Abstract
Background and Aims
Various studies and conservationist reports have warned about the contraction of the last subtropical Afro-Macaronesian forests. These relict vegetation zones have been restricted to a few oceanic and continental islands around the edges of Africa, due to aridification. Previous studies on relict species have generally focused on glacial effects on narrow endemics; however, little is known about the effects of aridification on the fates of previously widespread subtropical lineages.
Methods
Nuclear microsatellites and ecological niche modelling were used to understand observed patterns of genetic diversity in two emblematic species, widely distributed in these ecosystems: Canarina eminii (a palaeoendemic of the eastern Afromontane forests) and Canarina canariensis (a palaeoendemic of the Canarian laurel forests). The software DIYABC was used to test alternative demographic scenarios and an ensemble method was employed to model potential distributions of the selected plants from the end of the deglaciation to the present.
Key Results
All the populations assessed experienced a strong and recent population decline, revealing that locally widespread endemisms may also be alarmingly threatened.
Conclusions
The detected extinction debt, as well as the extinction spiral to which these populations are subjected, demands urgent conservation measures for the unique, biodiversity-rich ecosystems that they inhabit.
Keywords: Islands, nuclear microsatellites, subtropical flora, decline, aridification, extinction, genetic conservation
INTRODUCTION
Rare species are defined as those characterized by small population sizes, narrow geographic distributions and high habitat specificity, or any combination of these criteria (Rabinowitz, 1981). These species often display low genetic diversity, which makes them vulnerable to stochastic demographic phenomena (Kruckeberg and Rabinowitz, 1985; Hewitt, 2004). However, this relationship between rarity and genetic diversity is not an axiom: rare species may present high levels of genetic diversity (Stebbins, 1980; Gitzendanner and Soltis, 2000; Pérez de Paz and Caujapé-Castells, 2013; García-Verdugo et al., 2015), whereas some widespread and common species, such as exotic invasive plants, may exhibit low levels of genetic diversity (Tsutsui et al., 2000).
Relict species are rare species whose distribution ranges have been reduced to a few populations due to different processes (e.g. aridification), but with a degree of threat that gives them a great value in conservation biology (Habel and Assmann, 2009). Especially interesting are palaeoendemics, i.e. relict species that have become rare due to widespread extinction in their original distribution range (Stebbins and Major, 1965), and which frequently act as reservoirs of phylogenetic exclusivity, as the only surviving representatives of larger clades (Cronk, 1992; Faith, 1992). Biologists often characterize these species as ‘living fossils’, an irreplaceable heritage in the Tree of Life (Grandcolas et al., 2014). The need to conserve these rare and relict species and their habitat-restricted communities has increased in the current context of rapid human-induced climate change.
Studies on relict species have generally focused on glacial relicts, which are now constrained to some reduced ranges in mountain peaks or high latitudes (Petit et al., 2003; Hewitt, 2004). By contrast, relict species that have become rare as a result of aridification have received far less attention. Genetic studies on these species, however, are of particular relevance because they may provide insights into the capacity for resilience and the foreseeable evolutionary fates of species subject to global warming.
Here we focus on one such taxon: the angiosperm plant genus Canarina, belonging to the bellflower family, Campanulaceae (tribe Platycodoneae). This genus comprises only three species separated by >7000 km across the Sahara desert: C. canariensis, associated with the endemic laurel forests of the Canary Islands, and C. eminii and C. abyssinica, which are endemic to the relict Afromontane forests of East Africa. Canarina is the only African representative of Platycodoneae, an early-diverging tribe within family Campanulaceae (Roquet et al., 2008), while the remaining members of the tribe are distributed throughout Central and East Asia (Wang et al., 2013). Thus, Canarina complies with the criterion of phylogenetic exclusivity in a relict rare species (Faith, 1992).
Mairal et al. (2015a) reconstructed the biogeographic history of Canarina and showed that ancestors of this genus migrated from Asia to East Africa in the Middle Miocene. They linked the disjunct sister-group relationship between C. canariensis and C. eminii (dated ~7–6 Ma) to large-scale extinction and a gradual aridification process that would have started in North Africa in the Late Miocene (Sepulchre et al., 2006; Senut et al., 2009). This was followed by cycles of contraction and expansion affecting both subtropical and tropical taxa (Maley, 2000; Bonnefille, 2011), in which mountainous areas in the Sahara served as refuges during arid periods (Osborne et al., 2008). This Afro-Macaronesian biogeographic pattern has been observed in many other angiosperm genera, and is known as the Rand Flora pattern (Sanmartín et al., 2010; Pokorny et al., 2015; Mairal et al., 2017a). Thus, Canarina also fulfils the criterion of a palaeoendemic relict – the palaeoendemic character can also be seen in the Canarina populations of both species, in which reservoirs of ancient genetic diversity appear in the oldest massifs and palaeo-islands (Mairal et al., 2015b, 2017b).
Rare or relictual species are also generally characterized by high habitat specificity (Kruckeberg and Rabinowitz, 1985), either because the species have low environmental tolerance or a narrow niche, or because their habitats are currently under contraction. The habitats now occupied by Canarina (the Canarian laurel forest and the isolated patches of subtropical Afromontane forest in East African ‘sky islands’) have often been interpreted as the last remnants of a more humid vegetation that extended over the Mediterranean Basin and North Africa during the Early–Middle Miocene (Heald, 1951; Axelrod and Raven, 1978; White, 1983; Bramwell, 1985; McCormack et al., 2009; Fernández-Palacios et al., 2011); but see Kondraskov et al. (2015) for a different view on the relict character of the Canarian laurel forests. Plant lineages that took shelter in these enclaves have acquired a high degree of relictualism and contribute substantially to the exceptional endemicity of these areas (Fjeldså and Lovett, 1997; Juan et al., 2000), which are now classified as biodiversity hotspots; i.e. biogeographic regions whose great biodiversity is threatened by habitat loss (Myers et al., 2000; Mittermeier et al., 2004).
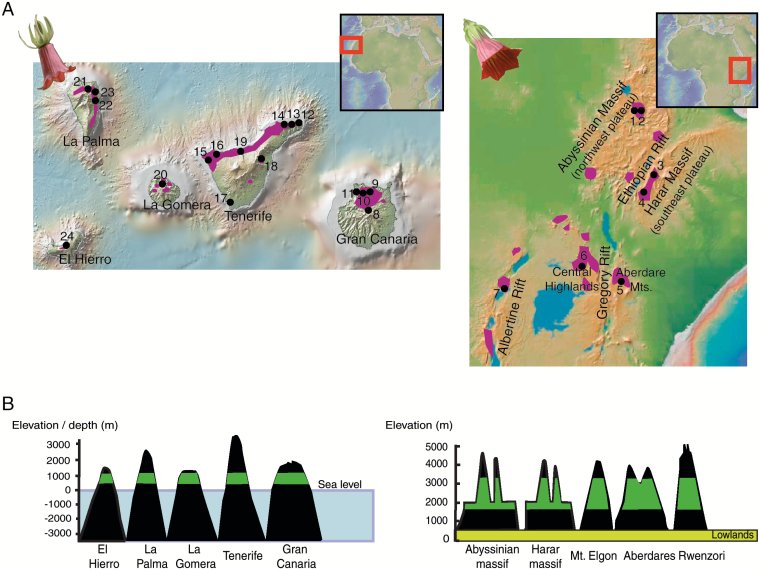
Both C. eminii and C. canariensis have widespread but disjunct distribution ranges and fulfil Rabinowitz’s (1981) ‘large and narrow’ criterion of rarity: species that are constantly sparse in a specific habitat but over a large range. Canarina canariensis is a terrestrial herb that propagates by seeds and shoots from its tuber (Bramwell and Bramwell, 2001); it occurs in the endemic Canarian laurel forests and (more rarely) in the adjacent thermophilous forests in the central and western islands of the Canarian Archipelago. Canarina eminii propagates from a long and thick root and grows as a terrestrial or (mainly) as an epiphyte of Afromontane forest trees (Podocarpus, Hagenia, Conopharyngia; Hedberg, 1961) in East Africa, from Ethiopia in the north to Malawi in the south (Fig. 1).
Fig. 1.
Distribution and scheme of the insular systems inhabited by the studied species. (A) Geographic distribution of C. canariensis in the Canary Islands (left) and C. eminii in the Afromontane forests (right). The distribution areas are shaded in purple. Sampled populations are represented with black dots accompanied by a population code number, as specified in Table 1. (B) Schematic representations of the insular systems and the subtropical vegetation belts inhabited by the studied species: oceanic islands and laurisilva forests for C. canariensis (left) and sky islands and Afromontane forests for C. eminii (right).
The steep topography and patchy distribution of the Eastern African forests (divided by deserts, savannahs and cultivated lands) and the deep ocean floor that lies between islands in the Canarian Archipelago have added to a high degree of geographic and ecological isolation (Fig. 1B), manifested by high genetic differentiation among populations and a marked geographic structure (Mairal et al., 2015a,b, 2017a). This great interpopulational diversity is threatened in C. eminii by human activities, such as logging and forest clearance (fires) linked to a rapid growth of the human population and agricultural development (Hedberg, 1961; Alemayehu, 2006), which have contributed to further fragmentation and isolation among forest patches (Fahrig, 2003). In the case of C. canariensis, although big patches of the laurisilva are currently under legal protection, this forest type has experienced a strong contraction during the last two centuries with respect to its estimated original range, due mostly to agricultural practices (Fernández-Palacios et al., 2011; but see de Nascimento et al., 2015 for an alternative view of Gran Canaria’s forests).
