + PP4 + PP5) | 41 | IIIC | serous | ND |
| K201 | 52 | IV | mesonephroid | + | ||||||
| D140 | 15 | c.4675G>A; r.[4665_4675del];[=] | p.(Glu1556Glyfs*13) | S | rs80356988 | Pathogenic (PS3 + PM1 + PM2 + PP1 + PP5) | 37 | IIIC | serous | ND |
| K73 K108 |
15 | c.4675+1G>A; r.[4485_4675del];[=] | p.(Ser1496Glyfs*14) | S | rs80358044 | Pathogenic (PVS1 + PS3 + PM1 + PM2 + PP1 + PP3 + PP5) | 44 41 |
IIIC IIIC |
serous clear cell |
ND - |
| D152 | 16 | c.4689C>G | p.(Tyr1563*) | N | rs80357433 | Pathogenic (PVS1 + PS4 + PM1 + PP1 + PP3 + PP4) | 38 | ND | serous | - |
| D72 | 16 | c.4986+6T>G, r.[4916+1_4916+65ins];[=] |
p.(Met1663Valfs*14) | S | rs80358086 | Pathogenic (PS3 + PS4 + PM1 + PM2 + PP1 + PP3 + PP5) | 47 | IIIC | serous | + |
| M50 | 20 | c.5266dup | p.(Gln1756Profs*74) | F | rs397507247 | Pathogenic (PVS1 + PM1 + PP5) | 52 | IIIC | serous | + |
| M108 | 63 | ND | serous | ND | ||||||
| M138 | 65 | ND | serous | + | ||||||
| M225 | 50 | IIID | serous | + | ||||||
| M226 | 51 | IIIC | serous | + | ||||||
| M227 | 58 | IIIC | serous | - | ||||||
| M314 | 46 | IIIC | endometrial | + | ||||||
| M323 | 37 | IIIC | serous | + | ||||||
| M368 | 60 | ND | serous | ND | ||||||
| M374 | 66 | ND | serous | ND | ||||||
| M378 | 36 | ND | serous | ND | ||||||
| D9 | 43 | IIIB | endometrial | + | ||||||
| D27 | 66 | IIIC | serous | ND | ||||||
| D66 | 47 | ND | serous | + | ||||||
| D83 | 50 | IIIC | serous | + | ||||||
| D99 | ND | IIIC | serous | ND | ||||||
| D105 | 50 | ND | serous | + | ||||||
| D144 | 56 | IIC | serous | ND | ||||||
| D149 | 40 | ND | serous | - | ||||||
| K53 | 60 | IIIC | serous | + | ||||||
| K121 | 52 | IIIB | clear cell | - | ||||||
| K197 | 43 | IIIC | endometrial | - | ||||||
| BRCA2(NM_000059.3; LRG_293t1) | ||||||||||
| M164 | 11 | c.2808_2811del | p.(Ala938Profs*21) | F | rs80359351 | Pathogenic (PVS1 + PS4 + PM2 + PP5) | 81 | ND | serous | + |
| D121 | 11 | c.3847_3848del | p.(Val1283Lysfs*2) | F | rs746229647 | Pathogenic (PVS1 + PM1 + PM2 + PP1 + PP5) | ND | ND | serous | ND |
| D114 | 11 | c.3860del | p.(Asn1287Ilefs*6) | F | rs80359406 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 54 | IIIC | serous | + |
| M8 | 11 | c.3975_3978dup | p.(Ala1327Cysfs*4) | F | rs764689249 | Pathogenic (PVS1 + PS4 + PM2 + PP5) | 45 | IV | serous | + |
| K183 | 56 | IV | low differentiated | + | ||||||
| K212 | 11 | c.4440T>G | p.(Tyr1480*) | N | rs397507719 | Pathogenic (PVS1 + PM1 + PM2 + PP1 + PP3 + PP5) | 54 | IIIC | serous | + |
| M178 | 11 | c.5042_5043del | p.(Val1681Glufs*7) | F | rs80359478 | Pathogenic (PVS1 + PS4 + PM1 + PM2 + PP1 + PP5) | 62 | IIIC | serous | - |
| D160 | 11 | c.5238dup | p.(Asn1747*) | F | rs80359499 | Pathogenic (PVS1 + PM1 + PM2 + PP1 + PP5) | 51 | IIIC | serous | - |
| K172 | 55 | IIIC | serous | - | ||||||
| K254 | 11 | c.5279C>A | p.(Ser1760*) | N | novel | Pathogenic (PVS1 + PM1 + PM2 + PP1 + PP3 + PP5) | ND | IV | mesonephroid | - |
| K93 | 11 | c.6393_6396del | p.(Lys2131Asnfs*5) | F | rs397507849 | Pathogenic (PVS1 + PM1 + PM2 + PP1 + PP5) | 76 | IIIC | serous | ND |
1 RS number based on the dbSNP Database (https://www.ncbi.nlm.nih.gov/projects/SNP/) (as of September 2018); 2 Interpretation of variants pathogenicity based on the American the College of Medical Genetics and Genomics (ACMG) recommendations [15], i.e., PVS1: null variant (nonsense, frameshift, canonical +/−1 or 2 splice sites, initiation codon, single or multi-exon deletion) in a gene where loss of function (LOF) is a known mechanism of a disease, PS1: same amino acid change as a previously established pathogenic variant regardless of nucleotide change, PS2: proven de novo (both maternity and paternity confirmed), PS3: well-established functional studies, PS4: the prevalence of the variant in affected individuals is significantly increased compared to the prevalence in controls, PM1: located in a mutational hot spot and/or in critical functional domain, PM2: absent from controls, PM3: for recessive disorders, detected in trans with a pathogenic variant, PM4: protein length changes due to in-frame or stop-loss variants, PM5: novel missense change at amino acid residue where a different pathogenic missense change has been seen before, PM6: assumed de novo, but without confirmation of paternity and maternity, PP1: co-segregation with disease in multiple affected family members, PP2: missense variant in a gene that has a low rate of benign missense variation, PP3: multiple lines of computational evidence support a deleterious effect on the gene or gene product, PP4: individual’s phenotype or family history is highly specific for a disease, PP5: reputable source reports variant as pathogenic. To classify a variant as pathogenic the following criteria need to be fulfilled: ≥ 2 strong (PS1–PS4) OR 1 strong (PS1–PS4) and ≥3 moderate (PM1–PM6) OR 1 strong (PS1–PS4) and 2 moderate (PM1–PM6) and ≥2 supporting (PP1–PP5) OR 1 strong (PS1–PS4) and 1 moderate (PM1–PM6) and ≥4 supporting (PP1-PP5). Abbreviations: FIGO: International Federation of Gynecology and Obstetrics; ND: no data; F: frameshift; N: nonsense; S: splicing; M: missense.
