Abstract
The circadian clock is an endogenous timekeeping network that integrates environmental signals with internal cues to coordinate diverse physiological processes. The circadian function depends on the precise regulation of rhythmic gene expression at the core of the oscillators. In addition to the well-characterized transcriptional feedback regulation of several clock components, additional regulatory mechanisms, such as alternative splicing, regulation of protein stability, and chromatin modifications are beginning to emerge. In this review, we discuss recent findings in the regulation of the circadian clock function in Arabidopsis thaliana. The involvement of chromatin modifications in the regulation of the core circadian clock genes is also discussed.
Keywords: circadian clock, oscillators, transcriptional and post-transcriptional regulation, chromatin modifications, Arabidopsis
1. Introduction
Plants, like animals, exhibit rhythmic biological activity, with a periodicity of 24 h. The rhythms are known as the circadian clock, which allows plants to coordinate the temporal organization of biological processes to specific times of the day or night [1]. The core components of the circadian system are oscillators, which are responsible for generating circadian rhythms. In addition to the central oscillators, the circadian system involves the input pathways that integrate oscillators’ functions with environmental timing stimuli, such as light and temperature, and the output pathways that provide a link between the oscillators and the diverse biological processes [2]. In the model plant Arabidopsis thaliana, more than 20 clock or clock-associated components are composed of interlocking transcription-translation feedback loops, which appears to be significantly more complicated than that of other model eukaryotes, such as Drosophila [3]. Different components of the circadian system act at distinct times throughout the day and night to reciprocally regulate expression of other clock genes at both the transcriptional and post-transcriptional levels (Figure 1):
Figure 1.
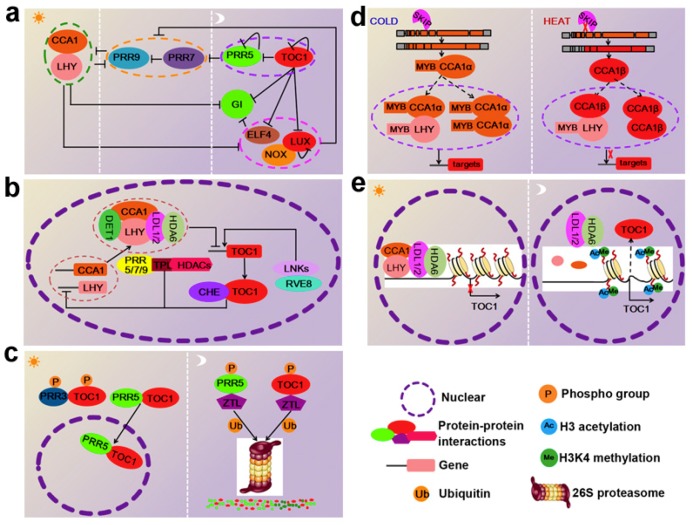
Multiple layers of regulation refine circadian clock activity in Arabidopsis thaliana. (a). Transcriptional feedback loops at the core of the circadian oscillator in Arabidopsis: At dawn, the expression of the pseudo-response regulator (PRR)-encoding genes; GIGANTEA (GI); timing of CAB expression 1 (TOC1); and the evening complex EC members LUX ARRHYTHMO (LUX), NOX, and early flowering 4 (ELF4) were repressed by circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY). PRR9, PRR7, PRR5, and TOC1 are sequentially expressed and repress the expression of CCA1 and LHY, as well as their own transcription. In the evening, all of the previously expressed components are repressed by TOC1. Subsequently, the EC maintains the repression of GI, PRR9 and PRR7. (b) Protein–protein interactions among clock components: The homo- or heterodimerization of CCA1 and LHY represses evening-phased genes by binding to an evening element in their promoters. To repress gene targets, circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY) require de-etiolated 1 (DET1), Histone deacetylase 6 (HDA6), and lysine-specific demethylase 1-like 1 and 2 (LDL1/2) as corepressors. Sequentially, the PRRs (PRR9, PRR7, and PRR5) bind to the CCA1 and LHY promoters and recruit TOPOLESS TPL and HDA6, thereby inhibiting the transcription of CCA1 and LHY. The interaction between TOC1 and CCA1 hiking expedition (CHE) helps TOC1 bind to the promoters of CCA1 and LHY. Additionally, night light-inducible and clock-regulated gene 1s (LNKs) interact with CCA1-like 5 (RVE8) and act as coactivators inducing the expression of TOC1. (c) Protein stability and turnover modulate the activity of oscillators: in the afternoon, the ZEITLUPE (ZTL)-mediated proteasomal degradation of TOC1 is interrupted by ZTL’s