Abstract
Background and Aims
In dioecious plants, sexual reproduction requires close proximity to potential mates, but clonal growth can increase this distance and, therefore, reduce the probability of mating. Reduction in sexual propagules can lead to decreased dispersal and gene flow between populations. Gene flow and clonal growth may be further influenced by the size of the habitat patch. The effects of habitat size and reproductive mode (sexual or asexual reproduction) on spatial genetic structure and segregation of the sexes were tested by quantifying the distributions of genotypes and the sexes using the dioecious liverwort Marchantia inflexa.
Methods
Plants were sampled from five pairs of small–large habitat patches to identify within- and among-population spatial genetic structure using 12 microsatellite markers. Spatial distributions were calculated as the likelihood that pairs of individuals were the same sex or genotype, and it was determined how that likelihood was affected by habitat patch size (small/large).
Key Results
Asexual reproduction dominates within populations, and asexual dispersal also occurred across populations. Spatial segregation of the sexes was observed within populations; males were more likely to be near individuals of the same sex than were females. Although the likelihood of both sexes being near members of the same sex was similarly greater on small habitat patches, on large habitat patches male genotypes were almost 15 % more likely to be near clonemates than were female genotypes.
Conclusions
The results show a sex difference in clonal clumping that was dependent upon habitat size, suggesting differential colonization and/or survival between males and females. The sexes and genotypes being structured differently within and among populations have implications for the persistence of populations and the interactions between them. This study demonstrates that studying only the sexes and not their genotypes (or vice versa) can limit our understanding of the extent to which reproductive modes (sexual or asexual) influence genetic structure both within and between populations.
Keywords: Bryophyte, dispersal, population dynamics, reproductive mode, sex ratio, spatial segregation of the sexes
INTRODUCTION
Many plant species use both clonal reproduction and sexual reproduction, where clonal reproduction allows plants to maintain and spread their populations and sexual reproduction ensures the advantages of the sexual process, particularly evolutionary potential (Frankham, 1995). Despite this benefit of clonal reproduction, extended periods of clonal growth can cause both spatial segregation of the sexes (SSS) and spatial genetic structure (SGS) within populations, leading to the reduction or complete loss of sexual reproduction (Barrett, 2015). As parental genotypes, or genets, increase in size through clonal expansion, their proximity to an individual of a different sex or a different genotype increases. This increase in distance can increase inbreeding among clones, or ramets, in self-compatible species (Eckert, 2000; Raabová et al., 2015) and can limit sexual reproduction in obligate outcrossing species, such as self-incompatible and dioecious species (Hu et al., 2017). The lack of sexual mates can enhance reliance on asexual reproduction, leading to a further reduction of genotype diversity and available mates. However, species that produce asexual propagules, such as bulbils or gemmae, may not experience the high clumping of genotypes caused by vegetative growth. Dispersal of asexual propagules within a population may allow genotypes to be interdigitated, thereby breaking up the clumps of genotypes that result from vegetative growth (Vallejo-Marín et al., 2010). For outcrossing species, identifying the spatial distribution of ramets and genets within a population can provide information on the prevalence of asexual vs. sexual reproduction. Assessing sexual reproduction is important to better predict population persistence and maintenance of genotypic diversity, and ultimately evolutionary potential.
Clonality can also limit interactions between nearby populations by reducing dispersal. Colonization and among-population dispersal mainly occur via sexual propagules (Starfinger and Stöcklin, 1996; Eriksson, 1997; Vanderpoorten et al., 2008), probably because sexual propagules are generally smaller than clonal fragments or asexual propagules (Tackenberg et al., 2003; Korpelainen et al., 2005). However, in aquatic habitats, long-distance dispersal of asexual propagules is possible and may allow gene flow between populations (Johansson and Nilsson, 1993), which may be the only dispersal occurring if sexual propagules are not being produced. Interactions between populations, specifically the prevalence of dispersal, can be quantified by identifying spatial distributions of genets and ramets between populations. If dispersal between populations occurs via sexually produced propagules, we predict that the same genotype would not be found across different populations and there would be low genetic differentiation between populations. If there is any dispersal by asexual propagules, then we expect identical genotypes in multiple, physically disconnected, populations.
Despite the importance of the sexes being near one another as a requirement for sexual reproduction, studies of SGS on dioecious species do not always include sex of an individual (Alberto et al., 2005; Ramaiya et al., 2010; Korpelainen et al., 2013; but see Eppley et al., 1998; Vandepitte et al., 2009; Mizuki et al., 2010; Dering et al., 2016). This omission is most probably because determining the sex of an individual is generally limited to observing individuals at the time when gametes are being produced, detecting physical connections of an individual of unknown sex with another individual of known sex or matching genotypes of known sex with genotypes of unknown sex. However, including sex as a variable is important because clonality can drive SSS within populations (Melampy, 1981) and lead to populations composed of only one sex (McLetchie et al., 2002; Crowley et al., 2005), preventing sexual reproduction even though there are multiple genotypes present. Quantifying the sex ratio instead of identifying the spatial distribution of the sexes can lead to incorrect conclusions regarding the potential for sexual reproduction within a population. For example, populations may contain the same number of males and females, but the sexes could be clumped within the populations, restricting sexual reproduction due to limited male gamete dispersal (Eppley, 2005).
Because dispersal and establishment differ between sexual and asexual propagules, we can identify the importance of reproductive mode (sexual or asexual) to population persistence or to gene flow by quantifying SGS. If the production of sexual or asexual propagules varies between populations that experience differing environmental variables, then propagule production, and any SGS that occurs as a result, can be linked to that environmental variable. SGS has been previously linked to environmental variables associated with specific sites, allowing an association to be made between that variable and the dispersal patterns of sexual and asexual propagules. These studies include self-compatible species (Chung et al., 2006; Arnaud-Haond et al., 2007a) and outcrossing species, including self-incompatible (Raabová et al., 2015) and dioecious (Dering et al., 2016) species, and across several environmental variables, including disturbance regimes (Kostrakiewicz-Gierałt, 2013), light (Vandepitte et al., 2010), elevation (Sun et al., 2001) and habitat size (Kettenring et al., 2010).
Habitat size can potentially have a large impact on between-population dynamics. If habitat quality is constant, a large habitat patch is associated with higher immigration rates when compared with smaller habitat patches (Johnson, 2005), and more immigrants could lead to more unique genotypes and potentially greater mating opportunities. Conversely, smaller habitat patches may have more limited immigration and reduced genetic variation (Biere et al., 2012), and there is a greater chance of losing one sex, therefore reducing or even eliminating mating potential. This reduced mating potential may cause greater reliance on asexual reproduction, which then limits among-population dynamics and affects the long-term persistence of the population particularly because smaller habitats can experience increased emigration rates (Thomas and Hanski, 1997; Johnson, 2005) and are more susceptible to demographic or environmental stochasticity (Lande, 1988).
Using the dioecious bryophyte Marchantia inflexa Nees et Mont., we quantified SGS of genotypes within and among populations in different size habitat patches (the environmental variable) and subsequently tested for (1) the signature of sexual vs. asexual reproduction on SGS; (2) the spatial association of the sexes across ramets within genets; and (3) the dependence of (1) and (2) on the size of the habitat patch. Within populations, if asexual reproduction is the main form of reproduction, we predict that overall most individuals within a population will consist of multiple copies of the same genotype. Additionally, if asexual reproduction is the main form of reproduction, then we predict significant genetic differentiation between populations and high SGS because asexual propagules generally stay within the source population. If sexual reproduction is the main form of reproduction within a population, then most individuals will be unique genotypes. Because sexual propagules could stay within the source population and would also disperse among populations, we would expect to see genetic mixing among populations. We also predict that there will be gene flow evidenced by the lack of significant genetic differentiation, so low SGS, between populations. Alternatively, both reproductive methods could be occurring and affecting SGS and gene flow. In this case, we expect to observe both unique genotypes and genotypes with multiple clones within and between habitat patches and that the level of genetic differentiation will indicate the prevalence of gene flow.