Figure 1.
Spectrum of the BRCA1/2 pathogenic variants identified in the studied cohort of 333 unselected ovarian cancer individuals. Each number in circle corresponds with the total number of individuals heterozygous for a specific variant. The figure was prepared using the ProteinPaint application (©Copyright 2015 St. Jude Children’s Research Hospital; 262 Danny Thomas Place, Memphis, TN 38105, USA) [16].
2.2. Pathogenic Variants in Moderate- and Low-Penetrance Genes
To further characterize this cohort, we analyzed 23 additional genes that might play a role in the predisposition to OC. Briefly, we identified 6% of carriers (n = 20/333) with 16 different pathogenic variants in one of the tested genes. The most frequently mutated genes were NBN and CHEK2, with pathogenic or likely pathogenic variants presented in six (n = 6/20; 30%) and four individuals (n = 4/20; 20%), respectively. Among six cases with the NBN pathogenic or likely pathogenic variants, four carried a well-known founder mutation, c.657_661del (p.Lys219Asnfs*16). In addition, we found two different pathogenic variants in the BLM and PALB2 genes, and in APC, ATM, BRIP1, MRE11A, RAD50, RAD51C (each variant observed in a single individual). Additionally, within this cohort, four cases were also carriers of the BRCA1 pathogenic variants (see details in Table 2).
Table 2.
Clinical, histopathological and molecular data of the ovarian cancer individuals heterozygous for the pathogenic or likely pathogenic variants in moderate- and low-penetrance genes.
| Case No. | Exon/Intron | Variant in Corresponding cDNA | Predicted Amino Acid Sequence | Variant Type | dbSNP ID 1 |
ACMG Classification 2 | Age, Years |
FIGO Stage | Histology | Family History |
BRCA1/2 Status |
|---|---|---|---|---|---|---|---|---|---|---|---|
| APC (NM_000038.4; LRG_130t1) | |||||||||||
| M33 | 16 | c.7927_7928del | p.(Leu2643Asnfs*10) | F | novel | Pathogenic (PVS1 + PM1 + PM2) |
41 | IIIC | serous | - | - |
| ATM(NM_000051.3, LRG_135t1) | |||||||||||
| D73 | 41 | c.6095G>A | p.(Arg2032Lys) | M | rs139770721 | Pathogenic (PS3 + PM2 + PP3 + PP5) |
59 | IC | serous/mucinous | - | - |
| BLM(NM_000057.2, LRG_20t1) | |||||||||||
| M66 | 2 | c.98 + 1G>C | p.(?) | S | rs750293380 | Pathogenic (PVS1 + PM2 + PP3) |
52 | ND | serous | - | BRCA1: c.3700_3704del |
| M71 | 7 | c.1642C>T | p.(Gln548*) | N | rs200389141 | Pathogenic (PVS1 + PP3 + PP5) |
61 | IIIC | serous | - | - |
| BRIP1 (NM_032043.2; LRG_300t1) | |||||||||||
| K190 | 11 | c.1510dup | p.(Ile504Asnfs*7) | F | rs775735278 | Pathogenic (PVS1 + PM2 + PP5) |
54 | IIB | endometrial | - | - |
| CHEK2(NM_007194.3) | |||||||||||
| M140 | 3 | c.444 + 1G>A, r.[ = ,444 + 1_444 + 2insATAG];[=] |
p.(Glu149Ilefs*41) | S | rs121908698 | Pathogenic (PVS1 + PS3 + PP3 + PP5) | 54 | ND | myxoid leiomyosarcoma | ND | - |
| M374 | 11 | c.1100del | p.(Thr367Metfs*15) | F | rs555607708 | Pathogenic (PVS1 + PS3 + PP3 + PP5) |
66 | IIIC | serous | ND | BRCA1: c.5266dup |
| D104 | 56 | ND | low differentiated | - | BRCA1: c.3700_3704del | ||||||
| M113 | 11 | c.1169A>C | p.(Tyr390Ser) | M | rs200928781 | Likely pathogenic (PS3 + PM1 + PP3 + PP5) | 27 | IIIB | serous | - | - |
| MRE11A(NM_005591.3; LRG_85) | |||||||||||
| M42 | 3 | c.77T>C | p.(Met26Thr) | M | rs372068015 | Likely pathogenic (PM2 + PM3 + PP1 + PP5) | 61 | IIB | serous | - | - |
| NBN (NM_002485.4; LRG_158t1) | |||||||||||
| K74 | 4 | c.373A>T | p.(Lys125*) | N | novel | Pathogenic (PVS1 + PM2 + PP3) |
45 | IV | serous | + | BRCA1: c.3756_3759del |
| K124 | 6 | c.643C>T | p.(Arg215Trp) | M | rs34767364 | Likely pathogenic (PS2 + PM1 + PM3) |
47 | IIIC | serous | - | - |
| M86 | 6 | c.657_661del | p.(Lys219Asnfs*16) | F | rs587776650 | Pathogenic (PVS1 + PS3 + PM2 + PP5) |
64 | IIIC | serous | - | - |
| D131 | 76 | ND | serous | ND | - | ||||||
| K135 | 55 | IIIC | serous | - | - | ||||||
| K208 | ND | IIIB | serous | - | - | ||||||
| PALB2 (NM_024675.3; LRG_308t1) | |||||||||||
| M407 | 5 | c.2456_2463del | p.(Lys819Thrfs*4) | F | novel | Pathogenic (PVS1 + PM2 + PP5) |
53 | IIB | serous | ND | - |
| K219 | 7 | c.2632G>T | p.(Glu878*) | N | novel | Pathogenic (PVS1 + PM1 + PM2 + PP3 + PP5) |
54 | IIIC | ND | - | - |
| RAD50 (NM_005732.3) | |||||||||||