In this study, we used molecular and ecological niche modelling approaches to explore the effects of historical climate change and recent anthropogenic activity on the demographic evolution of C. canariensis and C. eminii. We developed new molecular markers from a library of nuclear microsatellites (SSRs), and used these to document levels of genetic diversity within and among populations. We also tested the alternative scenarios of population dynamics of (1) constant size, (2) bottleneck, (3) demographic expansion and (4) decline, using approximate Bayesian computation techniques (Beaumont, 2010) based on SSR data. Finally, we employed ecological niche modelling to reconstruct the climatic requirements (niche breadth) of African and Canarian populations and project them back into the past (Last Glacial Maximum, Holocene) to understand the impact of recent climate change on the geographic distribution of populations. If climate change since the end of the deglaciation or human activities have driven the retreat of these forests, we should find a genetic signal linked to a recent demographic decrease; conversely, this signal would not appear if the demographic processes were older. A demographic decline would confirm the suspicions about the serious state of the threat to these forest patches. Accordingly, our main goals are: (1) to compare levels of population genetic diversity between these two species, and with those estimated by other markers [chloroplast sequences and amplified fragment length polymorphisms (AFLPs); Mairal et al., 2015b, 2017b]; (2) to search for the presence of population decline or bottlenecks that may indicate ongoing loss of genetic diversity; (3) to relate population dynamics to changes in the potential geographic distribution of Canarina populations over time; and (4) to provide a genetic basis to enable the design of suitable strategies for the genetic conservation of these taxa.
MATERIALS AND METHODS
Plant material, population sampling and DNA sequencing
Several sampling field campaigns were performed in East Africa and the Canary Islands (2009–14), which led to the collection of fresh material from 223 individuals in 24 different populations of Canarina: 17 of C. canariensis (N = 159) and seven of C. eminii (N = 64) (Table 1). For the largest populations, a minimum of ten samples was collected through the entire area of occupation to reduce the possible inflation in the genetic structure descriptors (Caujapé-Castells, 2010). DNA was extracted from silica-gel-dried leaves using the DNeasy Plant Mini Kit (Qiagen, CA, USA). The quality of DNA extractions was checked on 1 % agarose gels, and DNA concentration was measured using a NanoDrop spectrophotometer. All samples were diluted to 10 ng μL−1 prior to PCR amplification.
Table 1.
Genetic diversity descriptors and tests used for each of the populations of C. canariensis and C. eminii. For Tenerife, names of paleo-islands are indicated in parentheses
| Population | N | A | H e (s.e.) | N pa | M | |
|---|---|---|---|---|---|---|
| Canarina eminii, continental islands | ||||||
| 1. Gifta | Abyssinian massif | 8 | 1.58 | 0.167 (0.220) | 0.00 | 0.442 |
| 2. Dembecha | Abyssinian massif | 8 | 1.80 | 0.244 (0.270) | 0.07 | 0.349 |
| 3. Yirga | Harar massif | 8 | 2.00 | 0.301 (0.349) | 0.17 | 0.081 |
| 4. Harenna | Harar massif | 9 | 2.28 | 0.287 (0.218) | 0.33 | 0.299 |
| 5. Aberdare | Aberdare Mountains | 8 | 1.83 | 0.382 (0.222) | 0.01 | 0.128 |
| 6. Elgon | Mount Elgon | 13 | 1.99 | 0.339 (0.295) | 0.01 | 0.123 |
| 7. Rwenzori | Rwenzori Mountains | 10 | 2.24 | 0.410 (0.272) | 0.01 | 0.161 |
| Canarina canariensis, oceanic islands | ||||||
| 8. Cueva Corcho | Gran Canaria | 8 | 1.96 | 0.305 (0.34) | 0.14 | 0.111 |
| 9. Tilos de Moya | Gran Canaria | 12 | 2.04 | 0.325 (0.27) | 0.06 | 0.158 |
| 10. El Sao | Gran Canaria | 10 | 1.84 | 0.282 (0.26) | 0.00 | 0.229 |
| 11. Andenes de Guayedra | Gran Canaria | 12 | 1.72 | 0.292 (0.25) | 0.00 | 0.224 |
| 12. Camino Chamuscadas | Tenerife (Anaga) | 10 | 4.07 | 0.417 (0.29) | 0.37 | 0.235 |
| 13. El Bailadero | Tenerife (Anaga) | 11 | 3.89 | 0.378 (0.31) | 0.03 | 0.164 |
| 14. Tope del Carnero | Tenerife (Anaga) | 7 | 3.94 | 0.412(0.29) | 0.11 | 0.229 |
| 15. Teno Alto | Tenerife (Teno) | 8 | 2.37 | 0.400 (0.26) | 0.07 | 0.171 |
| 16. El Palmar | Tenerife (Teno) | 8 | 2.54 | 0.485 (0.27) | 0.25 | 0.198 |
| 17. Barranco del Infierno | Tenerife (Adeje) | 11 | 2.50 | 0.489 (0.23) | 0.00 | 0.184 |
| 18. Barranco de Badajoz | Tenerife | 5 | 3.50 | 0.316 (0.35) | 0.02 | 0.178 |
| 19. Barranco Ruiz | Tenerife | 6 | 2.20 | 0.411 (0.22) | 0.02 | 0.158 |
| 20. Tamargada | Gomera | 13 | 1.67 | 0.235 (0.26) | 0.04 | 0.250 |
| 21. Los Tilos | La Palma | 10 | 2.26 | 0.394 (0.25) | 0.14 | 0.202 |
| 22. Barranco del Agua | La Palma | 10 | 2.02 | 0.332 (0.24) | 0.01 | 0.188 |
| 23. Barranco La Barata | La Palma | 4 | 1.88 | 0.365 (0.26) | 0.00 | 0.258 |
| 24. El Hierro | El Hierro | 12 | 2.24 | 0.415 (0.20) | 0.17 | 0.183 |
A, allelic richness; Npa, number of private alleles standardized to sample size; M, Garza and Williamson’s M-ratio test.
Development of nuclear microsatellite markers and scoring
A microsatellite library was developed for C. canariensis at the Savannah River Ecology Laboratory (University of Georgia). The extracted plant DNA was enriched using magnetic streptavidin beads and biotinylated oligonucleotides representing CT and GT repetitions. The library was sequenced using a Titanium Sequencing Kit (Roche Life Sciences) on the Roche 454 GS-FLX Titanium platform. We selected tri- and tetra-nucleotide repetition motifs to reduce the risk of misinterpreting as homozygotes the heterozygotes with alleles differing by a single repeat. Primer design and PCR amplifications were performed in the laboratories of Jardín Botánico Canario ‘Viera y Clavijo’, Unidad Asociada CSIC del Cabildo de Gran Canaria (JBCVC-CSIC).
We selected and tested 26 primer pairs for C. canariensis, which were also tested for cross-amplification in C. eminii. Fifteen loci were discarded because of amplification failure in most samples, poor electrophoretic profiles or low levels of variation. Finally, 11 SSR loci were retained for C. canariensis and seven for C. eminii; these showed good quality profiles and were used to genotype all sampled individuals from both species. DNA amplification (PCR) was performed in a final volume of 20 μL, containing 1 μL of DNA, 12.55 μL of sterilized water, 1.5 μL of 10× buffer, 0.8 μL of MgCl2+, 0.3 μL of dNTP, 1 μL of BSA, 0.25 μL Taq DNA polymerase (Bioline), 1 μL of forward primer (0.2 μm), 0.8 μL of reverse primer (5 μm) and 0.8 μL of reverse-tailed primer. PCR products were labelled using the fluorescent dyes FAM, VIC, NED or PET (labels according to the original kit of Applied Biosystems), and an additional 19-bp fluorescently labelled M13 primer (5′-CACGACGTTGTAAAACGAC-3′) according to the methods of Boutin-Ganache et al., (2001). A tail sequence (5′-GTGTCTT-3′) was added to the 5′ end of the reverse primer to improve adenylation (addition of AMP) and to facilitate genotyping. Samples were incubated in a Verity 96 thermocycler using the following conditions: 2 min of denaturation at 94.4 °C, 30 cycles of 30 s of denaturation at 94.4 °C, 40 s at different annealing temperatures for each locus (Supplementary Data Table S1), 30 s of elongation at 72 °C and final extension for 8 min at 72 °C. Reactions were separated on 5 % polyacrylamide gels. The amplified fragments were scored using GeneMapper 4.0 software (Applied Biosystems) with the LIZ 500 size standard. We combined automatic detection of each allele with visual inspection of each sample, following Dewoody (2006) to reduce scoring errors. Finally, we identified peak profiles for each locus and allele, and we assigned a genotype to each individual.
To detect possible genotyping errors (stuttering, null alleles and allelic dropout), we analysed the genotyping matrix with the programs MICRO-CHECKER 2.2.1. (Van Oosterhout et al., 2004) and INEst 1.0 (Chybicki and Burczyk, 2009). A single locus in C. eminii and one locus in C. canariensis displayed null alleles with frequencies >0.05 and high FIS values, and were therefore removed from further analyses. Other loci were excluded due to low quality of electrophoretograms, especially in C. eminii, where several loci were removed due to the effect of cross-amplification. The final data matrices contained nine loci for C. canariensis and six loci for C. eminii.