In a dioecious species, there could be differential clustering between the sexes leading to SSS. Because females of the focal species have a higher vegetative growth rate and lower asexual propagule production rate than males (McLetchie and Puterbaugh, 2000; Fuselier and McLetchie, 2002), we predict that female ramets of the same genet will be more clustered than male ramets of the same genet. Lastly, relative to populations in large habitat patches, we predict that populations in small habitat patches will have low genetic diversity, which may be attributed to fewer resources (Kareiva, 1985), lower immigration rates (Johnson, 2005) and/or greater susceptibility to demographic stochasticity (Lande, 1988). These forces can also result in more variable sex ratios in smaller populations relative to large populations due to two factors: a pattern of reduced or no sexual reproduction producing new genotypes and the reliance on asexual reproduction for population persistence.
MATERIALS AND METHODS
Study organism
Marchantia inflexa is a thalloid liverwort (phylum Marchantiophyta) with separate male and female plants (dioecious). The distribution of M. inflexa ranges from the southern US to northern Venezuela (Bischler, 1984). Marchantia inflexa occurs as spatially separated populations along streams on distinct habitat patches of substrate (boulders and bedrock) separated from other habitat patches by water, creating distinct populations that are distant enough from each other to prevent fertilization between individuals from different populations (Garcia-Ramos et al., 2007; Stieha et al., 2016). The photosynthetic stage is haploid, and sex determination is under genetic control (Bischler, 1986). Due to the presence of heterogametic sex chromosomes, spores (sexual propagules) produced by the diploid sporophyte, which is matured on the female plant, are expected to have a 1:1 sex ratio (Bischler, 1986). Both sexes can reproduce asexually by both asexual propagules and extension by growth. Asexual propagules (gemmae within a cup) are produced on the thallus surface and are dispersed by water (Brodie, 1951). The gemmae can be dispersed aerially by water splashing in the cup (within-patch dispersal) or downstream by floating in flowing water (among-population dispersal) (C. Stieha, pers. obs.). Plants also grow horizontally by dichotomous branching of the thalli, and disintegration of older tissue results in fragmentation and production of physiologically independent plants. Male and female plants produce distinct sex structures seasonally that are elevated above the plant (antheridiophores and archegoniophores, respectively) permitting clear sex identification when present. However, when sex structures are absent, sex is not visually distinguishable, requiring plants to be grown in a greenhouse or growth chamber to develop sex structures. To identify genets (unique genotypes) and ramets (genotypic clones or clonemates) of genets, 12 polymorphic microsatellite markers have been previously developed (Brzyski et al., 2012).
Field sampling
Sampling took place in July 2011 along a 3 km section of the Quare River on the island of Trinidad, the Republic of Trinidad and Tobago. Representative specimens of M. inflexa were vouchered at the Missouri Botanical Garden (St. Louis, MO, USA, specimen numbers M0292113 and M092115) and at the National Herbarium of the Republic of Trinidad and Tobago (St. Augustine, Trinidad, specimen number TRIN34616, D.N. McLetchie, collector).
We quantified patterns of SGS in populations where both sexes co-occur but where the plant samples were initially of unknown sex and could be determined in the lab. This approach avoids possible biases of sampling only individuals with sex structures or only populations with individuals in the sexual phase. Five habitat patch pairs with M. inflexa were sampled, with each pair consisting of a small patch (1–3 m at its longest length) surrounded by water and a large patch (>7 m at its longest length) connected to the stream bank (Supplementary data File 1). The distance between each small and large patch pair ranged from 2.0 to 12.5 m (measured with a measuring tape, Table 1). Each patch pair was separated by at least 20 m from another patch pair, with pairs 1, 2 and 3 being within 200 m of each other and pairs 4 and 5 being approx. 1 km apart from the other pairs. Pairs are numbered in order from the most upstream pair (pair 1) to the most downstream pair (pair 5). Within a pair, the small patch and large patch were not necessarily upstream or downstream from each other.
Table 1.
Sizes and interpatch distances of the five small–large habitat patch pairs containing Marchantia inflexa sampled along Quare River
| Pair | Small (m) | Large (m) | Interpatch distance (m) |
|---|---|---|---|
| 1 | 2.5 | 16.0 | 12.5 |
| 2 | 3.5 | 25.0 | 5.0 |
| 3 | 2.1 | 7.0 | 11.0 |
| 4 | 2.6 | 7.0 | 6.0 |
| 5 | 2.0 | 12.0 | 2.0 |
Mean size ± s.e. for the small habitat and large habitat were 2.54 ± 0.27 and 13.4 ± 3.35 m, respectively.
Before sampling, we determined the longest transect length for each patch. A random point was located along this transect and a second transect (1 m long and within 1 m of the random point) was placed to intersect as many M. inflexa thalli as possible, and was located from the water edge to the interior of the patch. One exception was the small patch of pair 5 where the transect touched water on both ends. Sex expression was not occurring at the time of collection. At every 2 cm interval of the transect, if a plant was present we collected one growing thallus tip for a possible total of 50 plants per patch (Fig. 1). Most plants were small (<5 mm) and none of the transects intersected mats of overlapping thalli. Plants were returned to the USA and grown in a greenhouse at the University of Kentucky to determine sex by monitoring for sex expression and to obtain sufficient tissue for DNA extraction. Individuals that did not develop sex structures in the greenhouse were placed in a growth chamber (14 h at 20 °C and 10 h at 16 °C in 24 h light, fluorescent and incandescent light at approx. 35 μmol m–2 s–1) to promote sex expression. Nine plants died before expressing sex and four never expressed sex. These individuals were removed from all analyses.
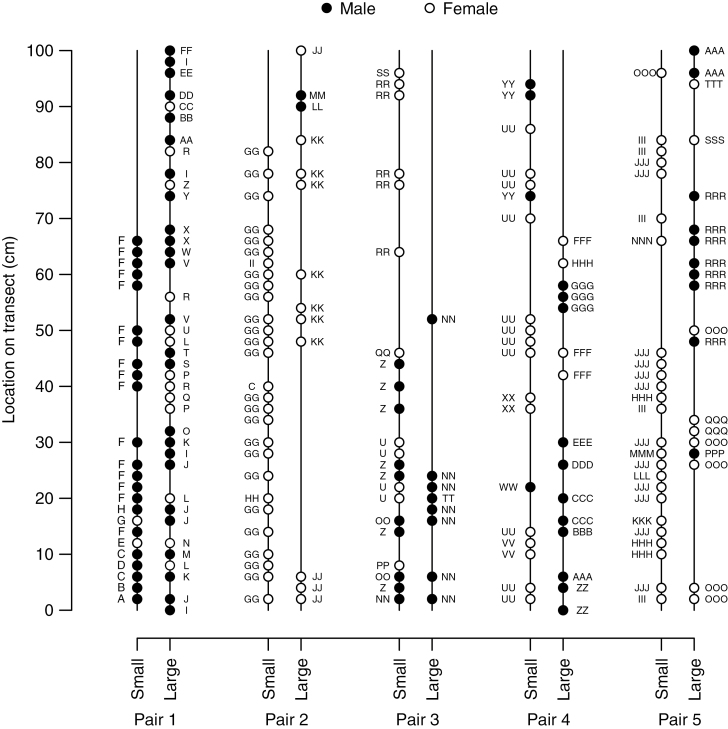
Fig. 1.