interaction with PRR3 (which hinders ZTL access) and PRR5 (which promotes TOC1 translocation to the nucleus). On the other hand, the phosphorylation of PRR5 and TOC1 enhances their binding to ZTL, which promotes their degradation later in the evening. (d) The alternative splicing regulates activity of CCA1: under high temperatures, the SNW/ski-interacting protein (SKIP) mediates CCA1 alternative splicing, leading to an aberrant spliced form, CCA1β, due to the fourth intron retention. CCA1β encodes a shorter protein lacking the DNA-binding Myb domain that is still able to homo/heterodimerize with the functional CCA1α and LHY. However, CCA1β/CCA1α and CCA1β/LHY dimers show reduced DNA binding activity to downstream targets. Under cold conditions, the accumulation of the correctly spliced variant CCA1α leads to increased CCA1 protein levels. Fully functional CCA1/CCA1 and CCA1/LHY dimers are thus able to bind to the promoters of targets. (e) Regulation of clock central oscillator TOC1 by chromatin modifications: in the morning, accumulated-CCA1/LHY, associated with the histone modification complex containing LDL1/2 and HDA6, attaches to the promoter of TOC1, thereby reducing the H3Ac and H3K4Me levels of TOC1. Consequently, TOC1 expression is low. In the evening, CCA1 and LHY expression are low, while TOC1 is highly expressed because the LDL1/2-HDA6 complex is released from the TOC1 promoter.
The orange star and light arch represent day and night, respectively. Ovals represent functional groups. Black bars indicate repression, and black arrows indicate the activation of transcription. Broken lines indicate relationships not proven to be direct or detected only under specific conditions
2. Transcriptional Regulation of the Circadian Clock Genes
In plants, the circadian oscillator comprises the morning and evening loops that have been proposed to form a negative feedback loop to generate circadian rhythms [4]. In the morning loop, two key single-MYB-domain-containing transcription factors, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) [5] and LATE ELONGATED HYPOCOTYL (LHY) [6], together with pseudo-response regulators 5, 7, and 9 (PRR5, PRR7, and PRR9) [7] interlock, and the core evening loop is composed of TIMING OF CAB EXPRESSION 1 (TOC1, also known as PRR1) [7,8]. Indeed, as two physically interacting and partially redundant proteins, the transcription and protein levels of CCA1 and LHY are highest in the morning [9,10]. CCA1 and LHY form a heterodimer in order to repress the expression of evening genes [11,12,13]. In addition, they also repress each other’s expression in an accurate timekeeping manner [5,6], since the single loss of function of either CCA1 or LHY causes a shortening of the period under free-running conditions, while the cca1 lhy double mutant is arrhythmic [14]. The first evening clock gene known to be repressed by CCA1 and LHY is TIMING OF CAB EXPRESSION 1 (TOC1, also known as PRR1), a member of the PRR family whose mutation shortens the period [8] by binding to a motif within the TOC1 promoter called the evening element (EE) [15]. Interestingly, the EE motif is always found in clock-regulated gene promoters [12], and also mediates cold responses [16]. The different signals converged at the EE possibly involve the binding of alternative transcription factor complexes, since CCA1 and LHY can form heterodimers [9,10], as well as the presence of protein–EE complexes, even in the absence of CCA1 and LHY [2]. In addition to TOC1, several other members of the PRR family, including PRR7 and PRR9, are also repressed by CCA1 and LHY [13,17]. Furthermore, several other evening genes, including GIGANTEA (GI), LUX ARRHYTHMO (LUX, an MYB-like transcription factor), and BROTHER OF LUX ARRHYTHMO (BOA, also known as NOX), as well as EARLY FLOWERING 3 (ELF3) and ELF4 (two unrelated novel nuclear proteins) are also repressed by CCA1 and LHY [18,19,20].