Genetic diversity and population structure
Deviations from Hardy–Weinberg equilibrium and linkage disequilibrium were tested using POPGENE version 4.2 (Yeh et al., 1997). Standard genetic diversity statistics, such as expected heterozygosity (He), were calculated using the software Arlequin (Excoffier et al., 2005). To account for sample size variation among populations, we estimated the allelic richness and the number of private alleles using rarefaction analysis implemented in the software HP-Rare v.1.0 (Kalinowski, 2005). To identify the signal of old or severe bottlenecks, we used Garza and Williamson’s M-ratio test (GW test) (2001), which is better suited to the detection of these events than methods based on the deficit of rare alleles (Williamson-Natesan, 2005). The GW test detects the presence of bottlenecks by determining the ratio between the number of alleles and the allele size range, according to the statistic M = k/r, where k is the number of alleles and r = Smax − Smin + 1, where S is allele size.
To detect the genetic composition of populations, we used the Bayesian clustering method implemented in STRUCTURE v.2.3 (Pritchard et al., 2000). This approach assumes that loci are in Hardy–Weinberg equilibrium and that there is linkage equilibrium within populations. Analyses were performed separately for each species using an admixture model with correlated allele frequencies among groups. We ran 500 000 Markov chain Monte Carlo iterations after a burn-in of 100 000 iterations for K values of 1–10, with ten repetitions for each K. The most probable value of K was determined with the method of Evanno et al. (2005) implemented in STRUCTURE Harvester (Earl, 2012). The matrices were adapted to the specific format of each of the programs using Transformer-4 (Caujapé-Castells et al., 2013). Microsatellite matrices are available in the DEMIURGE information system (http://www.demiurge-project.org/) with codes D-NMICR-122 (matrix of C. canariensis) and D-NMICR-123 (matrix of C. eminii). To quantify the amount of genetic variance attributable to geographic and population subdivision, a hierarchical analysis of molecular variance (AMOVA) was performed using the software ARLEQUIN v.3.0 (Excoffier et al., 2005). Exploratory analyses were performed considering, alternatively, the individual populations in palaeo-islands and sky islands separately, or grouped into geographic units as identified by STRUCTURE.
Demographic analysis
We used approximate Bayesian computation techniques as implemented in the software DIYABC v1.0.4.46beta (Cornuet et al., 2008, 2010) to statistically evaluate alternative scenarios for the demographic history of C. eminii and C. canariensis. DIYABC uses a coalescent framework to simulate complex evolutionary scenarios without the need to estimate the underlying likelihood function (Cornuet et al., 2008, 2010). Instead, simulated scenarios are summarized using variable estimates (summary statistics), which are then compared with observed values from the genetic data to estimate the posterior probability of the model parameters using Bayesian inference, and to compute measures of bias and precision for each scenario [95 % high posterior density (HPD) credibility intervals]. Due to the strong population structure observed in the two study species (Mairal et al., 2015b, 2017b) and the problems that can arise by assuming panmixia, we conducted separate analyses for each population, and we grouped only those populations that were geographically (<10 km) and genetically very close according to the STRUCTURE results, i.e. Gifta and Dembecha probably form an apomictic population, and were part of the same Afromontane forest patch, nowadays fragmented because of agriculture.
We considered four competing population history scenarios: (1) a null model of constant population size; (2) a bottleneck in the past with subsequent recovery; (3) a demographic expansion; and (4) a recent population decline without recovery (Fig. 1A). We assumed different effective population sizes for each historical scenario (Ne, N1, N3, N4). Table 2 shows the prior distributions of the demographic parameters for the four simulations, each simulating a change in the population size. We assumed that each locus followed a generalized stepwise mutation (GSM) model for microsatellite markers (Estoup et al., 2002), using a log-uniform distribution for the geometric distribution parameter (P = 10−1–30−1) and the mutation rate (μ = 10−4–10−3). In each of the tested scenarios, we simulated one million datasets and assumed absence of migration between populations. First, we estimated the posterior probability of each scenario by logistic regression, using the 1 % simulated datasets with summary statistics that were closest to the observed values. Bayesian credibility ranges (95 % HPD) of each simulated scenario were compared (Cornuet et al., 2008). We also estimated the posterior probability of the parameters from 1 % of the best-simulated datasets, using local linear regression and logit transformation of parameters. To make sure that the best scenario was not far off the observed data, we checked the goodness of fit by simulating 1000 data sets from the posterior predictive distribution of the parameter and compared SumStats with the observed data (Cornuet et al., 2010; Budde et al., 2013).
Table 2.
Prior distribution of demographic and historical parameters used as summary statistics for simulations of population size change in DIYABC (Fig. S1). Times of changes in effective population size are considered from the present (0) back in time: t1, t2, t3, t4. Mean μ is the mean mutation rate of microsatellites; Mean P is the mean of the geometric distribution parameter. All time events are expressed in numbers of generations. Conditions: N1 < Ne, N3 < Ne, N4 > Ne
| Parameter | Prior distribution |
|---|---|
| N e | Uniform (10–10 000) |
| t 1 | Uniform (1–10 000) |
| N 1 | Uniform (1–100) |
| t 2 | Uniform (1–10 000) |
| t 3 | Uniform (1–10 000) |
| N 3 | Uniform (1–100) |
| t 4 | Uniform (1–10 000) |
| N 4 | Uniform (4000–100 000) |
| Mean µ | Log-uniform (1.00E−004 to 1.00E−003) |
| Mean P | Log-uniform (1.00E−001 to 3.00E−001) |
N e, effective population size; N1 past effective population size in a bottleneck scenario; N3, past effective population size in an expansion scenario; N4 past effective population size in a decline scenario.
Ecological niche modelling
We modelled the current climatic niche occupied by C. eminii and C. canariensis using extant occurrence data together with climatic maps. Data points were obtained from published monographs and inventories (Hedberg, 1961), three online databases (www.jardincanario.org/flora-de-gran-canaria, www.gbif.org and www.anthos.es) and data compiled through field trips. In all, we used 108 records for C. canariensis and 80 records for C. eminii (Supplementary Data Tables S2, S3), covering the entire distributional ranges of the species. Climatic data for current conditions were obtained from WorldClim (www.worlclim.org;Hijmans et al., 2005). For past climate scenarios we downscaled WorldClim data from a 30 arc-second resolution to 1 km resolution and projected current and past climate conditions for several periods after the Last Glacial Maximum, since the end of the deglaciation to the present (18, 10, 5 and 0 kiloyears; for more information see Espíndola et al., 2012). Pleistocene sea-level changes [~18 kiloyears ago the sea levels were 110 m below the actual level (Rijsdijk et al., 2014)] strongly altered the surface area of the Canary Islands, and thus the extent of potentially suitable areas for C. canariensis. To account for these changes, we used a bathymetry layer and constructed new rasters using the sea levels modelled by Rijsdijk et al. (2014) and projected climate data, including sea levels (−110 m for 18 kiloyears, −46 m for 10 kiloyears, −4.7 m for 5 kiloyears). To model the distributions of the two species, we combined the available occurrences with a set of six bioclimatic variables that could be estimated for all past scenarios: total annual precipitation, maximum and minimum monthly precipitation, annual mean temperature, and maximum and minimum monthly temperature. Pseudoabsences were generated by selecting 5000 random points. We used ensemble modelling (Araújo and New, 2007) to generate our predictions. Three modelling techniques – generalized additive models (GAMs), the general boosting method (GBM) and random forests (RF) – were run and summarized using R packages (R Core Team, 2014): biomod2 (Thuiller et al., 2013), foreign, raster (Hijmans and van Etten, 2016), SDMTools (VanDerWal et al., 2011), rms (Harrell, 2016), gbm (Ridgeway, 2015), gam (Hastie, 2016), rJava (Urbanek, 2010), dismo (Hijmans et al., 2016) and randomForest (Liaw and Wiener, 2002) (references for R packages are given in the Supplementary Data; Mairal et al., 2017a). We used repeated split sampling to evaluate the performance of the models, successively splitting the dataset into 70 % for calibration and 30 % for evaluation by measuring the area under the curve (AUC).
RESULTS
Genetic diversity and population structure
Based on the nine SSR loci selected for C. canariensis and the six loci for C. eminii, we detected 54 and 23 alleles, respectively, in the two species. The greatest number of private alleles in C. canariensis was detected in populations from the three palaeo-islands of Tenerife (e.g. 0.37 in Camino Chamuscadas; 0.25 in El Palmar; Table 1), which predate the age of the island in its current configuration. In C. eminii, the greatest number of private alleles was detected in populations from the Harar massif (0.33; Table 1). These Canarian and East African populations were inferred as sources of allele migration events in previous studies based on AFLP and chloroplast sequence data (Mairal et al., 2015b, 2017b). The average number of alleles per locus ranged from 1.67 to 4.07 in C. canariensis and from 1.58 to 2.28 in C. eminii. Genetic diversity values, estimated as Nei’s heterozygosity (He), were relatively low compared with values generally obtained with microsatellite markers in endemic species (He = 0.42; Nybom, 2004): 0.167–0.41 in C. eminii and 0.235–0.489 in C. canariensis. Linkage disequilibrium was not significant (P > 0.05).