Spatial schematic of Marchantia inflexa plants as collected along ten 100 cm transects on five small–large habitat patch pairs. Plants, if present, were collected every 2 cm along the transect, with the location at 0 cm representing the water’s edge. Each unique genotype (genet and its genotypic clones) is represented by a letter combination. For example, genotype C is a different genotype from CC and CCC.
Genotyping
DNA was extracted from the thallus tissue of each plant sample using a modified cetyltrimethylammonium bromide (CTAB) method (adapted from Doyle and Doyle, 1987) and amplified using 12 polymorphic microsatellite primers specifically developed for M. inflexa (Brzyski et al., 2012). Primers were fluorescently labelled during PCR, and multiple loci were combined for fragment analysis. Genotyping took place at the University of Kentucky’s Advanced Genetic Technologies Center on a 3730 DNA Analyzer (Applied Biosystems, Fortune City, CA, USA) using the ROX 400HD internal size standard. Fragment analysis was performed with Peak Scanner version 1.0 (Applied Biosystems).
Descriptive genetic statistics
The majority of plants differed in alleles at two or more loci. There were 31 incidences where one plant differed from another at only one locus. Of these, 13 differed in their fragment lengths by 2 bp, but the others differed by up to 10 bp, suggesting that scoring error was not the cause of these single-allele differences. Therefore, for subsequent analyses, these 31 plants were treated as unique genotypes. To ensure that identical genotypes were of the same clone, we calculated Psex, which is the probability, based on the observed allele frequencies, that two individuals could have the same genotype by chance because of sexual reproduction (derived from Arnaud-Haond et al., 2007b). When the Psex value is <0.01, the identical genotypes can be considered to be of the same genet (Arnaud-Haond et al., 2007b).
Descriptive statistics of overall genetic variation were calculated using GDA 1.1 (Lewis and Zaykin, 2001) including allelic richness (number of alleles per polymorphic locus) and number of unique alleles. Because plant tissues are haploid, heterozygosity values and inbreeding coefficients could not be calculated. Instead, we calculated Shannon’s unbiased diversity measurement using GenAlEx 6.5 (Peakall and Smouse, 2012), which was also the program used to identify multilocus genotypes, done by matching across loci.
Genotypic richness of a patch was calculated as G/N where G is the number of genotypes (genets) and N is the total number of plants (ramets) sampled across all genotypes (Ellstrand and Roose, 1987). Sex ratios for both genets and ramets were calculated as the proportion of males. Habitat patch size was treated as a categorical variable designated as either small or large. To determine the relationship of habitat size to genotypic richness, allelic richness, unique alleles and sex ratio, we used paired t-tests, where we paired small and large patches based upon our sampling procedure described above. To test for a relationship of genotypic richness and sex ratio (genet and ramet) between ramets and genets, we used the Pearson correlation coefficient (r). For the analyses involving the number of unique alleles and the sex ratios, we used the non-parametric Spearman’s coefficient (ρ) due to violation of the normality assumption for the residuals. Lastly, overall biases in sex ratios were tested at the genet and ramet levels. To test if the sex ratios of populations from small patches were more variable than populations from large patches, we used Levene’s test to detect if the variances differed between these two groups (Vandepitte et al., 2010) at the genet and ramet level.
Patterns of spatial distributions of the sexes and genotypes within habitat patches
To study the spatial distribution of the sexes and spatial genetic structure within a habitat patch, we quantified how distance, sex and habitat patch size (small/large) affected the likelihood that pairs of individuals were the same sex or genotype. To determine the likelihood that two individuals were the same sex, which corresponded to the level of SSS within a patch, the patches needed to contain both sexes; therefore, we excluded the three single-sex patches from the analysis of the spatial distribution of the sexes (the small patch in patch pairs 2 and 5, and the large patch in patch pair 3). To determine the likelihood that two randomly selected individuals were the same genotype, which measured the SGS within a patch, we used individuals from all genotypes within all patches. To quantify the clump size of a genotype based on sex and patch size, we only used individuals that were from genotypes with multiple individuals in a single patch.
To perform our analyses and account for the SGS of individuals within the patches, we first converted the transect data to pairwise comparisons between individuals within a patch. We included the pairwise comparisons of an individual with itself, i.e. when the distance between two individuals equals 0 cm; removing these comparisons did not qualitatively affect the results. Because each pairwise comparison has two individuals, we used each comparison twice when analysing our data. In one instance, one individual was the focal individual. In the next instance, the other individual was the focal individual. For each pairwise comparison, we computed the spatial distance (in centimetres) between the two individuals. For the focal individual, we included its sex, genotype, individual identification number related to its spatial location within the patch, the patch size and the patch pair identification number. For each pairwise comparison, we determined the binary response variables of whether or not the two individuals were the same sex or genotype. Therefore, to analyse our data, we employed logistic regression with both fixed and random effects using the lme4 library (Bates et al., 2014) in R (R Core Team, 2013). Besides analysing our data focusing on whether two individuals were the same genotype, we also analysed our data by computing the genetic distance between individuals within a patch, which can account for mutation within a clone and the history of sexual reproduction (Supplementary data File 2; Table S1, Fig. S1). However, this analysis gave similar results to the above analysis; therefore, we focus on the analysis of whether two individuals were the same genotype.
In our statistical model, we included the fixed effects of the sex of the focal individual, habitat patch size and distance between the two sampling units, and the random effects of patch pair identification number, genotype and individual identification code. Patch pair identification number accounted for similarities between the two patches within a patch pair. Genotype as a random effect encompassed the potential life history differences among genotypes. We included an individual identification code nested within genotype to account for the fact that each sampled individual had multiple pairwise comparisons. We employed both random intercepts and random slopes with respect to the relationship between genotype, individual and distance. Different genotypes could have life history differences (such as more vegetative growth or more asexual reproduction) that would affect how far individuals of that genotype could be dispersed from other individuals of that genotype. By including the interaction between genotype, individual and distance, we can focus on the sex of the genotype, the distance between two individuals and the patch size that was measured, and their effects on whether or not the neighbouring individual is the same sex or genotype.
To determine which fixed effects significantly explained the probability that two individuals were the same sex or genotype, we performed model reduction (Crawley, 2007). We started with a full model containing all the random effects, the interactions between distance and individual nested within genotype, all the fixed effects and interactions among fixed effects. To determine the significance of the fixed effects, we computed the deviance between a model with the term and a model without the term and tested for significance in the difference in the deviance using a χ2 test. When the interaction terms were significant, we used a z-test to determine differences among the combination of the fixed effects. We performed these comparisons by setting one combination as the reference and testing whether the coefficients for the other combinations (and therefore the change in the coefficient from the reference level) were significantly different from the reference.
Patterns of genetic differentiation among habitat patches
We calculated genetic differentiation between the five patch pairs as well as among all patches with Shannon’s Mutual Information Index (sHua) using GenAlEx 6.5 (Peakall and Smouse, 2012). This index provides the same information as FST (Wright, 1951) but with the advantages of being more sensitive to rare alleles, incorporating sample sizes and providing a more robust estimate of dispersal than other methods (Sherwin et al., 2006; Sherwin, 2010). Genetic differentiation values range from zero to one, with zero indicating complete overlap of genotypes across all habitat patches, and one indicating that no alleles are shared between patches. We also performed an analysis of molecular variance (AMOVA) to quantify hierarchical genetic differentiation within and among populations using GenAlEx 6.5 (Peakall and Smouse, 2012).