The expression of CCA1 and LHY is also repressed by TOC1, PRR7, and PRR9 [21,22,23]. After dawn, PRR9 is the first to be expressed, followed consecutively by PRR7 and PRR5 in the afternoon and finally by TOC1 in the evening [24]. Thus, this mechanism ensures that CCA1 and LHY are only expressed during a narrow window of time near dawn [25]. The repression of CCA1 caused by TOC1 involves the interaction between TOC1 and CCA1 HIKING EXPEDITION (CHE) in an as-yet undefined manner [26]. Furthermore, ELF4, ELF3, and LUX form an “evening complex” to repress the expression of the day-phased clock gene PRR9 [27,28,29,30]; mutation of any member of this complex causes plants to become arrhythmic [20]. Several genes homologous to CCA1 and LHY, such as REVEILLE8 (RVE8, also known as LHY and CCA1-like 5, or LCL5) are also repressed by PRRs [31,32]. However, unlike the repressors CCA1 and LHY, RVE8 promotes hundreds of evening genes that contain EEs in their promoters, such as PRR5, TOC1, GI, ELF4, and LUX [32,33]. Furthermore, RVE4 and RVE6, two close homologs of RVE8, also play partially redundant roles in the circadian system, since the rve4 rve6 rve8 triple mutant not only exhibits a more extreme long-period phenotype than the rve8 single mutant, but also loses the predominantly afternoon-phased EE binding activity [33].
3. Post-Transcriptional Regulation of the Circadian Clock Genes
In addition to transcriptional regulation, many post-transcriptional regulatory mechanisms, such as alternative splicing (AS) and regulation of protein stability, are key to plant circadian oscillators. Many clock genes, including CCA1, LHY, RVE8, TOC1, ELF3, and GI are regulated by alternative splicing [34,35,36]. The abundance of the different splice variants is regulated by temperature [34,37], photoperiod, salt stress, and light [36,38], supporting the idea that AS is an important mechanistic link between environmental signals and clock performance. Indeed, the splice variant of CCA1 with a retained fourth intron produces a protein called CCA1β, which lacks the MYB-like DNA-binding domain and is suppressed under cold conditions [37]. CCA1β can interrupt the function of full-length CCA1 (CCA1α) by competing with CCA1α and LHY in the formation of nonfunctional homo- and heterodimers [37]. In turn, an LHY splice variant with a premature stop codon accumulates at low temperatures [34], indicating a role for AS in maintaining an appropriate balance between CCA1 and LHY during cold acclimation. Moreover, spliceosome regulators that affect plant clock function have been reported. For example, protein arginine methyltransferease 5 (PRMT5), a conserved methyltransferase involved in the methylation of histones, regulates the alternative splicing of PRR9 [39,40], presumably via PRMT5-dependent methylation of splicing factors [41]. Another protein involved in mRNA splicing is SNW/Ski-interacting protein (SKIP), a component of the spliceosome. The SKIP also regulates the AS of PRR9 and other clock genes including PRR7, CCA1, LHY, and TOC1 [35], and loss of SKIP causes a long period phenotype [35]. Additionally, the mutation of SPLICEOSOMAL TIMEKEEPER LOCUS 1 (STIPL1), a homolog of a human spliceosomal protein involved in spliceosome disassembly, causes a long-period phenotype [42]. In stipl1 mutants, the accumulation of splicing variants of CCA1, LHY, TOC1, and PRR9 transcript is altered [42].