| M161 | 21 | c.3266_3273delinsT | p.(Lys1089Ilefs*17) | F | novel | Pathogenic (PVS1 + PM2 + PP5) |
46 | IA | serous | - | - |
| RAD51C(NM_058216.1; LRG_314t1) | |||||||||||
| K237 | 4 | c.706-2A>G, r.[706_837del];[=] |
p.(Arg237_Val280del) | S | rs587780259 | Pathogenic (PVS1 + PM2 + PP3 + PP5) |
ND | IIC | serous | + | - |
1 RS number based on the dbSNP Database (https://www.ncbi.nlm.nih.gov/projects/SNP/) (as of September 2018); 2 Interpretation of variants pathogenicity based on the American the College of Medical Genetics and Genomics (ACMG) recommendations [15], i.e., PVS1: null variant (nonsense, frameshift, canonical +/−1 or 2 splice sites, initiation codon, single or multi-exon deletion) in a gene where loss of function (LOF) is a known mechanism of a disease, PS1: same amino acid change as a previously established pathogenic variant regardless of nucleotide change, PS2: proven de novo (both maternity and paternity confirmed), PS3: well-established functional studies, PS4: the prevalence of the variant in affected individuals is significantly increased compared to the prevalence in controls, PM1: located in a mutational hot spot and/or in critical functional domain, PM2: absent from controls, PM3: for recessive disorders, detected in trans with a pathogenic variant, PM4: protein length changes due to in-frame or stop-loss variants, PM5: novel missense change at amino acid residue where a different pathogenic missense change has been seen before, PM6: assumed de novo, but without confirmation of paternity and maternity, PP1: co-segregation with disease in multiple affected family members, PP2: missense variant in a gene that has a low rate of benign missense variation, PP3: multiple lines of computational evidence support a deleterious effect on the gene or gene product, PP4: individual’s phenotype or family history is highly specific for a disease, PP5: reputable source reports variant as pathogenic. To classify a variant as pathogenic the following criteria need to be fulfilled: ≥2 strong (PS1–PS4) OR 1 strong (PS1–PS4) and ≥3 moderate (PM1–PM6) OR 1 strong (PS1–PS4) and 2 moderate (PM1–PM6) and ≥2 supporting (PP1–PP5) OR 1 strong (PS1–PS4) and 1 moderate (PM1–PM6) and ≥4 supporting (PP1–PP5). Abbreviations: FIGO: International Federation of Gynecology and Obstetrics; ND: no data; F: frameshift; N: nonsense; S: splicing; M: missense.
The average age of onset in this group of individuals was 53.9 (range 27–76). All identified variants that were classified as potentially causative (pathogenic or likely pathogenic) are displayed in Table 2 and Figure 2.
Figure 2.
Spectrum of the pathogenic or likely pathogenic variants identified in moderate- and low-penetrance genes in the studied cohort of 333 unselected ovarian cancer individuals. Each number in circle corresponds with the total number of individuals heterozygous for a specific variant. The figure was prepared using the ProteinPaint application [16].
2.3. Variants of Uncertain Significance (VUS) in Low-Penetrance Genes
Detailed list of all identified VUS is shown in Table S1. The most common alteration was a missense variant in CHEK2, c.470T>C (p.Ile157Thr), with a prevalence of 2.7% (n = 9/333), including two individuals being recognized as carriers of the BRCA1 pathogenic variants. The second most frequently observed variant was c.511A>G (p.Ile171Val) in the NBN gene, found in six individuals (n = 6/333; 1.8%), including two carriers of BRCA1 and PALB2 pathogenic variants. Additionally, in two patients, we identified two different variants located in consensus splice site region (c.38-3C>G and c.30G>A) of the NBN gene, resulting possibly in an abnormal splicing. However, as no source of RNA was available for these individuals, the missplicing cannot be confirmed.
Finally, we identified an individual classified as a compound heterozygous for BARD1, with an in-frame deletion in exon 4, c.1075_1095del (p.Leu359_Pro365del), and a silent mutation c.1977A>G, both variants reside in the trans position. The proband, diagnosed with OC at the age of 65, had a positive family history for this type of cancer. The proband’s sister died of OC at the age of 36 over three decades ago; therefore, no biological material was available to perform further genetic testing (Figures S1 and S2).