RESULTS
Descriptive genetic statistics
We sampled 128 plants from small patches and 98 plants from large patches for a total of 226 plants (Table 2). After genotyping, 13 individuals contained at least one locus that did not successfully amplify and were removed from all analyses. Of the remaining 213, there were 79 multilocus genotypes, with 34 having multiple ramets and 45 being single unique genotypes (Fig. 1). The probability of an identical genotype being produced by distinct sexual reproductive events was significantly low (Psex < 0.001), allowing us to conclude that individuals of the same genotype are the products of asexual reproduction.
Table 2.
Genetic descriptive statistics of Marchantia inflexa collected within each habitat patch pair, including sample size, the average number of alleles per locus and the number of total alleles unique to that patch, Shannon’s unbiased diversity measurement, genotypic richness (G/N) and sex ratios (proportion of males) for genets and ramets
| Substrate pair | Substrate size | n | Alleles/locus | Unique alleles | Diversity | G/N | Genet sex ratio | Ramet sex ratio |
|---|---|---|---|---|---|---|---|---|
| 1 | Small | 24 | 2.90 | 3 | 0.336 | 0.33 | 0.75 | 0.92 |
| Large | 38 | 2.73 | 1 | 0.343 | 0.63 | 0.67 | 0.63 | |
| 2 | Small | 28 | 2.14 | 0 | 0.339 | 0.14 | 0 | 0 |
| Large | 13 | 2.56 | 0 | 0.088 | 0.31 | 0.50 | 0.15 | |
| 3 | Small | 22 | 2.45 | 0 | 0.277 | 0.36 | 0.38 | 0.45 |
| Large | 8 | 2.00 | 0 | 0.330 | 0.25 | 1.00 | 1.00 | |
| 4 | Small | 20 | 2.33 | 3 | 0.522 | 0.25 | 0.40 | 0.20 |
| Large | 16 | 3.12 | 2 | 0.260 | 0.56 | 0.78 | 0.75 | |
| 5 | Small | 25 | 2.81 | 1 | 0.266 | 0.32 | 0 | 0 |
| Large | 19 | 2.00 | 0 | 0.271 | 0.37 | 0.43 | 0.58 |
The overall average allelic richness was 2.5 alleles/locus per patch (range 2.00–3.12; Table 2). Small patches had, on average, higher numerical values for allelic richness (2.53 compared with 2.48), unique alleles (seven compared with three) and diversity (0.35 compared with 0.26) than large patches, but none of these variables was statistically different or correlated with one another (ρ = 0.37–0.57, d.f. = 9, P > 0.2). Average patch genotypic richness was 0.35 (range 0.14–0.63; Table 2). In all but one patch pair, small patches had lower genotypic richness (average = 0.28) than large patches (average = 0.42), but this was not statistically significant (paired t-test, d.f. = 4, P = 0.14).
Three populations contained only one sex, one large male-only patch and two small female-only patches, and all contained more than one genotype (Fig. 1; Table 2). Overall, genet sex ratio (0.52, proportion of males) did not differ from 0.5 (χ2 = 0.1139, d.f. =1, P = 0.736) and ramet sex ratio (0.44) tended to be female bias (χ2 = 3.4318, d.f. =1, P = 0.064). Based on patch pairs, genet sex ratios were more male biased on large patches than on small patches (paired t-test, d.f. = 4, P = 0.04), but ramet sex ratio was not associated with patch size (paired t-test, d.f. = 4, P = 0.14). Variance in sex ratio did not differ between the populations from the large and small habitat patches (genet sex ratio F = 0.6543, d.f. =1, P = 0.4420; ramet sex ratio F = 0.5251, d.f. =1, P = 0.4893). Over all patches, ramet and genet sex ratios were highly correlated with each other (r = 0.9072, d.f. = 9, P < 0.001).
Patterns of spatial distributions of sex and genotypes within habitat patches
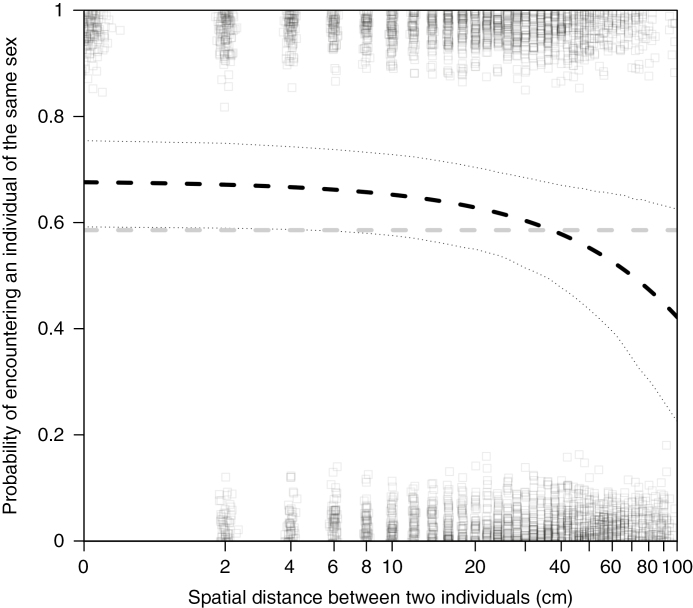
The likelihood that two individuals were the same sex was affected by the sex of the focal individual, the patch size and the distance between the two individuals, but was not affected by the interactions between these main effects (Table 3). As the distance between the two individuals increased, the probability that they were the same sex decreased from about 0.7 at 0 cm to about 0.4 at 100 cm between individuals (Fig. 2), which suggests spatial segregation of the sexes. However, the spatial segregation of the sexes was asymmetrical with respect to the sexes and the patch size. Males were about 28 % more likely to be near other males than females were to be near other females (significant main effect of the sex of the focal individual; Table 3). Individuals on small patches were about 46 % more likely to be near individuals of the same sex than were individuals on large patches (significant main effect of patch size; Table 3).
Table 3.
Statistical tests for both the comparison between the sexes and the comparison among genotypes
| Term | Base model | Δdeviance | d.f. | P | |
|---|---|---|---|---|---|
| Comparison between the sexes | Distance × sex × size | Distance + sex + size + distance × sex + distance × size + sex × size | 1.0756 | 1 | 0.2997 |
| Sex × size | Distance + sex + size + distance × sex + distance × size | 0.2151 | 1 | 0.6428 | |
| Distance × sex | Distance + sex + size + distance × size | 1.0556 | 1 | 0.3042 | |
| Distance × size | Distance + sex + size | 1.6034 | 1 | 0.2054 | |
| Size | Distance + sex | 7.4035 | 1 | 0.00651 | |
| Ssex | Distance + size | 7.6285 | 1 | 0.005745 | |
| Distance | Sex + size | 5.0022 | 1 | 0.02531 | |
| Comparison among genotypes | Distance × sex × size† | Distance + sex + size + distance × sex + distance × size + sex × size | 10.735 | 1 | 0.0011 |
| Distance × sex | Distance + sex + size + distance × size + sex × size | 0.8148 | 1 | 0.3667 | |
| Distance × size | Distance + sex + size + sex × size | 15.188 | 1 | <0.0001 | |
| Sex × size | Distance + sex + size + distance × size | 5.5102 | 1 | 0.0189 |
We performed a model reduction by starting with a full model containing all fixed effects and all interaction terms among the fixed effects, and removed one term at a time until we were left with the simplest model. To determine whether or not a term was significant, we computed the change in deviance (∆deviance) when we compared the statistical fit of the base model with the statistical fit of the base model and the term. If the change in deviance was significant based on a χ2 test, we left the term in. These models also included the effect of genotype and individual identification number nested within genotype as random intercepts, and the interaction between distance, genotype and individual nested within genotype as a random slope. Terms in bold are the highest order terms included in the final statistical model. If the interaction term was significant, all the main effects of that interaction term were also included. For example, in the ‘Comparison among genotypes’, the sex × size interaction is significant, so we retained the interaction term as well as sex and size by themselves.