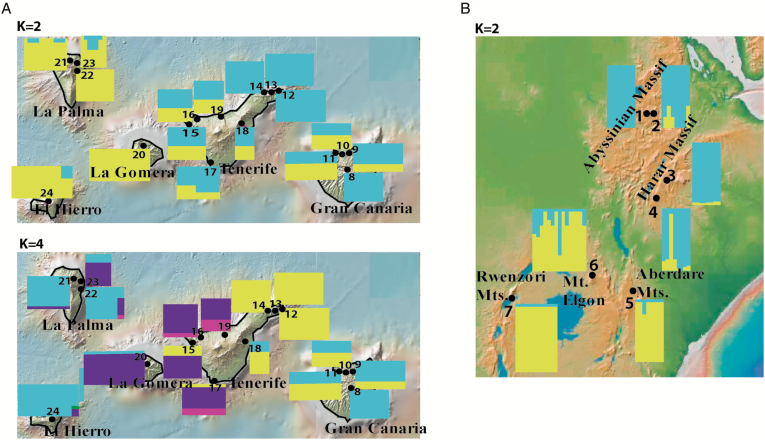
The method of Evanno et al. (2005) indicated that the most likely number of genetic groups was K = 2 for C. canariensis (ΔK = 1301) and K = 2 for C. eminii (ΔK = 108) (Supplementary Data Tables S4, S5, Fig. S2). A more complex genetic structure was revealed for C. canariensis by K = 4, delimiting two additional groups [ΔK = 60 (Supplementary Data Table S4)]. In general, populations of C. canariensis displayed a clear east/west structure, with a stronger admixture within the islands of Tenerife and Gran Canaria (Fig. 2A). In C. eminii, the structure of K = 2 supported a north/south division among populations (Fig. 2B).
Fig. 2.
Results of the genetic structure analyses for (A) C. canariensis and (B) C. eminii. Histograms show the Bayesian clustering of individuals within populations (STRUCTURE). Colours represent the individual proportion for each of the inferred Bayesian groups. All other conventions as in Fig. 1.
Hierarchical AMOVA analyses showed the largest proportion of genetic variation among groups, when each group was considered separately (23.22 % in C. eminii; 22.21 % in C. canariensis; Supplementary Data Table S6). Additionally, the STRUCTURE sublevels among groups (K = 2 for C. eminii and K = 2, K = 4 for C. canariensis) were consistent with the AMOVA analyses (Supplementary Data Table S6).
Population dynamics
All values for the GW test were M <0.68, the critical value below which populations are assumed to have suffered a recent population decline (with datasets of seven loci or more; Garza and Williamson, 2001). Though we could only reliably score six loci in C. eminii (see the Materials and methods section), the results of the demographic analysis (below) and the low values obtained with the GW test (Table 1) support a demographic decline in all populations of C. canariensis and C. eminii.
In both species, the ‘recent population decline’ scenario (Supplementary Data Fig. S1) was associated with the highest posterior probabilities and the narrowest 95 % HPD intervals in the DIYABC analysis (Table 3). For some populations, we could not discriminate among alternative hypotheses, as we did not obtain robust statistics for any given scenario [e.g. Gifta, Harenna, Yirga and La Gomera (Tamargada); Table 3, Supplementary Data Table S7]. The current estimated effective population size Ne was estimated between 500 and 1300 across populations, and was about two orders of magnitude smaller than the effective population size before the decline. Population decline was estimated to have started ~2000–5000 generations ago.
Table 3.
Results of DIYABC analyses. In each population the best supported scenario is indicated according to its posterior probability [PP; mean and 95 % Bayesian highest posterior density interval (HPD)], obtained by logistic regression from 1 % of the simulations that best approximate the observed values. NA (not available) is shown for populations for which it was not possible to discriminate a statistically robust scenario
| Population | Population | No. of individuals | Best supported scenario | PP (HPD) | N e | N 4 | t 4 | N eµmic |
|---|---|---|---|---|---|---|---|---|
| Canarina eminii, continental islands | ||||||||
| 1. Gifta and Dembecha | Abyssinian massif | 16 | NA | − | − | − | − | − |
| 3. Yirga | Harar massif | 13 | NA | − | − | − | − | − |
| 4. Harenna | Harar massif | 9 | NA | − | − | − | − | − |
| 5. Aberdare | Aberdare Mountains | 8 | Decline | 0.7511 (0.6710, 0.8312) | 5.17E+02 | 6.33E+E04 | 2.03E+03 | 9.73E−02 |
| 6. Elgon | Mount Elgon | 8 | Decline | 0.6912 (0.6544, 0.7280) | 5.35E+02 | 5.94E+E04 | 2.89E+03 | 1.39E−01 |
| 7. Rwenzori | Rwenzori Mountains | 10 | Decline | 0.6986 (0.6631, 0.7342) | 1.03E+03 | 5.84E+E04 | 3.56E+03 | 1.92E−01 |
| Canarina canariensis, oceanic islands | ||||||||
| 8. Cueva Corcho | Gran Canaria | 8 | Decline | 0.9293 (0.9066, 0.9519) | 5.50E+02 | 6.59E+E04 | 1.72E+03 | 9.91E−02 |
| 9. Tilos de Moya | Gran Canaria | 12 | Decline | 0.9400 (0.7882, 1.0000) | 6.60E+02 | 6.33E+E04 | 2.01E+03 | 1.22E−01 |
| 10. El Sao | Gran Canaria | 10 | Decline | 0.3902 (0.3325, 0.4478) | 6.83E+02 | 5.11E+E04 | 4.51E+03 | 1.05E−01 |
| 11. Andenes de Guayedra | Gran Canaria | 12 | Decline | 0.6930 (0.6497, 0.7363) | 5.00E+02 | 5.77E+E04 | 2.90E+03 | 9.07E−02 |
| 12. Camino Chamuscadas | Tenerife (Anaga) | 10 | Decline | 0.5972 (0.5646, 0.6298) | 1.25E+03 | 5.06E+E04 | 4.96E+03 | 4.01E−01 |
| 13. El Bailadero | Tenerife (Anaga) | 11 | Decline | 0.9265 (0.7775, 1.0000) | 1.03E+03 | 6.07E+E04 | 4.02E+03 | 2.10E−01 |
| 14. Tope del Carnero | Tenerife (Anaga) | 7 | Decline | 0.6000 (0.1706, 1.0000) | 1.40E+03 | 7.48E+E03 | 3.51E+03 | 2.78E−01 |
| 15. Teno Alto | Tenerife (Teno) | 8 | Decline | 0.9523 (0.9360, 0.9686) | 1.06E+03 | 6.90E+E04 | 2.87E+03 | 2.09E−01 |
| 16. El Palmar | Tenerife (Teno) | 8 | Decline | 0.9511 (0.9354, 0.9668) | 1.34E+03 | 6.25E+E04 | 3.46E+03 | 2.61E−01 |
| 17. Barranco del Infierno | Tenerife (Adeje) | 11 | Decline | 0.9940 (0.9880, 1.0000) | 9.31E+02 | 6.89E+E04 | 1.87E+03 | 1.73E−01 |
| 18. Barranco de Badajoz | Tenerife | 5 | Decline | 0.8499 (0.8096, 0.8902) | 7.41E+02 | 6.65E+E04 | 3.39E+03 | 1.56E−01 |
| 19. Barranco Ruiz | Tenerife | 6 | Decline | 0, 8775 (0.8423, 0.9127) | 8.70E+02 | 6.18E+E04 | 2.35E+03 | 1.45E−01 |
| 20. Tamargada | La Gomera | 13 | NA | − | − | − | − | − |
| 21. Los Tilos | La Palma | 10 | Decline | 0.8024 (0.7579, 0.8469) | 9.00E+02 | 5.07E+E04 | 2.83E+03 | 1.64E−01 |
| 22. Barranco del Agua | La Palma | 10 | Decline | 0.7253 (0.6768, 0.7738) | 7.70E+02 | 5.71E+E04 | 2.37E+03 | 1.43E−01 |
| 24. El Hierro | El Hierro | 12 | Decline | 0.7498 (0.6942, 0.8054) | 9.18E+02 | 5.43E+E04 | 2.44E+03 | 1.51E−01 |
N e, current effective population size; N4, effective population size at the beginning of the decline; t4, start time of decline, measured in number of generations; Neμmic, current effective population size × micros mutation rate.
Ecological niche modelling
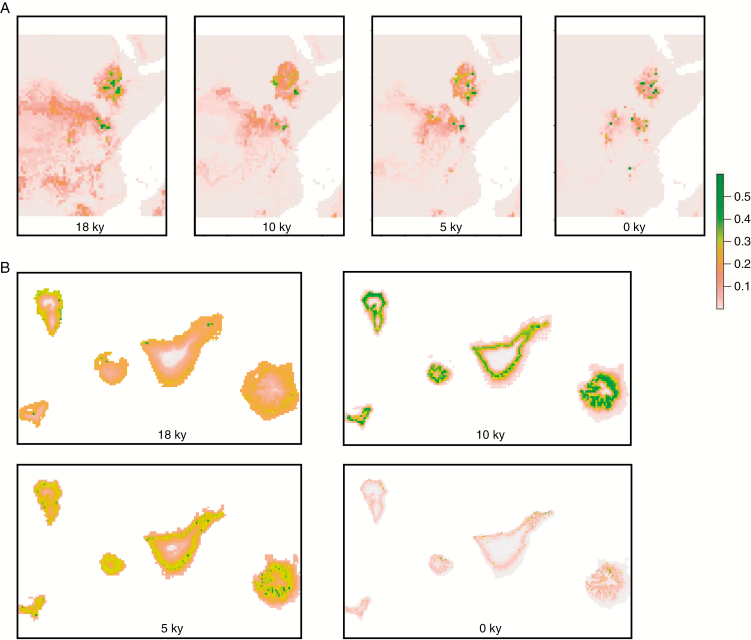
Species distribution models indicated that the inferred potential distribution for the present largely coincided with the extant distribution of both species. The AUC values were generally high (with values of 0.71 and 0.96 for C. canariensis and C. eminii respectively), suggesting that the models are consistent. Our hindcast climate niche projections showed that the areas with favourable climatic conditions for C. eminii and C. canariensis experienced a reduction from the start of the deglaciation (around 18 kiloyears ago) to the present (Fig. 3). Present projections show patches of climatic suitability (climatic refugia) that largely coincide with the extant distribution of the species (Fig. 3).