3. Discussion
Constitutional loss-of-function pathogenic variants in the tumor suppressor genes BRCA1 and BRCA2 are widely known to confer an elevated risk of breast and/or ovarian cancer, and their mutational screening has become a standard in routine diagnostic practice. It has been estimated that the lifetime risk for developing OC varies from 20% to 50%, depending on the gene in which the pathogenic variant occurs [17]. As only ~15% of OC individuals are heterozygous for the BRCA1/2 pathogenic variants [18,19,20], the identification of other genes associated with inherited susceptibility to OC is of great clinical importance. So far, several studies aimed at the evaluation of germline alterations in other genes using multi-gene panel testing in a large series of well-characterized OC individuals have been performed, and the potential moderate- and low-penetrance genes have been reported, but their elevated risk of OC is less well characterized [9,10,21,22,23]. Importantly, considering the ethnicity-specific differences in the variants spectrum and frequency, their assessment in all ethnic groups, including the Polish population, is required before making any clinical conclusions. Therefore, in this report, we investigated the prevalence and spectrum of the germline pathogenic and likely pathogenic variants in 25 cancer-related genes through a comprehensive NGS analysis in a large cohort of 333 unselected OC individuals from northern Poland.
Overall, we identified 91 pathogenic or likely pathogenic variant in 87 OC individuals (n = 87/333; 26.1%;) (Figure 3), in agreement with the previous reports [23,24,25]. Besides the BRCA1/2 genes, the most frequently mutated were NBN and CHEK2, with the alterations observed in 1.8% (n = 6/333) and 1.2% (n = 4/333) of cases, respectively (Table 2). Both genes encode for proteins involved in nonhomologous end joining (NHEJ) or homologous recombination (HR), which are critical mechanisms for the accurate repair of DNA double-strand breaks (DSBs). Particularly, nibrin, the product of the NBN gene, regulates the activity of the MRE11A/RAD50/NBN protein complex, while checkpoint kinase 2, the product of the CHEK2 gene, is activated upon DNA damage, and is responsible for transducing the DNA damage signal to downstream repair proteins, such as p53 [21]. According to the recent National Comprehensive Cancer Network (NCCN) guidelines, germline pathogenic variants in both these genes confer to an elevated risk of breast cancer, with the recommendation of the annual mammogram beginning at the age of 40; for OC individuals, however, these variants are not proven to result in an increased risk of OC, or rather, insufficient evidence have been so far reported [26]. In line with the previous studies, suggesting that the specific CHEK2 variant c.1100del (p.Thr367Metfs*15) is more prevalent in the Northern and Eastern Europe countries compared with North America [27,28], we identified the presence of this alteration in two cases from the studied cohort (Table 2 and Figure 2). However, both individuals carried simultaneously a pathogenic variant in the BRCA1 gene; therefore, our findings likely confirmed the previous observations indicating a lack of c.1100del association with an increased risk of OC [29]. Regarding the NBN gene, several studies clearly shown the elevated risk of breast cancer with a positive NBN testing result, however, this risk assessment was limited to a specific NBN pathogenic variant, c.657_661del (p.Lys219Asnfs*16), observed frequently in the Slavic population [30,31]. In this report, variant c.657_661del was present in 4/6 NBN mutation-positive individuals (Table 2 and Figure 2). Other potentially causative variants in the NBN gene included novel nonsense and missense alterations, c.373A>T (p.Lys125*) and c.643C>T (p.Arg215Trp), respectively; the remaining was previously described in twin brothers with Nijmegen breakage syndrome (MIM: 251260) [32]. All but one individual with a positive NBN screening result had a negative family history for cancer, with the negative result of the BRCA1/2 screening. As there is clear evidence that alterations in this gene predispose to an increased risk of breast cancer, but for determining the risk assessment for OC further studies are required, the clinicians involved in the case of OC individuals should be aware of this potential association.
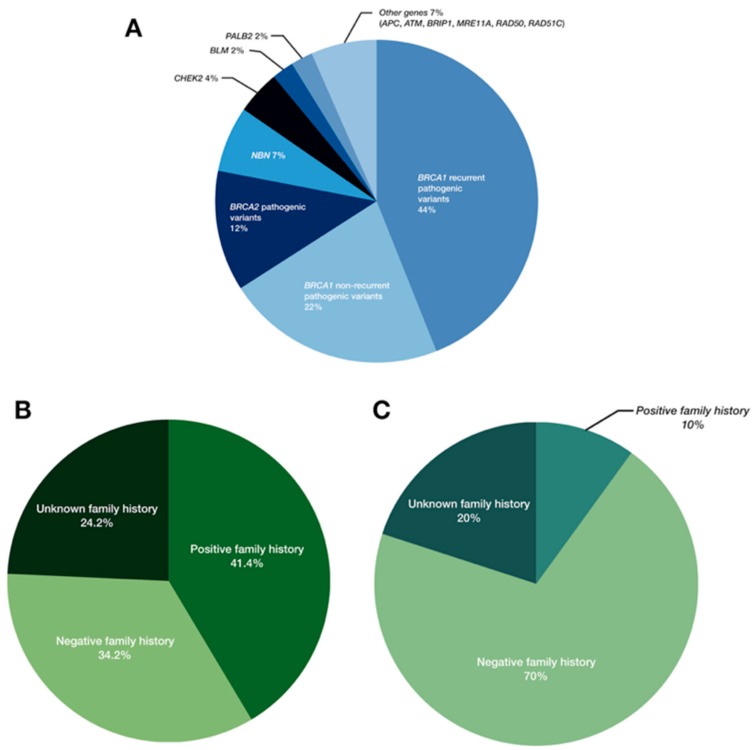
Figure 3.