†Differences between statistical models with and without the term were significant, However, the coefficient for the term was not significantly different from 0.
Fig. 2.
As the distance between two individuals increased, the probability that they were the same sex decreased. In our populations, we observed clumping of the sexes, where individuals in close proximity were more likely to be the same sex (dashed black line) than expected in a completely mixed population (dashed grey line). We controlled for the size of the habitat patch and the sex of the focal individual, but these factors did not interact with distance (see Table 3). The black dotted lines are the bootstrapped upper and lower 95 % confidence intervals computed based on 1000 simulations using the bootMer function in the lme4 package (Bates et al., 2014). Boxes are the raw data on whether pairs of individuals were the same sex (y = 1) or the opposite sex (y = 0). We added jitter along both axes to make all the data visible. To be able to plot distance = 0 on a log10 graph, we computed log10(distance + 1) and adjusted the x-axis labels to account for this transformation.
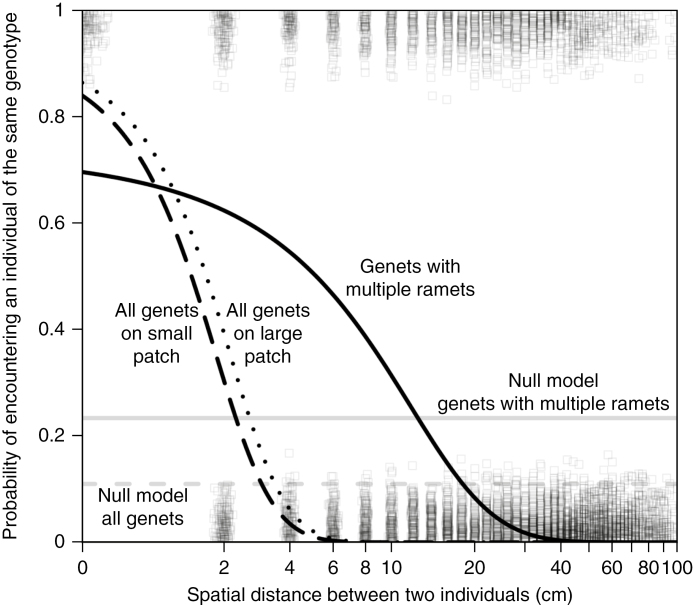
Although the likelihood that two individuals were the same sex was affected only by the main effects, the likelihood that two individuals were the same genotype depended on the interaction of patch size with both distance and the sex of the focal individual (Table 4). The probability that two individuals were the same genotype decreased to zero as the distance between the two individuals became greater than 6 cm; this probability decreased faster on small patches than on large patches (Fig. 3). The effects of patch size also depended on the sex of the focal individual. Males on large patches were about 13 % more likely to be next to the same genotype relative to males on small patches (z = –1.88, P = 0.060) and about 15 % more likely than females on large patches (z = –2.23, P = 0.026). Females on small patches were not significantly more or less likely to be next to individuals of the same genotype than individuals from different combinations of sex and patches (vs. female on large patches, vs. males on small patches and vs. male on large patches; all P > 0.19).
Table 4.
Genetic differentiation (sHua) of Marchantia inflexa populations
| sHua | 1S | 1L | 2S | 2L | 3S | 3L | 4S | 4L | 5S | 5L |
|---|---|---|---|---|---|---|---|---|---|---|
| 1S | ||||||||||
| 1L | 0.227 | |||||||||
| 2S | 0.255 | 0.193 | ||||||||
| 2L | 0.280 | 0.227 | 0.082 | |||||||
| 3S | 0.268 | 0.140 | 0.196 | 0.232 | ||||||
| 3L | 0.441 | 0.323 | 0.421 | 0.502 | 0.383 | |||||
| 4S | 0.243 | 0.167 | 0.173 | 0.257 | 0.200 | 0.371 | ||||
| 4L | 0.262 | 0.234 | 0.290 | 0.316 | 0.238 | 0.570 | 0.248 | |||
| 5S | 0.281 | 0.246 | 0.145 | 0.186 | 0.206 | 0.465 | 0.253 | 0.293 | ||
| 5L | 0.259 | 0.132 | 0.168 | 0.232 | 0.163 | 0.459 | 0.165 | 0.348 | 0.215 |
The comparisons between small–large habitat pairs are in bold.
Fig. 3.
The probability that two individuals (ramets) were the same genotype decreased as the distance between the two individuals increased. The patch size affected this relationship, where individuals on small patches (black, dashed line) were less likely to be near genetically identical individuals than individuals on large patches (black, dotted line). If individuals were randomly distributed within a patch (null model), the probabilty that any two individuals were the same genotype would be about 0.1 despite the distance between individuals. This result means that, across all individuals (ramets) and genotypes (genets), clumping of genotypes occurs and the spatial extent of the clumping of genets is smaller for small patches than for large patches. Because many genotypes are represented by multiple individuals within a patch, we can account for the genotypes that are found in only one individual and focus on the spatial clumping of a genotype. These individuals are not dispersed equally throughout the patch (null model is grey, solid line), but form clumps up to 40 cm wide (black, solid line). Boxes show whether the two individuals in each pairwise comparison are the same genotype (y = 1) or are not the same genotype (y = 0). We added jitter along both the x-axis and y-axis so that all the data points could be observed. Distance is graphed as log10(distance + 1) to allow us to plot a distance of 0 cm; the labels on the x-axis correct for the transformation.
When we determined the size of a genotype clump by focusing on focal individuals whose genotype was observed more than once within a patch, we found that the likelihood that two individuals were the same genotype reached zero as the distance between the two individuals approached 40 cm (Δdeviance = 23.98, d.f. = 1, P < 0.0001; Fig. 3), compared with the 6 cm distance when all focal individuals were used. Neither patch, sex nor any interaction terms was significant (P > 0.16 for all interactions and main effects besides distance; analysis not shown).
Patterns of genetic differentiation among habitat patches
Of the 53 total alleles identified, ten (18.9 %) were shared among all habitat patches. Five genotypes were found in more than one patch with no consistent pattern (one male and one female genotype were found in both patches within a pair, while three genotypes, two females and one male, were found across patch pairs), but there were no genotypes found in more than two patches.
Genetic differentiation between patches within patch pairs averaged 0.23 and, when averaged across all patches, was 0.27 (range = 0.08–0.57; Table 4). The results of the AMOVA indicated that 64 % of the existing genetic variation was found within populations and 36 % among populations (P < 0.001).
DISCUSSION
In the clonal, dioecious system of Marchantia inflexa, we documented both spatial segregation of the sexes (SSS) and spatial genetic structuring (SGS) of genotypes that differed by sex. Among habitat patches, we observed segregation of both the genotypes and the sexes (see the variation in sex ratio among the patches). Within habitat patches, sexes were spatially segregated, with males being more clumped than females, whereas genotypes were also clumped but the degree of clumping depended on the interactions with habitat patch size and sex. From our results, we found that among populations, the distribution of genotypes is influenced by both sexual and asexual propagules, while within-population dynamics were strongly influenced by asexual reproduction.