Fig. 3.
Geographic projections of the climatic niche model of (A) Canarina eminii and (B) Canarina canariensis over four time periods: 18, 10, 5 and 0 kiloyears (ky) (modelling sea-level changes). Colour scale indicates climatic suitability values; brown colour indicates low values and dark green indicates high values.
DISCUSSION
Low genetic diversity and strong population structure in Canarina endemics
Population genetic theory predicts that species subject to habitat fragmentation, e.g. due to habitat loss, are more susceptible to stochastic demographic fluctuations, such as genetic bottlenecks or declines in effective population size. This is often accompanied by an upward surge of inter-population genetic differentiation, higher inbreeding and loss of genetic variability (Stebbins, 1980; Kruckeberg and Rabinowitz, 1985; Lande, 1993), which can eventually drive a species to extinction. This ‘fragmentation syndrome’ (Lande, 1993) seems to be evidenced in Canarina. Our ecological niche models indicate that both C. eminii and C. canariensis have experienced a contraction of their climatic suitability areas since 18 kiloyears ago, which eventually led to the fragmented, patchy distribution observed today (Fig. 3). The low levels of genetic variability detected at the population level (Table 1) are in agreement with those obtained with other types of molecular marker (chloroplast DNA sequences and AFLPs; Mairal et al., 2015b, 2017b). They are also generally below those obtained with microsatellites in endemic species (Nybom, 2004), and specifically below those reported in other taxa distributed in the study areas (Table 4). Note, however, that comparing genetic statistics across distantly related phylogenetic groups is problematic because their values can be influenced by taxon-specific biological differences (Gitzendanner and Soltis, 2000).
Table 4.
Average values of microsatellite diversity statistics for several species studied in the Canary Islands and Afromontane forests
| Species | A | H e | Reference |
|---|---|---|---|
| Canary Islands | |||
| Canarina canariensis | 2.5 | 0.367 | – |
| Bencomia exstipulata | 3.50 | 0.440 | González-Pérez et al., 2009a |
| Bencomia caudata | 5.13 | 0.620 | González-Pérez et al., 2009a |
| Myrica rivas-martinezii | 6.50 | 0.560 | González-Pérez et al., 2009b |
| Neochamalea pulverulenta | 7.85 | 0.425–0.812 | Rigueiro et al., 2009 |
| Myrica faya | 9.30 | 0.570 | González-Pérez et al., 2009b |
| Olea europaea subsp. guanchica | 14.00 | 0.710 | García-Verdugo et al., 2010 |
| Ilex perado subsp. lopezlilloi | 2.13 | 0.480 | Sosa et al., 2010a |
| Ilex perado subsp. platyphylla | 5.37 | 0.600 | Sosa et al., 2010a |
| Ilex canariensis | 2.38 | 0.420 | Sosa et al., 2010a |
| Sambucus palmensis | 6.80 | 0.499 | Sosa et al., 2010b |
| Silene nocteolens | 15.83 | 0.780 | Sosa et al., 2010a |
| Pinus canariensis | 22.80 | 0.771 | López de Heredia et al., 2010 |
| Parolinia ornata | 4.38 | 0.515 | González-Pérez and Caujapé-Castells, 2014 |
| Ruta oreojasme | 7.62 | 0.687 | Meloni et al., 2015 |
| Phoenix canariensis | 4.41 | 0.476 | Saro et al., 2015 |
| Micromeria rivas-martinezii | 9.80 | 0.500 | Puppo et al., 2016 |
| Micromeria glomerata | 4.27 | 0.310 | Puppo et al., 2016 |
| Afromontane forests | |||
| Canarina eminii | 1.93 | 0.300 | – |
| Coffea arabica | 2–3.20 | 0.050–0.242 | Silvestrini et al., 2007 |
| Acacia senegal | 6.90 | 0.667 | Omondi et al., 2010 |
| Senegalia mellifera | 7.00 | 0.620 | Ruiz Guajardo et al., 2010 |
| Prunus africana | 2.0 – 4.00 | 0.105–0.728 | Kadu et al., 2011 |
A, allelic richness.
The loss of genetic diversity is also accompanied in Canarina by a large among-population differentiation (see AMOVA in Supplementary Data Table S6) and great genetic exclusivity in some populations, especially those that may have acted as refugia or genetic reservoirs in the past (e.g. the palaeo-islands of Tenerife or Mount Elgon in Kenya), though this diversity, detected also by AFLPs and plastid sequences, seems to be of older origin than the one reported here (Mairal et al., 2015b, 2017a).
In C. canariensis, our SSR analyses detected a strong population structure akin to the one revealed by AFLPs and DNA sequences: an east/west genetic split, with the strongest difference found within Tenerife (Fig. 2A; Mairal et al., 2015b, 2017a). The only difference is that SSRs detected higher levels of genetic admixture in the population of the paleo-island of Adeje than the other markers (Fig. 2A; this had been predicted in Mairal et al., 2015b). In C. eminii, SSRs detect a north/south genetic diversity structure, which was not detected by AFLPs or sequences (Mairal et al., 2017b). This might reflect more recent population dynamics in agreement with similar contact patterns found in other Afromontane plants (Assefa et al., 2007; Masao et al., 2013). Alternatively, it could be an artefact of the lack of specificity in our SSR markers, which were designed for C. canariensis. Additionally, we found genetic admixture in Mount Elgon (Fig. 2B), which agrees well with its purported role as a crossroad among populations at both sides of the Rift Valley (Mairal et al., 2017b), and with the descent of vegetation belts during the Last Glacial Maximum (Chala et al., 2017).
Historical population declines in widespread rare species
The time lag between range reduction and the collapse of populations is known as ‘extinction debt’ (Tilman et al., 1994). Initially, if studies are conducted shortly after habitat loss has occurred, the evolutionary cost of the extinction debt is not obvious and thus difficult to detect (Triantis et al., 2010). Several generations are needed to detect the total impact of habitat loss and fragmentation (Tilman et al., 1994; Tilman, 1999). This seems to be the case in Canarina: although this endemic genus of evergreen subtropical forests is still widely distributed in regions of East Africa and Macaronesia, it also shows a worrying population decline scenario. Indeed, demographic analyses based on SSRs (Tables 1 and 3) and ecological niche model projections (Fig. 3) suggest that both C. eminii and C. canariensis underwent a strong and recent population decline.
When did this decline start and what were its causes? Our previous studies showed that the current disjunct distribution of genus Canarina across the Sahara Desert can be explained by the climatic aridification during the Late Miocene/Pliocene and the resulting contraction in the distribution ranges of subtropical forests in Africa and Macaronesia (Mairal et al., 2015a, b, 2017a, b; Pokorny et al., 2015). Although this hypothesis may explain the current continental disjunction, the population decline detected here is probably more recent. The DIYABC analysis detected a population decline ~2000–5000 generations ago, which could not be recovered with coalescent-based Bayesian skyline plots (Drummond et al., 2005; Mairal et al., 2015b). This is in agreement with observations that SSR markers detect more recent signals of genetic diversity than chloroplast DNA and AFLP markers (Chakraborty et al., 2014). Since the generation time of Canarina is not known precisely due to its tuberous habit (it can sometimes behave as a biennial or even show longer generation times), it is difficult to assign an absolute temporal framework for this decline; also, the time scale here differed slightly depending on the population. However, the temporal range of the decline detected here was consistent within the past 10 000 years (if we consider the plants as biennial) or 50 000 years (if we consider generation times of 10 years), i.e. it probably took place during the last glacial cycle (Pleistocene–Holocene).
This decline coincides with palaeoclimate projections from our ecological niche models, which support a contraction of the distribution ranges of both species from 18 kiloyears ago to the present (Fig. 3). Caution should be exercised in the interpretation of these models due to the low-resolution climate datasets available, which could underestimate potential microrefugia (Gavin et al., 2014). However, though geographic locations may be biased, the models are highly congruent with the extant distribution of the species and furnish evidence of a population decline. Furthermore, a recent study (Chala et al., 2017) supports our models by projecting the descent of vegetation belts during the Last Glacial Maximum.
Interestingly, DIYABC detected a less pronounced population decline in the populations with the highest levels of genetic diversity (Table 1 and N4 versus Ne in Table 3). These populations are located in areas that served as refugia during climatic oscillations or tectonic and volcanic events, such as Mount Elgon in Africa or the palaeo-islands of Tenerife, thereby supporting their role as reservoirs of genetic diversity against extinction, both during the Pleistocene and in more recent times (Mairal et al., 2015b, 2017b; this study).