Frequency of pathogenic and likely pathogenic variants in all analyzed genes (A), the family history status in the BRCA1 and BRCA2 mutation-positive individuals (B) and in the individuals heterozygous for the pathogenic or likely pathogenic variants in other breast/ovarian cancer susceptibility genes (C).
Several other proteins, coded by one of the following genes: ATM, BLM, BRIP1, MRE11A, PALB2, RAD50 or RAD51C, interact with BRCA1/2 proteins in the DBs repair process by HR or NHEJ mechanism, and therefore, they are considered as alternative candidates for OC-susceptibility genes. Indeed, it has been reported that the BRIP1 and RAD51 pathogenic variants confer at least a 6-fold increased risk for OC [29,33,34], but not for breast cancer, in contrast to the NBN and CHEK2 genes. Although the pathogenic variants in these genes were identified in the current study only in two cases (each in a single individual), from a clinical point of view, these results had a great impact on the management of these individuals, as they should be offered risk-reducing salpingo-oophorectomy (RRSO) at the age of 45–50, in line with the current NCCN recommendations [26]. We also found the pathogenic or likely pathogenic variants in other genes, such as ATM, BLM, MRE11A, PALB2, or RAD50, occurring sporadically in 1–2 individuals (Table 2), but their role in the predisposition to OC is still unclear [29,35,36,37].
The most striking finding was the paucity of the pathogenic variants in any of the MMR genes, resulting in Lynch syndrome (MIM: 120435), that is, MLH1, MSH2, MSH6, or PMS2, that are clearly considered as the major causes of hereditary epithelial OC in addition to BRCA1/2 alterations, with a lifetime risk of OC estimated at 6–10% [10]. One of the possible explanations might be that OC in Lynch syndrome is mostly of endometrial or clear cell histology [29,38,39], while ~70% of cases in the studied cohort represent the high-grade serous OC subtype. Nevertheless, similar to the BRIP1 and RAD51 pathogenic variants carriers, individuals heterozygous for a pathogenic variant within one of the aforementioned MMR genes may consider prophylactic RRSO and/or total abdominal hysterectomy [26]; thus, it is critical to identify early those women to provide them with accurate genetic counseling and personalized surveillance.
In the current study, the pathogenic and likely pathogenic variants in the BRCA1/2 genes were identified in 18% (n = 60/333) and 3.3% (n = 11/333) of cases for BRCA1 and BRCA2, respectively, together affecting ~21% of women (Table 1 and Figure 1) who are eligible for the poly (ADP-ribose) polymerase 1 inhibitors (iPARP1) -targeted therapy [40]. Comparing with our preliminary data reported on 134 OC individuals with 20 BRCA1/2 mutation-positive carriers [11], the prevalence here was higher than previously observed (21% versus 14.9%); however, this is still in line with the recent large-scale study performed on 21,401 families, with the BRCA1/2 overall frequency of 24% [41]. As expected, the most recurrent pathogenic variants were Polish founder mutations, that is, c.5266dup (p.Gln1756Profs*74) and c.3700_3704del (p.Val1234Glnfs*8), accounting for 6.6% (n = 22/333) and 4.5% of all cases (n = 15/333), respectively. Indeed, Rebbeck et al. (2018) clearly confirmed based on the analysis performed on 29,700 BRCA1/2-mutation positive families that these two alterations are the most commonly-observed variants in Eastern Europe, including Polish population [19].
The natural consequence of comprehensive molecular screening is a growing number of VUS, for which interpretation is often challenging and clinically problematic. In the present study, we described an individual being a compound heterozygous for BARD1, with an in-frame deletion in exon 4, c.1075_1095del (p.Leu359_Pro365del), and a silent mutation c.1977A>G, reside in trans position (Figures S1 and S2). Both alterations lead to missplicing and overexpression of alternative isoforms, named gamma and eta, and consequently, to significant telomere instability [42]. Taking into account a positive family history for OC, potential actionability of those variants cannot be excluded.
A considerable percentage of mutation-positive individuals for whom the personal data was available was diagnosed at the age of ≥60 (n = 18/85; 21%). Numerous carriers presented a negative family history for cancer, confirming prior observations of the limited significance of age on onset and family history when selecting patients for genetic screening (Table 1 and Table 2 and Figure 3B,C) [11,20,43].
In conclusion, we reported here the spectrum and prevalence of the pathogenic and likely pathogenic variants in OC-susceptibility genes in a Polish population. Through a comprehensive NGS analysis of the high-penetrance BRCA1/2 genes and 23 additional moderate- and low-penetrance genes in a large cohort of 333 unselected OC women from northern Poland, we estimated the overall ethnicity-specific pathogenic variants’ frequency in these genes at ~26%, including ~21% of the BRCA1/2 mutation-positive individuals and ~5% of cases with pathogenic alterations in the other cancer-related genes.
Although the mutational screening using multi-gene panel testing have not yet had therapeutic implications, as only the individuals confirmed with an advanced high-grade serous OC and heterozygous for the germline or somatic BRCA1/2 pathogenic variant are eligible for the targeted therapy with iPARP1 (The Polish National Health Program), the identification of the alterations in other OC-susceptibility genes may significantly improve the personalized risk assessment for OC in these individuals. Patients with RAD51C and RAD51D pathogenic variants who were enrolled in the ARIEL 3 trial showed a significant response to the treatment, with an average progression-free survival time (PFS) of 16.4 months (range, 5.4–30.4 months) [25]. In addition, reversion mutations leading to regained function of HR-proteins (e.g., BRCA1, RAD51C and RAD51D) in tumor cells result in PARP resistance [44]. These results indicate that the target group of OC patients that may benefit from iPARP therapy is more heterogeneous, and as a consequence, that targeted therapies programs inclusion criteria should be amended accordingly.