Among-population dynamics, specifically gene flow between patches, was demonstrated genetically to be minimal. Our data suggest limited gene flow as indicated by the presence of significant genetic variation among patches (36 %) and relatively high levels of genetic differentiation (sHua) among patches (average = 0.27 ± 0.02). Large habitat patches tended to have higher genotypic diversity than small patches, which suggests that sexual reproduction occurs more often in large patches or that large patches receive more immigrants than small patches. Because the spatial distance between genets is shorter in small patches, suggesting better sexual reproduction potential in smaller than in larger patches, we suspect immigration is the most likely explanation for this trend. Asexual propagules are generally considered not to contribute to long-distance dispersal and are more important for within-population dynamics of seed plants (Starfinger and Stöcklin, 1996; Santamaría, 2002). As reported for other bryophytes (Laaka-Lindberg et al., 2003; Pohjamo et al., 2006), our study showed that these propagules can actually influence among-population dynamics by dispersing between populations via the water as evidenced by identical genotypes observed across populations. We found five incidences of asexual dispersal, occurring among either patch pairs 1, 2 and 3, or between patch pairs 4 and 5. Patches 1–3 were all located within a 200 m river stretch and were separated from patches 4 and 5 by approx. 1 km. These distances suggest a potentially large dispersal distance of asexual propagules of up to 200 m but <1 km. Rarefaction curves (data not shown) indicated that our sampling protocol was adequate to quantify genetic variation but, because only a sub-set of the population was sampled, additional identical genotypes between populations are possible. There are minimal data on the actual distance travelled by asexual propagules in bryophytes (Korpelainen et al., 2005; but see Pohjamo et al., 2006), but simulations have suggested that the lower limit of the average long-distance travel of asexual propagules in M. inflexa is expected to be 2–5 m (Stieha et al., 2014). When gene flow is measured in other bryophytes (Korpelainen et al., 2005; Paasch et al., 2015), clonality does not appear to limit dispersal potential as long as the habitat is aquatic. However, the movement of asexual propagules in our experiment is probably not a common occurrence given the significant genetic differentiation among populations. When compared with other values of genetic differentiation in liverworts, which have been reported to be as high as 0.928 (average = 0.614 in Korpelainen et al., 2005; 0.480 in Bączkiewicz, 2013), our value is much lower but it is important to note that these studies have mostly been on a large scale of hundreds of kilometres.
Within populations, asexual reproduction was prevalent and had a strong influence on SGS. Genotypic diversity, though variable, was low on average, suggesting prevalent asexual reproduction. Both clonal growth and asexual propagules, which fall near the parent plant, would result in the genotype clumping observed in this study, where individuals were likely to interact with other individuals of the same genotype up to 6 cm away. This amount of clumping alone reduces the potential for sexual reproduction (Wyatt, 1977; Reynolds, 1980; McLetchie, 1996). However, clumping was also observed in the sexes, causing spatial segregation of males and females and even further reducing proximity to mates. Interestingly, SSS differed by sex and by habitat patch size. On larger patches, which were male biased, male genotypes were more clumped than female genotypes, whereas in populations on small habitat patches male and female genotypes did not differ with respect to clumping. This patch difference in the clumping of genotypes between the sexes indicates the effects of environmental conditions (including patch age) on the dynamics of our study organism. Identifying these specific environmental conditions will require further studies.
Environmental effects have been shown to drive aggregation of the sexes in many seed plant species. For example, in the dioecious perennial Mercurialis perennis, male genets grew more in high light conditions than females, which led to strong segregation of the sexes in high light areas due to large male genets, but no segregation of the sexes in low light areas (Vandepitte et al., 2010). Our results demonstrate a sex difference based on the size of the habitat, and this can be readily seen when the average population-level parameters of the large and small patches are compared (Table 2). On average, male-biased sex ratios occur on large habitat patches relative to small habitat patches (0.622 ± 0.139 vs. 0.314 ± 0.176, means ± s.e., respectively) and are linked to more male genotypes and not more ramets per genet in males. On average, large habitat patches contained twice as many male genotypes as female genotypes (6.01 ± 2.67 vs. 3.18 ± 1.34), but females were favoured in the overall numbers of ramets per genet (overall males had 2.36 ± 0.61 and females had 2.82 ± 0.89 ramets per genet). On the other hand, small habitat patches contained on average half as many male genotypes as female genotypes (2.19 ± 1.10 vs. 4.36 ± 1.03), and females were favoured in the overall numbers of ramets per genet (overall males had 3.00 ± 0.51 and females had 3.81 ± 1.09 ramets per genet). Additionally, the two single-sex populations in small patches are female and the one single-sex population in a large habitat patch was male. Assuming similar spore colonization rates by the sexes, these patterns suggest higher establishment or survival rates for male genotypes relative to females in large relative to small habitat patches, and females that do survive tend to have more ramets per genet than males. In seed plants, male-biased sex ratios have been linked to higher cost for sexual reproduction in females where, as populations age, the sex ratio becomes more male biased due to lower female survival (Geber, 1999; Obeso, 2002; Field et al., 2013). In M. inflexa, higher female cost to sexual reproduction assumes that sperm is not limiting but it actually might be because sperm dispersal distance is on the order of centimetres in bryophytes (Wyatt, 1977; Reynolds, 1980; McLetchie, 1996), and we know that at these small distances individuals are more likely to be near individuals of the same sex than individuals of the opposite sex, causing SSS and females to be unfertilized. Sperm limitation coupled with higher pre-fertilization cost in males relative to females has been hypothesized to contribute to female-biased sex ratios among bryophytes (McLetchie, 1992; Stark et al., 2000).
We propose that the high number of female genets in small habitat patches may be the result of a high establishment rate by either spores (sexual propagule) or gemmae (asexual propagule). Females and males are likely to have similar colonization rates via sexual propagules due to the 1:1 spore sex ratio. However, because there is low gemma production in females relative to males (McLetchie and Puterbaugh, 2000), females are less likely to colonize via gemmae than males. Thus the presence of a female-biased sex ratio in populations on small habitat patches might better reflect higher levels of establishment and persistence of females rather than colonization rates. Relative to males, higher female survival can occur as a result of higher dehydration tolerance in females at the gemma (Stieha et al., 2014) and thallus (Marks et al., 2016) stages. Higher dehydration tolerance in females coupled with more variability in moisture conditions in small vs. large habitat patches that might occur in the drier periods of the year would favour females over males. However, variation in moisture of different habitat sizes has not yet been documented.
Due to its self-perpetuating cycle of mate reduction through clonal growth, asexual reproduction often becomes the dominant form of reproduction in mixed reproductive mode (sexual and asexual) species (i.e. Brzyski and Culley, 2011), thereby having a potentially large influence on both within- and among-patch dynamics. In dioecious species, spatial segregation of the sexes may greatly increase the distance to the nearest potential mate because even though a neighbouring individual may be a different genotype, it may be the same sex. Therefore, it is imperative to identify both the sex and the genotype of individuals. In doing so, we were able to show that SSS occurs both within populations, which depends on sex and habitat patch size, and among populations because some populations are of only one sex, and the sex ratio pattern we observed is consistent with sex differences in physiology. Lastly, the sex ratio pattern corroborates the intuitive prediction of a positive relationship between ramet and genet sex ratio inferred for clonal seed plants (Eppley et al., 1998; Vandepitte et al., 2009; Petzold et al., 2013; and see McLetchie and García-Ramos, 2017 for the explicit model of this relationship in clonal dioecious plants that includes a non-linear component). Quantifying the distribution of the sexes and the distribution of genotypes both within and between populations provides a more complete picture of individual and population dynamics.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. File 1: a schematic diagram representing a map of population pairs and their position relative to each other and the stream bank. File 2: Methods and Results of computing the genetic distance between individuals within a patch. Table S1: likelihood ratio test analysis. Figure S1: effect of spatial distance on genetic distance between two individuals.