By contrast, a much sharper decline is observed in the most fragmented laurel forest populations in the Canaries, i.e. Barranco de Badajoz and Barranco Ruiz in Tenerife (Table 3). In C. eminii, the sky islands located at lower latitudes (Rwenzori Mountains, Mount Elgon, Aberdare Mountains) harbour greater genetic diversity than populations in the massif of Abyssinia, which were probably more strongly affected by glaciations and human activities (e.g. Gifta and Dembecha in Table 1; cf. Mairal et al., 2017b). Loss of ecological interactions in these areas, such as deficiency of pollinators, may also have had a direct impact on genetic diversity in rare species (Lavergne et al. 2004).
Conservation biology of Canarina
Our ecological niche models may render an approximation of the effect of climate change on habitat loss and the associated loss of genetic diversity during the last glacial/interglacial cycles. Nowadays, however, human activities constitute an additional threat to the maintenance of genetic diversity in Canarina. Afro-Macaronesian forests have been logged in the past centuries for timber, agriculture and human settlements, bringing them to the verge of extinction, especially in topographically less complex areas, which are more easily exploited.
At present, the Afromontane forests have disappeared almost entirely from the Ethiopian highlands (estimated loss of up to of 35 % of originally forested areas) (EFAP, 1994; Kebede et al., 2007). This situation is aggravated by the decrease in soil quality due to changes in water resources (Kloos and Legesse, 2010), which has caused a dramatic decline in these forests, and even their disappearance in large areas (Reusing, 2000; FAO, 2001).
In the Canary Islands, the area of laurel forests has recently declined to 12.5 % of their potential distribution (Fernández-Palacios et al., 2011); this includes the laurel forest contractions of Gran Canaria and Tenerife to 1 % and 10 %, respectively, of their original distributions (Santos Guerra, 1990; Del Arco et al., 2010; Fernández-Palacios et al., 2011). The areas of adjacent suitable areas, such as thermophilous woodlands, have also declined considerably (de Nascimento et al., 2015).
One of the biggest threats to rare widespread species is the sensitivity of their populations to demographic stochasticity (Kruckeberg and Rabinowitz, 1985; Barrett et al., 1991). This could be a critical factor for the survival of Canarina in African and Macaronesian evergreen forest habitats, given the detected current decline in size of many populations (Table 3). Furthermore, the generally low levels of genetic diversity detected here make these taxa more susceptible to the effects of inbreeding (especially in small populations). Intraspecific genetic variation is the most fundamental level of biodiversity, providing the basis for evolutionary change, and is crucial for maintaining the capacity of species to adapt to new environmental conditions (Frankham et al., 2002; Yannic et al., 2013).
Although C. canariensis is locally abundant in some areas (e.g. Anaga, Teno and Tilos de Moya), greater efforts should be made to protect populations that contain exclusive genetic diversity. This is the case for populations in the three palaeo-islands of Tenerife (which also harbour pristine areas of laurisilva) and of Mount Elgon in the case of C. eminii. These areas act not only as reservoirs of unique genetic diversity, but also as cradles and sources of new allele migration events to nearby areas (Mairal et al., 2015b, 2017b; Caujapé-Castells et al., 2017). Though the results reported here refer only to two rare species (C. eminii and C. canariensis), similar patterns are expected (and have been observed) in other rare species, e.g. the Afroalpine plant Lobelia rhynchopetalum (Chala et al., 2016).
At present, a large area covered by the endemic laurel forest (laurisilva) across the Canary Archipelago is under legal protection (Fernández-Palacios et al., 2011), but this is not the case for the isolated patches of Afromontane forests in East Africa. Though some areas are preserved as National Parks (e.g. Harenna Forest, Rwenzori Mountains and Mount Elgon), conservation measures are not strictly enforced. Given the political and difficult economic situation of these countries, which makes it even more challenging to extend protection areas to preserve new populations, one crucial action in this current context would be ex situ conservation, via seed banks. Priority should be given to the conservation of the different geographically structured areas, covering in each of them those populations exhibiting the highest levels of ancient and endemic phylogenetic diversity. Genetic evidence gathered for Canarina (DNA, AFLPs and SSRs) further suggests that certain populations should be established as shrine areas in order to conserve the greater genetic variation ex situ: in the case of C. eminii, one population at each side of the Rift Valley and the population of Mount Elgon; for C. canariensis, one population in each palaeo-island in Tenerife (home to the best-preserved areas of laurisilva in Tenerife). Ex situ conservation plans should collect seeds covering these priority areas and avoid gene flow among them in order to favour potential speciation in the future. Thus, while the different areas should be managed separately (avoiding mixed reinforcements, i.e. without introducing propagules from different source areas), management within areas should stress the preservation of several populations, in order to safeguard a significant proportion of genetic variability of the species.
CONCLUSIONS
Genetic conservation studies often focus on rare species with a reduced distribution area (‘narrow endemics’; Kruckeberg and Rabinowitz, 1985; Oostermeijer et al., 2003; Fernández-Mazuecos et al., 2014). However, geographically widespread species can also be considered rare (relict) and subject to the same status of threat, especially if habitat loss has led to isolation of populations and small population size (‘large size and narrow distribution’; Kruckeberg and Rabinowitz, 1985). Here we show that this is the case for C. eminii and C. canariensis, two endemic bellflower species associated with the last remnants of subtropical forest in Africa and Macaronesia. Although both are locally widespread (C. canariensis is present in most of the Canarian islands and C. eminii occurs over a large part of East Africa), they feature the genetic signals of recent population decline due to habitat loss. The latter seems to be an effect of both historical climate change and rapid, human-induced, forest fragmentation in recent times. Especially for the Afromontane forests, loss of habitat due to increasing aridity and the alarming rates of forecast urban growth make these threatened ecosystems areas of priority interest for the conservation community (Seto et al., 2012).
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: primers used for PCR amplification for each locus. Table S2: geographic coordinates used in the ecological niche modelling of Canarina canariensis. Table S3: geographic coordinates used in the ecological niche modelling of Canarina eminii. Table S4: estimated probability of the likelihood function according to the Evanno method for the STRUCTURE analyses of C. canariensis. Table S5: estimated probability of the likelihood function according to the Evanno method for the STRUCTURE analyses of C. eminii. Table S6: hierarchical analysis of molecular variance (AMOVA). Table S7: probabilities for each scenario performed with DIYABC. Confidence intervals are shown in square brackets. Figure S1: graphical representation of the four scenarios simulating changes in population size with DIYABC. Supplementary references. Figure S2: Estimated probability of the likelihood function according to the Evanno method.
ACKNOWLEDGEMENTS
Field work could not have been conducted without the cooperation of the staff at the Jardín Botánico Canario ‘Viera y Clavijo’ − Unidad Asociada, CSIC (Cabildo de Gran Canaria). We thank the Cabildo de Tenerife, Jacinto Leralta, Ángel Fernández (La Gomera), Félix Manuel Medina from the Cabildo of La Palma and the Cabildo of El Hierro for help with accommodation and sampling logistics during field expeditions. We thank Juan Jose Aldasoro, Mª Luisa Alarcón and Andrea Sánchez-Meseguer for providing African samples, and Alicia Agudo, Carlos García-Verdugo, CNAE Members, Paloma Torroba and Victoria Culshaw for help during different stages of this work. This work was supported by a PhD research grant (BES-2010–037261) to M.M., including short research stays at JBCVC-CSIC (J.C.C.), University of Lausanne (N.A.) and University of Fribourg (L.P. and M.H.) funded by MINECO (grants CGL2009-1332-C03-01 (MICINN), CGL2012-40129-C02-01 and CGL2015-67849-P) (MINECO/FEDER) to I.S.; grant CGL2012-40129-C02-02 (MINECO) and a Marie Curie Intra-European Fellowship (PIEF-GA-2012-329088) to M.H. and project grant DEMIURGO (MAC/1/C20) to J.C.C.