The application of this approach will allow early detection in those women with pathogenic variants in the genes that clearly predispose them to an elevated risk of OC, providing them accurate genetic counseling with prophylactic management options. Finally, as several of these OC-susceptibility genes, i.e., NBN, ATM or BRIP1, are associated with the development of autosomal recessive disorders, such as Nijmegen breakage syndrome, ataxia-teleangiectasia (MIM: 208900) or Fanconi anemia (MIM: 609054), respectively, genetic counselling for carriers of pathogenic variants in any of these genes should also include a discussion of reproductive and prenatal screening possibilities.
4. Materials and Methods
4.1. Individuals and Sample Collection
The study comprised 333 unselected for age or family history ovarian cancer individuals that were referred to the University Hospital in Gdansk and the Red Cross Hospital in Gdynia between 2007 and 2013.
The histological diagnosis of ovarian cancer was evaluated by an expert pathologist; the most representative subtypes were as follows: serous ovarian cancer, accounting for 70.3% (n = 234/333) of cases, followed by endometrial (n = 32/333; 9.6%;), mucinous (n = 18/333; 5.4%;), undifferentiated (n = 18/333; 5.4%;), and clear cell subtype (n = 13/333; 3.9%;). The remaining 22 cases included tumors of mixed type (n = 8), unspecified tumors (n = 5), carcinosarcomas (n = 3), leiomyosarcomas, and adult granulosa cell tumors (each observed in a single individual). The average age at diagnosis was 58.6 years (27–87).
Informed consent was obtained from all the individuals and the study was approved by the medical review board of the Medical University of Gdansk (NKEBN/399/2011-2012; NKBBN/446/2015).
4.2. DNA Extraction
Genomic DNA was extracted from the whole blood using a red-blood-cell lysis buffer followed by standard phenol-chloroform procedure.
4.3. Molecular Analysis
All samples enrolled to the study were screened for BRCA1 (MIM: 113705) and BRCA2 (MIM: 600185) pathogenic variants using NGS with BRCA MASTR assay v1.2 (Multiplicom, Niel, Belgium). Molecular analysis of additional 23 genes, including APC (MIM: 611731), ATM (MIM: 607585), BARD1 (MIM: 601593), BLM (MIM: 604610), BRIP1 (MIM: 605882), CDH1 (MIM: 192090), CDKN2A (MIM: 600160), CHEK2 (MIM: 604373), MLH1 (MIM: 120436), MRE11A (MIM: 600814), MSH2 (MIM: 609309), MSH6 (MIM: 600678), NBN (MIM: 602667), PALB2 (MIM: 610355), PMS2 (MIM: 600259), PTEN (MIM: 601728), RAD50 (MIM: 604040), RAD51 (MIM: 179617), RAD51B (MIM: 602948), RAD51C (MIM: 602774), RAD51D (MIM: 602954), STK11 (MIM: 602216), TP53 (MIM: 191170), was performed using a combination of TruSeq Custom Amplicon assay (Illumina Inc., San Diego, CA, USA) and the HEAT-Seq Oncology Panel (Roche Sequencing), followed by MiSeq targeted re-sequencing at minimum of 99× coverage (Illumina, Inc., San Diego, CA, USA). The cut-off of 20% was applied. The analysis was performed with Illumina Variant Studio Software v3.0 (Illumina Inc., San Diego, CA, USA), Sequence Pilot (JSI Medical Systems, Ettenheim, Germany) and Geneious Software v9.05 (Biomatters Ltd., Auckland, New Zealand). Presence of pathogenic or likely pathogenic variants detected by NGS analysis was confirmed by bi-directional Sanger Sequencing (ABI PRISM 3130, Life Technologies, Inc., Carlsbad, CA, USA). Interpretation of variants pathogenicity was performed based on the American College of Medical Genetics and Genomics (ACMG) recommendations [15].
5. Conclusions
In conclusion, we propose obligatory BRCA1/2 screening in all OC patients in the Polish population. In addition, in the negative cases analysis of the additional cancer-related genes should be considered, including MMR genes, BRIP1, RAD51C, and RAD51D. Unfortunately, given the potential costs of such expanded diagnostics, it will probably need to be limited to the selected group of patients with specific clinical and histopathological characterization.
To establish recommendations for the expansion of mutational analysis (including other cancer-related genes) in OC-individuals, further studies in larger OC series with detailed family histories are required.
Acknowledgments
We thank the individuals and their families for participating in this study. Magdalena Koczkowska, PhD is also affiliated with the Department of Genetics, University of Alabama at Birmingham, AL, USA.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6694/10/11/442/s1. Figure S1: A pedigree of the family with proband being a compound heterozygote for two BARD1 variants: an in-frame deletion in exon 4, c.1075_1095del (p.Leu359_Pro365del) and a silent variant in exon 10, c.1977A>G, resulting in the missplicing, [r.159\_1903del], [=] (p.Cys53_Trp635delinsfs*12); Figure S2: Extended analysis result for variants: c.1075_1095del and c.1977A>G; Table S1: Variants of uncertain significance (VUS) identified in the studied cohort of 333 unselected ovarian cancer individuals.
Author Contributions
Conceptualization, M.R. and M.K.; Methodology, M.R. and M.K.; Software, M.R.; Validation, M.K., A.K., N.K., P.K., and M.R.; Formal Analysis, M.K., and M.R.; Investigation, M.K., A.K., N.K., P.K., and M.R.; Resources, M.S. (Maciej Stukan), I.B., M.S. (Marcin Sniadecki), J.D. and D.W.; Data Curation, M.R.; Writing-Original Draft Preparation, M.K. and M.R.; Writing-Review & Editing, M.K., W.B., B.W. and M.R.; Visualization, N.K., M.K. and M.R.; Supervision, M.R. and J.L.; Project Administration, M.R. and J.L.; Funding Acquisition, M.R. and J.L.