ACKNOWLEDGEMENTS
The authors thank the Wildlife Section of the Forestry Division of the Republic of Trinidad and Tobago for collection and export permits, and the Water and Sewage Authority for access to Quare River. We also thank the Department of Plant and Soil Sciences at the University of Kentucky for greenhouse space. Funding was provided by a Research Postdoctoral Fellowship from the University of Kentucky awarded to J.R.B.
LITERATURE CITED
- Alberto F, Gouveia L, Arnaud-Haond S, Pérez-Lloréns JL, Duarte CM, Serrao EA. 2005. Within-population spatial genetic structure, neighbourhood size and clonal subrange in the seagrass Cymodocea nodosa. Molecular Ecology 14: 2669–2681. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Migliaccio M, Diaz-Almela E, et al. . 2007. a Vicariance patterns in the Mediterranean Sea: east–west cleavage and low dispersal in the endemic seagrass Posidonia oceanica. Journal of Biogeography 34: 963–976. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. 2007b Standardizing methods to address clonality in population studies. Molecular Ecology 16: 5115–5139. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. 2015. Influences of clonality in plant sexual reproduction. Proceedings of the National Academy of Sciences, USA 112: 8859–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bączkiewicz A. 2013. Genetic diversity of leafy liverwort species (Jungermanniidae, Marchantiophyta) in Poland: diversity of leafy liverwort species with various reproductive modes. Biodiversity: Research and Conservation 27: 3–54. [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: linear mixed-effects models using Eigen and S4. R package version 1.1–7 http://CRAN.R-project.org/package=lme4.
- Biere A, Van Andel J, Van de Koppel J. 2012. Populations: ecology and genetics. In: van Andel J, Aronson J, eds. Restoration ecology: the new frontier, 2nd edn. Chichester, UK: Blackwell Publishing, 73–86. [Google Scholar]
- Bischler H. 1984. Marchantia L. The New World species. Bryophytorum Bibliotheca 26: 1–228. [Google Scholar]
- Bischler H. 1986. Marchantia polymorpha L.s. lat. karyotype analysis. Journal of the Hattori Botanical Laboratory 60: 105–117. [Google Scholar]
- Brodie HJ. 1951. The splash-cup mechanism in plants. Canadian Journal of Botany 29: 224–234. [Google Scholar]
- Brzyski JR, Culley TM. 2011. Genetic variation and clonal structure of the rare shrub Spiraea virginiana (Rosaceae). Conservation Genetics 12: 1323–1332. [Google Scholar]
- Brzyski JR, Adams KJ, Walter CM, Gale KH, McLetchie DN. 2012. Characterization of 12 polymorphic microsatellite markers in the liverwort Marchantia inflexa (Marchantiaceae). American Journal of Botany Primer Notes and Protocols 99: e440–e442. [DOI] [PubMed] [Google Scholar]
- Chung JM, Lee BC, Kim JS, Park CW, Chung MY, Chung MG. 2006. Fine-scale genetic structure among genetic individuals of the clone-forming monotypic genus Echinosophora koreensis (Fabaceae). Annals of Botany 98: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley MJ. 2007. The R book. Chichester, UK: John Wiley and Sons. [Google Scholar]
- Crowley PH, Stieha CR, McLetchie DN. 2005. Overgrowth competition, fragmentation and sex-ratio dynamics: a spatially explicit, sub-individual-based model. Journal of Theoretical Biology 233: 25–42. [DOI] [PubMed] [Google Scholar]
- Dering M, Rączka G, Szmyt J. 2016. Sex-specific pattern of spatial genetic structure in dioecious and clonal tree species, Populus alba L. Tree Genetics and Genomes 12: 70. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Eckert CG. 2000. Contributions of autogamy and geitonogamy to self-fertilization in a mass-flowering, clonal plant. Ecology 81: 532–542. [Google Scholar]
- Ellstrand NC, Roose ML. 1987. Patterns of genotypic diversity in clonal plant species. American Journal of Botany 74: 123–131. [Google Scholar]
- Eppley SM. 2005. Spatial segregation of the sexes and nutrients affect reproductive success in a dioecious wind-pollinated grass. Plant Ecology 181: 179–190. [Google Scholar]
- Eppley SM, Stanton ML, Grosberg RK. 1998. Intrapopulation sex ratio variation in the salt grass Distichlis spicata. American Naturalist 152: 659–670. [DOI] [PubMed] [Google Scholar]
- Eriksson O. 1997. Clonal life histories and the evolution of seed recruitment. In: de Kroon H, van Groenendael J, eds. The ecology and evolution of clonal plants. Leiden, The Netherlands: Backhuys Publishers, 211–226. [Google Scholar]
- Field DL, Pickup M, Barrett SCH. 2013. Comparative analyses of sex-ratio variation in dioecious flowering plants. Evolution 67: 661–672. [DOI] [PubMed] [Google Scholar]
- Frankham R. 1995. Conservation genetics. Annual Review of Genetics 29: 305–327. [DOI] [PubMed] [Google Scholar]
- Fuselier L, McLetchie DN. 2002. Maintenance of sexually dimorphic pre-adult traits in Marchantia inflexa (Marchantiacae). American Journal of Botany 89: 592–601. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramos G, Stieha C, McLetchie DN, Crowley PH. 2007. Persistence of the sexes in metapopulations under intense asymmetric competition. Journal of Ecology 95: 937–950. [Google Scholar]
- Geber MA. 1999. Theories of the evolution of sexual dimorphism. In: Geber MA, Dawson TE, Delph LF, eds. Gender and sexual dimorphisms in flowering plants. Berlin: Springer, 97–122. [Google Scholar]
- Hu A, Gale SW, Kumar P, Saunders RMK, Sun M, Fischer GA. 2017. Preponderance of clonality triggers loss of sex in Bulbophyllum bicolor, an obligately outcrossing epiphytic orchid. Molecular Ecology 26: 3358–3372. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Nilsson C. 1993. Hydrochory, population dynamics and distribution of the clonal aquatic plant Ranunculus lingua. Journal of Ecology 81: 81–91. [Google Scholar]
- Johnson DM. 2005. Metapopulation models: an empirical test of model assumptions and evaluation methods. Ecology 86: 3088–3098. [Google Scholar]
- Kareiva P. 1985. Finding and losing host plants by Phyllotreta: patch size and surrounding habitat. Ecology 66: 1809–1816. [Google Scholar]
- Kettenring KM, McCormick MK, Baron HM, Whigham DF. 2010. Phragmites australis (Common reed) invasion in the Rhode River subestuary of the Chesapeake Bay: disentangling the effects of foliar nutrients, genetic diversity, patch size, and seed viability. Estuaries and Coasts 33: 118–126. [Google Scholar]
- Korpelainen H, Pohjamo M, Laaka-Lindberg S. 2005. How efficiently does bryophyte dispersal lead to gene flow?Journal of the Hattori Botanical Laboratory 97: 195–205. [Google Scholar]
- Korpelainen H, von Cräutlein M, Kostamo K, Virtanen V. 2013. Spatial genetic structure of aquatic bryophytes in a connected lake system. Plant Biology 15: 514–521. [DOI] [PubMed] [Google Scholar]
- Kostrakiewicz-Gierałt K. 2013. The impact of disturbance gradient on recruitment of clonal plant species in Molinietum caeruleae meadows. Polish Journal of Ecology 61: 519–533. [Google Scholar]
- Laaka-Lindberg S, Korpelainen H, Pohjamo M. 2003. Dispersal of asexual propagules in bryophytes. Journal of the Hattori Botanical Laboratory 93: 319–330. [Google Scholar]
- Lande R. 1988. Genetics and demography in biological conservation. Science 241: 1455–1460. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D. 2001. Genetic data analysis: computer program for analysis of allelic data, version 1.1 http://www.eeb.uconn.edu/people/plewis/software.php.