LITERATURE CITED
- Alemayehu T. 2006. Diversity and ecology of vascular epiphytes in Harenna Afromontane forest, Bale, Ethiopia. PhD thesis, Addis Ababa University. [Google Scholar]
- Araújo MB, New M. 2007. Ensemble forecasting of species distributions. Trends in Ecology and Evolution 22: 42–47. [DOI] [PubMed] [Google Scholar]
- del Arco Aguilar MJ, González-González R, Garzón-Machado V, Pizarro-Hernández B. 2010. Actual and potential natural vegetation on the Canary Islands and its conservation status. Biodiversity and Conservation 19: 3089–3140. [Google Scholar]
- Assefa A, Ehrich D, Taberlet P, Nemomissa S, Brochmann C. 2007. Pleistocene colonization of afro-alpine ‘sky islands’ by the arctic-alpine Arabis alpina. Heredity 99: 1–10. [DOI] [PubMed] [Google Scholar]
- Axelrod DI, Raven PH. 1978. Late Cretaceous and Tertiary vegetation history of Africa. In: Werger MJA, ed. Biogeography and ecology of Southern Africa. Dordrecht: Springer, 77–130. [Google Scholar]
- Barrett SC, Kohn JR, Falk DA, Holsinger KE. 1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. Genetics and Conservation of Rare Plants 29: 3–30. [Google Scholar]
- Beaumont MA. 2010. Approximate Bayesian computation in evolution and ecology. Annual Review of Ecology, Evolution, and Systematics 41: 379–406. [Google Scholar]
- Bonnefille R. 2011. Rainforest responses to past climate changes in tropical Africa. In: Bush M, Flenley J, Gosling W, eds. Tropical rainforest responses to climate change, 2nd edn. Berlin: Springer, 125–184. [Google Scholar]
- Bramwell D. 1985. Contribución a la biogeografía de las Islas Canarias. Botánica Macaronésica 14: 3–34. [Google Scholar]
- Bramwell D, Bramwell ZI. 2001. Flores silvestres de las Islas Canarias. Madrid: Rueda. [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF. 2001. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques 31: 24–28. [PubMed] [Google Scholar]
- Budde KB, González-Martínez SC, Hardy OJ, Heuertz M. 2013. The ancient tropical rainforest tree Symphonia globulifera L. f. (Clusiaceae) was not restricted to postulated Pleistocene refugia in Atlantic Equatorial Africa. Heredity 111: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caujapé-Castells J. 2010. General GST and θ inflation due to biased intra-population sampling, and its consequences for the conservation of the Canarian flora. Conservation Genetics 11: 709–720. [Google Scholar]
- Caujapé-Castells J, Sabbagh I, Castellano JJ, et al. 2013. Transformer-4 version 2.0.1, a free multi-platform software to quickly reformat genotype matrices of any marker type, and archive them in the Demiurge information system. Molecular Ecology Resources 13: 484–493. [DOI] [PubMed] [Google Scholar]
- Caujapé-Castells J, García-Verdugo C, Marrero-Rodríguez A, Fernández-Palacios JM, Crawford DJ, Mort ME. 2017. Island ontogenies, syngameons, and the origins of genetic diversity in the Canarian flora. Perspectives in Plant Evolution, Ecology and Systematics 27: 9–22. [Google Scholar]
- Chakraborty D, Sinha A, Ramakrishnan U. 2014. Mixed fortunes: ancient expansion and recent decline in population size of a subtropical montane primate, the Arunachal macaque Macaca munzala. PLoS ONE doi: 10.1371/journal.pone.0097061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chala D, Brochmann C, Psomas A, et al. 2016. Goodbye to tropical alpine plant giants under warmer climates? Loss of range and genetic diversity in Lobelia rhynchopetalum. Ecology and Evolution 6: 8931–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chala D, Zimmermann NE, Brochmann C, Bakkestuen V. 2017. Migration corridors for alpine plants among the ‘sky islands’ of eastern Africa: do they, or did they exist?Alpine Botany 127: 133–144. [Google Scholar]
- Chybicki IJ, Burczyk J. 2009. Simultaneous estimation of null alleles and inbreeding coefficients. Journal of Heredity 100: 106–113. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Santos F, Beaumont MA, et al. 2008. Inferring population history with DIYABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24: 2713–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Ravigné V, Estoup A. 2010. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinformatics 11: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk QCB. 1992. Relict floras of Atlantic islands: patterns assessed. Biological Journal of the Linnean Society 46: 91–103. [Google Scholar]
- Dewoody J, Nason JD, Hipkins VD. 2006. Mitigating scoring errors in microsatellite data from wild populations. Molecular Ecology Notes 6: 951–957. [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution 22: 1185–1192. [DOI] [PubMed] [Google Scholar]
- Earl DA. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- EFAP 1994. Ethiopia Forestry Action Program. Final report. Addis Ababa: Ministry of Natural Resources Development and Environmental Protection. [Google Scholar]
- Espíndola A, Pellissier L, Maiorano L, Hordijk W, Guisan A, Alvarez N. 2012. Predicting present and future intra-specific genetic structure through niche hindcasting across 24 millennia. Ecology Letters 15: 649–657. [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. 2002. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology 11: 1591–1604. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1: 117693430500100003. [PMC free article] [PubMed] [Google Scholar]
- Fahrig L. 2003. Effects of habitat fragmentation on biodiversity. Annual Review of Ecology, Evolution, and Systematics 34: 487–515. [Google Scholar]
- Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biological Conservation 61: 1–10. [Google Scholar]
- FAO 2001. Forest resource assessment 2000. Forestry Paper 140. Rome: Food and Agricultural Organization of the United Nations. [Google Scholar]
- Fernández-Mazuecos M, Jiménez-Mejías P, Rotllan-Puig X, Vargas P. 2014. Narrow endemics to Mediterranean islands: moderate genetic diversity but narrow climatic niche of the ancient, critically endangered Naufraga (Apiaceae). Perspectives in Plant Ecology, Evolution and Systematics 16: 190–202. [Google Scholar]
- Fernández-Palacios JM, de Nascimento L, Otto R, et al. 2011. A reconstruction of Palaeo-Macaronesia, with particular reference to the long-term biogeography of the Atlantic island laurel forests. Journal of Biogeography 38: 226–246. [Google Scholar]
- Fjeldså J, Lovett JC. 1997. Geographical patterns of old and young species in African forest biota: the significance of specific montane areas as evolutionary centres. Biodiversity and Conservation 6: 325–346. [Google Scholar]
- Frankham R, Briscoe D, McInnes K, Ballou J. 2002. Introduction to conservation genetics. Cambridge, UK: Cambridge University Press. [Google Scholar]
- García-Verdugo C, Forrest AD, Fay MF, Vargas P. 2010. The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, Olea europaea subsp. guanchica. Evolution 64: 3525–3536. [DOI] [PubMed] [Google Scholar]
- García-Verdugo C, Sajeva M, La Mantia T, Harrouni C, Msanda F, Caujapé-Castells J. 2015. Do island plant populations really have lower genetic variation than mainland populations? Effects of selection and distribution range on genetic diversity estimates. Molecular Ecology 24: 726–741. [DOI] [PubMed] [Google Scholar]
- Garza JC, Williamson EG. 2001. Detection of reduction in population size using data from microsatellite loci. Molecular Ecology 10: 305–318. [DOI] [PubMed] [Google Scholar]
- Gavin DG, Fitzpatrick MC, Gugger PF, et al. 2014. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytologist 204: 37–54. [DOI] [PubMed] [Google Scholar]
- Gitzendanner MA, Soltis PS. 2000. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany 87: 783–792. [PubMed] [Google Scholar]
- González-Pérez MA, Lledo MD, Lexer C, et al. 2009. a Genetic diversity and differentiation in natural and reintroduced populations of Bencomia exstipulata and comparisons with B. caudata (Rosaceae) in the Canary Islands: an analysis using microsatellites. Botanical Journal of the Linnean Society 160: 429–441. [Google Scholar]
- González-Pérez MA, Sosa PA, Rivero E, González-González EA, Naranjo A. 2009b Molecular markers reveal no genetic differentiation between Myrica rivas-martinezii and M. faya (Myricaceae). Annals of Botany 103: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pérez MÁ, Caujapé-Castells J. 2014. Development and characterization of nuclear microsatellite markers for Parolinia ornata Webb (Brassicaceae), and cross-species amplification in all species described in the Canarian endemic genus Parolinia. Conservation Genetics Resources 6: 705–706 [Google Scholar]
- Grandcolas P, Nattier R, Trewick S. 2014. Relict species: a relict concept?Trends in Ecology & Evolution 29: 655–663. [DOI] [PubMed] [Google Scholar]
- Habel JC, Assmann T, eds 2009. Relict species: phylogeography and conservation biology. Heidelberg, Dordrecht, London, New York:Springer Science & Business Media. [Google Scholar]
- Hedberg O. 1961. Monograph of the genus Canarina L. (Campanulaceae). Svensk Botanisk Tidskrift 55: 17–62. [Google Scholar]
- Heald WF. 1951. Sky islands of Arizona. Natural History 60: 95–96. [Google Scholar]
- Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London B: Biological Sciences 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Juan C, Emerson BC, Oromí P, Hewitt GM. 2000. Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends in Ecology & Evolution 15: 104–109. [DOI] [PubMed] [Google Scholar]
- Kadu CAC, Schueler S, Konrad H, et al. 2011. Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands. Molecular Ecology 20: 165–178. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. 2005. HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Molecular Ecology Notes 5: 187–189. [Google Scholar]
- Kebede M, Ehrich D, Taberlet P, Nemomissa S, Brochmann C. 2007. Phylogeography and conservation genetics of a giant lobelia (Lobelia giberroa) in Ethiopian and Tropical East African mountains. Molecular Ecology 16: 1233–1243. [DOI] [PubMed] [Google Scholar]
- Kloos H, Legesse W, eds 2010. Water resources management in Ethiopia: implications for the Nile basin. Amherst, NY: Cambria Press. [Google Scholar]