Funding
This research was funded by National Science Center grant number 2011/02/A/NZ2/00017.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:e359–e386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Noone A.M., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., Chen H.S., et al. SEER Cancer Statistics Review, 1975–2015. [(accessed on April 2018)]; Available online: https://seer.cancer.gov/csr/1975_2015/
- 3.Farley J., Ozbun L.L., Birrer M.J. Genomic analysis of epithelial ovarian cancer. Cell Res. 2008;18:538–548. doi: 10.1038/cr.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat J. Ovarian carcinomas: Five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012;460:237–249. doi: 10.1007/s00428-012-1203-5. [DOI] [PubMed] [Google Scholar]
- 5.Jayson G.C., Kohn E.C., Kitchener H.C., Ledermann J.A. Ovarian cancer. Lancet. 2014;384:1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 6.Moore K., Colombo N., Scambia G., Kim B.G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018 doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 7.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 8.Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 9.Walsh T., Lee M.K., Casadei S., Thornton A.M., Stray S.M., Pennil C., Nord A.S., Mandell J.B., Swisher E.M., King M.C. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H.M., Li S., Black M.H., Lee S., Hoiness R., Wu S., Mu W., Huether R., Chen J., Sridhar S., et al. Association of Breast and Ovarian Cancers with Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol. 2018 doi: 10.1001/jamaoncol.2018.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratajska M., Krygier M., Stukan M., Kuzniacka A., Koczkowska M., Dudziak M., Sniadecki M., Debniak J., Wydra D., Brozek I., et al. Mutational analysis of BRCA1/2 in a group of 134 consecutive ovarian cancer patients. Novel and recurrent BRCA1/2 alterations detected by next generation sequencing. J. Appl. Genet. 2015;56:193–198. doi: 10.1007/s13353-014-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brozek I., Cybulska C., Ratajska M., Piatkowska M., Kluska A., Balabas A., Dabrowska M., Nowakowska D., Niwinska A., Pamula-Pilat J., et al. Prevalence of the most frequent BRCA1 mutations in Polish population. J. Appl. Genet. 2011;52:325–330. doi: 10.1007/s13353-011-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brozek I., Ochman K., Debniak J., Morzuch L., Ratajska M., Stepnowska M., Stukan M., Emerich J., Limon J. High frequency of BRCA1/2 germline mutations in consecutive ovarian cancer patients in Poland. Gynecol. Oncol. 2008;108:433–437. doi: 10.1016/j.ygyno.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 14.Gorski B., Byrski T., Huzarski T., Jakubowska A., Menkiszak J., Gronwald J., Pluzanska A., Bebenek M., Fischer-Maliszewska L., Grzybowska E., et al. Founder mutations in the BRCA1 gene in Polish families with breast-ovarian cancer. Am. J. Hum. Genet. 2000;66:1963–1968. doi: 10.1086/302922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X., Edmonson M.N., Wilkinson M.R., Patel A., Wu G., Liu Y., Li Y., Zhang Z., Rusch M.C., Parker M., et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016;48:4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 18.Pal T., Permuth-Wey J., Betts J.A., Krischer J.P., Fiorica J., Arango H., LaPolla J., Hoffman M., Martino M.A., Wakeley K., et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 19.Rebbeck T.R., Friebel T.M., Friedman E., Hamann U., Huo D., Kwong A., Olah E., Olopade O.I., Solano A.R., Teo S.H., et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018;39:593–620. doi: 10.1002/humu.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Risch H.A., McLaughlin J.R., Cole D.E., Rosen B., Bradley L., Kwan E., Jack E., Vesprini D.J., Kuperstein G., Abrahamson J.L., et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am. J. Hum. Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eoh K.J., Kim J.E., Park H.S., Lee S.T., Park J.S., Han J.W., Lee J.Y., Kim S., Kim S.W., Kim J.H., et al. Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2. Cancer Res. Treat. 2018;50:917–925. doi: 10.4143/crt.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minion L.E., Dolinsky J.S., Chase D.M., Dunlop C.L., Chao E.C., Monk B.J. Hereditary predisposition to ovarian cancer, looking beyond BRCA1/BRCA2. Gynecol. Oncol. 2015;137:86–92. doi: 10.1016/j.ygyno.2015.01.537. [DOI] [PubMed] [Google Scholar]
- 23.Norquist B.M., Harrell M.I., Brady M.F., Walsh T., Lee M.K., Gulsuner S., Bernards S.S., Casadei S., Yi Q., Burger R.A., et al. Inherited Mutations in Women with Ovarian Carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh T., Casadei S., Lee M.K., Pennil C.C., Nord A.S., Thornton A.M., Roeb W., Agnew K.J., Stray S.M., Wickramanayake A., et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swisher E.M., Lin K.K., Oza A.M., Scott C.L., Giordano H., Sun J., Konecny G.E., Coleman R.L., Tinker A.V., O’Malley D.M., et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial. Lancet. Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 26.Daly M.B., Pilarski R., Berry M., Buys S.S., Farmer M., Friedman S., Garber J.E., Kauff N.D., Khan S., Klein C., et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J. Natl. Compr. Cancer Netw. 2017;15:9–20. doi: 10.6004/jnccn.2017.0003. [DOI] [PubMed] [Google Scholar]