- Marks RA, Burton JF, McLetchie DN. 2016. Sex differences and plasticity in dehydration tolerance: insight from a tropical liverwort. Annals of Botany 118: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLetchie DN. 1992. Sex ratio from germination through maturity and its reproductive consequences in the liverwort Sphaerocarpos texanus.Oecologia 92: 273–278. [DOI] [PubMed] [Google Scholar]
- McLetchie DN. 1996. Sperm limitation and genetic effects on fecundity in the dioecious liverwort Sphaerocarpos texanus. Sexual Plant Reproduction 9: 87–92. [Google Scholar]
- McLetchie DN, García-Ramos G. 2017. A predictive relationship between population and genetic sex ratios in clonal species. Acta Oecologia 80: 18–23. [Google Scholar]
- McLetchie DN, Puterbaugh MN. 2000. Population sex ratios, sex-specific clonal traits and trade-offs among these traits in the liverwort Marchantia inflexa. Oikos 90: 227–237. [Google Scholar]
- McLetchie DN, García-Ramos G, Crowley PH. 2002. Local sex-ratio dynamics: a model for the dioecious liverwort Marchantia inflexa. Evolutionary Ecology 15: 231–254. [Google Scholar]
- Melampy MN. 1981. Sex-linked niche differentiation in two species of Thalictrum. American Midland Naturalist 106: 325–334. [Google Scholar]
- Mizuki I, Ishida K, Tani N, Tsumura Y. 2010. Fine-scale spatial structure of genets and sexes in the dioecious plant Dioscorea japonica, which disperses by both bulbils and seeds. Evolutionary Ecology 24: 1399–1415. [Google Scholar]
- Obeso JR. 2002. The costs of reproduction in plants. New Phytologist 155: 321–348. [DOI] [PubMed] [Google Scholar]
- Paasch AE, Mischler BD, Nosratinia S, Stark LR, Fisher K. 2015. Decoupling of sexual reproduction and genetic diversity in the female-biased Mojave desert moss Syntrichia caninervis (Pottiaceae). International Journal of Plant Science 176: 751–761. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold A, Pfeiffer T, Jansen F, Eusemann P, Schnittler M. 2013. Sex ratios and clonal growth in dioecious Populus euphratica Oliv., Xinjiang Prov., Western China. Trees 27: 729–744. [Google Scholar]
- Pohjamo M, Laaka-Lindberg S, Ovaskainen O, Korpelainen H. 2006. Dispersal potential of spores and asexual gemmae in an epixylic hepatic Anastrophyllum hellerianum. Evolutionary Ecology 20: 415–430. [Google Scholar]
- Raabová J, van Rossum F, Jacquemart AL, Raspé O. 2015. Population size affects genetic diversity and fine-scale spatial genetic structure in the clonal distylous herb Menyanthes trifoliata. Perspectives in Plant Ecology, Evolution and Systematics 17: 193–200. [Google Scholar]
- Ramaiya M, Johnson MG, Shaw B, et al. . 2010. Morphologically cryptic biological species within the liverwort Frullania asagrayana. American Journal of Botany 97: 1707–1718. [DOI] [PubMed] [Google Scholar]
- R Core Team 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reynolds DN. 1980. Gamete dispersal in Mnium ciliare. The Bryologist 83: 73–77. [Google Scholar]
- Santamaría L. 2002. Why are most aquatic plants widely distributed? Dispersal, clonal growth and small-scale heterogeneity in a stressful environment. Acta oecologica 23: 137–154. [Google Scholar]
- Sherwin WB. 2010. Entropy and information approaches to genetic diversity and its expression: genomic geography. Entropy 12: 1765–1798. [Google Scholar]
- Sherwin WB, Jabot F, Rush R, Rossetto M. 2006. Measurement of biological information with applications from genes to landscapes. Molecular Ecology 15: 2857–2869. [DOI] [PubMed] [Google Scholar]
- Starfinger U, Stöcklin J. 1996. Seed, pollen, and clonal dispersal and their role in structuring plant populations. Progress in Botany 57: 337–355. [Google Scholar]
- Stark LR, Mishler BD, McLetchie DN. 2000. The cost of realized sexual reproduction: assessing patterns of reproductive allocation and sporophyte abortion in a desert moss. American Journal of Botany 87: 1599–1608. [PubMed] [Google Scholar]
- Stieha C, García-Ramos G, McLetchie DN, Crowley P. 2016. Maintenance of the sexes and persistence of a clonal organism in spatially complex metapopulations. Evolutionary Ecology 31: 363–386. [Google Scholar]
- Stieha CR, Middleton AR, Stieha JK, Trott SH, McLetchie DN. 2014. The dispersal process of asexual propagules and the contribution to population persistence in Marchantia (Marchantiaceae). American Journal of Botany 101: 348–356. [DOI] [PubMed] [Google Scholar]
- Sun S, Gao X, Cai Y. 2001. Variations in sexual and asexual reproduction of Scirpus mariqueteer along an elevational gradient. Ecological Research 16: 263–274. [Google Scholar]
- Tackenberg O, Poschlod P, Bonn S. 2003. Assessment of wind dispersal potential in plant species. Ecological Monographs 73: 191–205. [Google Scholar]
- Thomas CD, Hanski I. 1997. Butterfly metapopulations. In: Hanski I, Gilpin ME, eds. Metapopulation biology. San Diego: Academic Press, 359–386. [Google Scholar]
- Vallejo-Marín M, Dorken ME, Barrett SCH. 2010. The ecological and evolutionary consequences of clonality for plant mating. Annual Review of Ecology, Evolution, and Systematics 41: 193–213. [Google Scholar]
- Vandepitte K, Roldán-Ruiz I, Leus L, Jacquemyn H, Honnay O. 2009. Canopy closure shapes clonal diversity and fine-scale genetic structure in the dioecious understory perennial Mercurialis perennis. Journal of Ecology 97: 404–414. [Google Scholar]
- Vandepitte K, Honnay O, De Meyer T, Jacquemyn H, Roldán-Ruiz I. 2010. Patterns of sex ratio variation and genetic diversity in the dioecious forest perennial Mercurialis perennis. Plant Ecology 206: 105–114. [Google Scholar]
- Vanderpoorten A, Devos N, Goffinet B, Hardy OJ, Shaw AJ. 2008. The barriers to oceanic island radiation in bryophytes: insights from the phylogeography of the moss Grimmia montana. Journal of Biogeography 35: 654–663. [Google Scholar]
- Wright S. 1951. The genetical structure of populations. Annals of Eugenics 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wyatt R. 1977. Spatial pattern and gamete dispersal distances in Atrichum angustatum. The Bryologist 80: 284–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.