- Kondraskov P, Schuetz N, Schuessler C, et al. 2015. Biogeography of Mediterranean hotspot biodiversity: reevaluating the ‘Tertiary’ relict hypothesis of Macaronesian laurel forests. PLoS ONE doi: 10.1371/journal.pone.0132091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruckeberg AR, Rabinowitz D. 1985. Biological aspects of endemism in higher plants. Annual Review of Ecology and Systematics 16: 447–479. [Google Scholar]
- Lande R. 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. American Naturalist 142: 911–927. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Thompson JD, Garnier E, Debussche M. 2004. The biology and ecology of narrow endemic and widespread plants: a comparative study of trait variation in 20 congeneric pairs. Oikos 107: 505–518. [Google Scholar]
- López de Heredia U, Venturas M, López RA, Gil L. 2010. High biogeographical and evolutionary value of Canary Island pine populations out of the elevational pine belt: the case of a relict coastal population. Journal of Biogeography 37: 2371–2383. [Google Scholar]
- Mairal M, Pokorny L, Aldasoro JJ, Alarcón M, Sanmartín I. 2015a Ancient vicariance and climate-driven extinction explain continental-wide disjunctions in Africa: the case of the Rand Flora genus Canarina (Campanulaceae). Molecular Ecology 24: 1335–1354. [DOI] [PubMed] [Google Scholar]
- Mairal M, Sanmartín I, Aldasoro JJ, Culshaw V, Manolopoulou I, Alarcón M. 2015b Paleo-islands as refugia and sources of genetic diversity within volcanic archipelagos: the case of the widespread endemic Canarina canariensis (Campanulaceae). Molecular Ecology 24: 3944–3963. [DOI] [PubMed] [Google Scholar]
- Mairal M, Sanmartín I, Pellissier L. 2017a Lineage-specific climatic niche drives the tempo of vicariance in the Rand Flora. Journal of Biogeography 44: 911–923. [Google Scholar]
- Mairal M, Sanmartín I, Herrero A, et al. 2017. b Geographic barriers and Pleistocene climate change shaped patterns of genetic variation in the Eastern Afromontane biodiversity hotspot. Scientific Reports 7: e45749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley J. 2000. Last Glacial Maximum lacustrine and fluviatile formations in the Tibesti and other Saharan mountains, and large-scale climatic teleconnections linked to the activity of the Subtropical Jet Stream. Global and Planetary Change 26: 121–136. [Google Scholar]
- Masao CA, Gizaw A, Piñeiro R, et al. 2013. Phylogeographic history and taxonomy of some afro-alpine grasses assessed based on AFLPs and morphometry: Deschampsia cespitosa, D. angusta and Koeleria capensis. Alpine Botany 123: 107–122. [Google Scholar]
- McCormack JE, Huang H, Knowles LL. 2009. Sky islands. In: Gillespie RG, Clague DA, eds. Encyclopedia of islands. Berkeley: University of California Press, 841–843. [Google Scholar]
- Meloni M, Reid A, Caujapé-Castells J, Soto M, Fernández-Palacios JM, Conti E. 2015. High genetic diversity and population structure in the endangered Canarian endemic Ruta oreojasme (Rutaceae). Genetica 143: 571–580. [DOI] [PubMed] [Google Scholar]
- Mittermeier RA, Robles Gil P, Hoffmann M, et al. 2004. Hotspots revisited. Mexico City: CEMEX. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- de Nascimento L, Nogué S, Criado C, et al. 2015. Reconstructing Holocene vegetation on the island of Gran Canaria before and after human colonization. Holocene 26: 113–125. [Google Scholar]
- Nybom H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143–1155. [DOI] [PubMed] [Google Scholar]
- Omondi SF, Kireger E, Dangasuk OG, et al. 2010. Genetic diversity and population structure of Acacia senegal (L) Willd. in Kenya. Tropical Plant Biology 3: 59–70. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538. [Google Scholar]
- Oostermeijer JGB, Luijten SH, Den Nijs JCM. 2003. Integrating demographic and genetic approaches in plant conservation. Biological Conservation 113: 389–398. [Google Scholar]
- Osborne AH, Vance D, Rohling EJ, Barton N, Rogerson M, Fello N. 2008. A humid corridor across the Sahara for the migration of early modern humans out of Africa 120,000 years ago. Proceedings of the National Academy of Sciences of the USA 105: 16444–16447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Paz J, Caujapé-Castells J. 2013. A review of the allozyme data set for the Canarian endemic flora: causes of the high genetic diversity levels and implications for conservation. Annals of Botany 111: 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu JL, et al. 2003. Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300: 1563–1565. [DOI] [PubMed] [Google Scholar]
- Pokorny L, Riina R, Mairal M, et al. 2015. Living on the edge: timing of Rand Flora disjunctions congruent with ongoing aridification in Africa. Frontiers in Genetics 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo P, Curto M, Meimberg H. 2016. Genetic structure of Micromeria (Lamiaceae) in Tenerife, the imprint of geological history and hybridization on within-island diversification. Ecology and Evolution 6: 3443–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz D. 1981. Seven forms of rarity. In: Synge H, ed. The biological aspects of rare plant conservation. Chichester: Wiley, 205–217. [Google Scholar]
- Reusing M. 2000. Change detection of natural high forests in Ethiopia using remote sensing and GIS techniques. International Archives of Photogrammetry and Remote Sensing 33: 1253–1258. [Google Scholar]
- Rigueiro C, Arroyo JM, Valido A, Jordano P. 2009. Isolation and characterization of 13 microsatellite loci for Neochamaelea pulverulenta (Cneoraceae). Molecular Ecology Resources 9: 1497–1500. [DOI] [PubMed] [Google Scholar]
- Rijsdijk KF, Hengl T, Norder SJ, Otto R, et al. 2014. Quantifying surface-area changes of volcanic islands driven by Pleistocene sea-level cycles: biogeographical implications for the Macaronesian archipelagos. Journal of Biogeography 41: 1242–1254. [Google Scholar]
- Roquet C, Sáez L, Aldasoro JJ, Susanna A, Alarcón ML, Garcia-Jacas N. 2008. Natural delineation, molecular phylogeny and floral evolution in Campanula. Systematic Botany 33: 203–217. [Google Scholar]
- Ruiz Guajardo JC, Schnabel A, Ennos R, Preuss S, OteorArnaiz A, Stone G. 2010. Landscape genetics of the key African acacia species Senegalia mellifera (Vahl) – the importance of the Kenyan Rift Valley. Molecular Ecology 19: 5126–5139. [DOI] [PubMed] [Google Scholar]
- Sanmartín I, Anderson CL, Alarcon M, Ronquist F, Aldasoro JJ. 2010. Bayesian island biogeography in a continental setting: the Rand Flora case. Biology Letters 6: 703– 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Guerra AS. 1990. Bosques de laurisilva en la región macaronésica. Serie Naturaleza y Medio Ambiente, No. 49. Strasbourg: Council of Europe. [Google Scholar]
- Saro I, González-Pérez MA, García-Verdugo C, Sosa PA. 2015. Patterns of genetic diversity in Phoenix canariensis, a widespread oceanic palm (species) endemic from the Canarian archipelago. Tree Genetics & Genomes 11: 1–13. [Google Scholar]
- Senut B, Pickford M, Ségalen L. 2009. Neogene desertification of Africa. Comptes Rendus Geoscience 341: 591–602. [Google Scholar]
- Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tierceli JJ, Brunet M. 2006. Tectonic uplift and Eastern Africa aridification. Science 313: 1419–1423. [DOI] [PubMed] [Google Scholar]
- Seto K, Guneralp B, Hutyra L. 2012. Global forecasts of urban expansion to 2030 and direct impacts on biodiversity and carbon pools. Proceedings of the National Academy of Sciences of the USA 109: 16083–16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestrini M, Junqueira MG, Favarin AC, et al. 2007. Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellites markers. Genetic Resources and Crop Evolution 54: 1367–1379. [Google Scholar]
- Sosa PA, González-Pérez MA, González-González E, et al. 2010. a Biología de la conservación de endemismos vegetales de los Parques Nacionales canarios: caracterización genética y demográfica. In: Ramírez L, Asensio B, eds. Proyectos de investigación en parques nacionales 2006–2009. Madrid: Organismo Autónomo Parques Nacionales, 225–248. [Google Scholar]
- Sosa PA, González-Pérez MÁ, Moreno C, Clarke JB. 2010b Conservation genetics of the endangered endemic Sambucus palmensis Link (Sambucaceae) from the Canary Islands. Conservation Genetics 11: 2357–2368. [Google Scholar]
- Stebbins GL. 1980. Rarity of plant species: a synthetic viewpoint. Rhodora 82: 77–86. [Google Scholar]
- Stebbins GL, Major J. 1965. Endemism and speciation in the California flora. Ecological Monographs 35: 1–35. [Google Scholar]
- Tilman D. 1999. The ecological consequences of changes in biodiversity: a search for general principles. Ecology 80: 1455–1474. [Google Scholar]
- Tilman D, May RM, Lehman CL, Nowak MA. 1994. Habitat destruction and the extinction debt. Nature 371: 65–66. [Google Scholar]
- Triantis KA, Borges PA, Ladle RJ, et al. 2010. Extinction debt on oceanic islands. Ecography 33: 285–294. [Google Scholar]
- Tsutsui ND, Suarez AV, Holway DA, Case TJ. 2000. Reduced genetic variation and the success of an invasive species. Proceedings of the National Academy of Sciences of the USA 97: 5948–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhou SL, Hong DY. 2013. Molecular phylogeny of the platycodonoid group. (Campanulaceae s. str.) with special reference to the circumscription of Codonopsis. Taxon 62: 498–504. [Google Scholar]
- White F. 1983. The vegetation of Africa: a descriptive memoir to accompany the UNESCO/AETFAT/UNSO vegetation map of Africa. Natural Resources Research Report No. 20. Paris: UNESCO. [Google Scholar]
- Williamson-Natesan EG. 2005. Comparison of methods for detecting bottlenecks from microsatellite loci. Conservation Genetics 6: 551–562. [Google Scholar]
- Yannic G, Pellissier L, Ortego J, et al. 2013. Genetic diversity in caribou linked to past and future climate change. Nature Climate Change 4: 132–137. [Google Scholar]
- Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX. 1997. POPGENE, the user-friendly shareware for population genetic analysis. https://sites.ualberta.ca/~fyeh/popgene.html. Molecular Biology and Biotechnology Centre, University of Alberta, Canada. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.