- 27.Iniesta M.D., Gorin M.A., Chien L.C., Thomas S.M., Milliron K.J., Douglas J.A., Merajver S.D. Absence of CHEK2*1100delC mutation in families with hereditary breast cancer in North America. Cancer Genet. Cytogenet. 2010;202:136–140. doi: 10.1016/j.cancergencyto.2010.07.124. [DOI] [PubMed] [Google Scholar]
- 28.Kuusisto K.M., Bebel A., Vihinen M., Schleutker J., Sallinen S.L. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 2011;13:e20. doi: 10.1186/bcr2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toss A., Tomasello C., Razzaboni E., Contu G., Grandi G., Cagnacci A., Schilder R.J., Cortesi L. Hereditary ovarian cancer: Not only BRCA 1 and 2 genes. Biomed. Res. Int. 2015;2015:e341723. doi: 10.1155/2015/341723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffen J., Nowakowska D., Niwinska A., Czapczak D., Kluska A., Piatkowska M., Wisniewska A., Paszko Z. Germline mutations 657del5 of the NBS1 gene contribute significantly to the incidence of breast cancer in Central Poland. Int. J. Cancer. 2006;119:472–475. doi: 10.1002/ijc.21853. [DOI] [PubMed] [Google Scholar]
- 31.Zhang B., Beeghly-Fadiel A., Long J., Zheng W. Genetic variants associated with breast-cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet. Oncol. 2011;12:477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seemanova E., Sperling K., Neitzel H., Varon R., Hadac J., Butova O., Schrock E., Seeman P., Digweed M. Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability. J. Med. Genet. 2006;43:218–224. doi: 10.1136/jmg.2005.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loveday C., Turnbull C., Ruark E., Xicola R.M., Ramsay E., Hughes D., Warren-Perry M., Snape K., Eccles D., Evans D.G., et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat. Genet. 2012;44:475–476. doi: 10.1038/ng.2224. [DOI] [PubMed] [Google Scholar]
- 34.Ramus S.J., Song H., Dicks E., Tyrer J.P., Rosenthal A.N., Intermaggio M.P., Fraser L., Gentry-Maharaj A., Hayward J., Philpott S., et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women with Ovarian Cancer. J. Natl. Cancer Instig. 2015;107:e214. doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anisimenko M.S., Kozyakov A.E., Paul G.A., Kovalenko S.P. The frequency of the BLM p.Q548X (c.1642C>T) mutation in breast cancer patients from Russia is no higher than in the general population. Breast Cancer Res. Treat. 2014;148:689–690. doi: 10.1007/s10549-014-3187-0. [DOI] [PubMed] [Google Scholar]
- 36.Bogdanova N., Togo A.V., Ratajska M., Kluzniak W., Takhirova Z., Tarp T., Prokofyeva D., Bermisheva M., Yanus G.A., Gorodnova T.V., et al. Prevalence of the BLM nonsense mutation, p.Q548X, in ovarian cancer patients from Central and Eastern Europe. Fam. Cancer. 2015;14:145–149. doi: 10.1007/s10689-014-9748-x. [DOI] [PubMed] [Google Scholar]
- 37.Imyanitov E., Prokofyeva D., Bogdanova N., Dork T. The frequency of the BLM*p.Q548X (c.1642C>T) mutation in breast cancer patients from Russia. Breast Cancer Res. Treat. 2014;148:695–696. doi: 10.1007/s10549-014-3198-x. [DOI] [PubMed] [Google Scholar]
- 38.Chui M.H., Ryan P., Radigan J., Ferguson S.E., Pollett A., Aronson M., Semotiuk K., Holter S., Sy K., Kwon J.S., et al. The histomorphology of Lynch syndrome-associated ovarian carcinomas: Toward a subtype-specific screening strategy. Am. J. Surg. Pathol. 2014;38:1173–1181. doi: 10.1097/PAS.0000000000000298. [DOI] [PubMed] [Google Scholar]
- 39.Pal T., Permuth-Wey J., Kumar A., Sellers T.A. Systematic review and meta-analysis of ovarian cancers: Estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin. Cancer Res. 2008;14:6847–6854. doi: 10.1158/1078-0432.CCR-08-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Audeh M.W., Carmichael J., Penson R.T., Friedlander M., Powell B., Bell-McGuinn K.M., Scott C., Weitzel J.N., Oaknin A., Loman N., et al. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 41.Kast K., Rhiem K., Wappenschmidt B., Hahnen E., Hauke J., Bluemcke B., Zarghooni V., Herold N., Ditsch N., Kiechle M., et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J. Med. Genet. 2016;53:465–471. doi: 10.1136/jmedgenet-2015-103672. [DOI] [PubMed] [Google Scholar]
- 42.Pilyugin M., Andre P.A., Ratajska M., Kuzniacka A., Limon J., Tournier B.B., Colas J., Laurent G., Irminger-Finger I. Antagonizing functions of BARD1 and its alternatively spliced variant BARD1delta in telomere stability. Oncotarget. 2017;8:9339–9353. doi: 10.18632/oncotarget.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brozek I., Ratajska M., Piatkowska M., Kluska A., Balabas A., Dabrowska M., Nowakowska D., Niwinska A., Rachtan J., Steffen J., et al. Limited significance of family history for presence of BRCA1 gene mutation in Polish breast and ovarian cancer cases. Fam. Cancer. 2012;11:351–354. doi: 10.1007/s10689-012-9519-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dockery L.E., Gunderson C.C., Moore K.N. Rucaparib: The past, present, and future of a newly approved PARP inhibitor for ovarian cancer. Onco. Targets Ther. 2017;10:3029–3037. doi: 10.2147/OTT.S114714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.