Protein–protein interaction networks are also critical for the regulation of circadian clock functions. Indeed, the repressive activity of CCA1 and LHY on evening genes depends on DE-ETIOLATED1 (DET1), a repressor of photomorphogenesis [43]. Similarly, PRR9, PRR7, and PRR5 physically interact with a transcriptional co-repressor, TOPLESS, and this interaction is required for the transcriptional repression of CCA1 and LHY [44]. The activity of this repressed complex seems to be required for TPL-dependent recruitment of histone deacetylase 6 (HDA6) [44]. A recent report demonstrated that lysine-specific demethylase 1 (LSD1)-like histone demethylases LDL1 and LDL2 form a histone modification complex with the histone deacetylase HDA6. The LDL1/LDL2–HDA6 histone modification complex is functionally associated with CCA1/LHY in the regulation of circadian clock genes [45]. Additionally, two clock-regulated and light-induced proteins, night light-inducible and clock-regulated 1 and 2 proteins (LNK1 and LNK2), can interact with CCA1, LHY, RVE4, and RVE8 [46,47]. In fact, the recruitment of LNK1 to the PRR5 and TOC1 promoters happens via interaction with RVE4 and RVE8, and the activation of PRR5 as well as TOC1 transcription by RVE8 requires LNK1 and LNK2 as transcriptional coactivators [46].
As a common mechanism for modulating protein stability, ubiquitination is also involved in the degradation of clock proteins. The F-box protein ZEITLUPE (ZTL) and its homologs—flavin binding, Kelch repeat, F-box (FKF1), and LOV KELCH protein 2 (LKP2)—have been characterized as circadian clock regulators in Arabidopsis. ZTL, with a blue light photosensor LOV (light, oxygen, and voltage) domain and a KELCH protein–protein interaction domain, targets TOC1 and PRR5 for ubiquitination and subsequent proteasome degradation [48,49]. Similarly, FKF1 and LKP2 also contribute to shaping PRR5 and TOC1 protein oscillations through direct interaction and degradation [48,50]. GI physically interacts with ZTL and stabilizes it, while ZTL reciprocally controls GI stability and nucleocytoplasmic partitioning [51]. Finally, the degradation of GI is promoted via ELF3-mediated interaction with the E3-ubiquitin ligase constitutive photomorphogenic 1 (COP1), thereby also triggering the ubiquitination and degradation of the substrate adaptor ELF3 [52].
Phosphorylation has also been demonstrated to affect clock protein stability and interactions with other proteins. Indeed, elevated phosphorylation of all PRR proteins, leading to the degradation of many PRRs, has been observed [53]. Phosphorylation of PRR5 and TOC1 enhances each protein’s interactions with ZTL, thereby promoting each one’s subsequent degradation [53]. In contrast, PRR5 interacting with TOC1 enhances PRR5’s accumulation in the nucleus, and prevents it from being targeted for degradation by ZTL, which is exclusively found in the cytoplasm [50].
4. Involvement of the Circadian Clock Regulation by Chromatin Modifications
In eukaryotic cells, genomic DNA is packaged with histones to form a complex structure known as chromatin. Thus, the chromatin structure is directly linked to the regulation of gene expression in response to developmental and environmental cues, through the modulation and accessibility of transcriptional regulatory proteins [54]. Multiple chemical and reversible modifications of DNA and histones, such as DNA methylation, histone acetylation, and histone methylation regulate chromatin activity and function [54,55]. To date, accumulating evidence reveals that changes in chromatin structure modulate circadian function (Table 1).
Table 1.
Regulation of the core circadian clock genes by chromatin modifications in Arabidopsis.
| Process | Histone Mark | Chromatin Modifier | Core Clock Component | References |
|---|---|---|---|---|
| DNA Methylation | Unknown | CCA1 | [56] | |
| Histone Acetylation | H3K9/H3K27ac | Unknown | CCA1, LHY, TOC1, PRR5, GI | [59] |
| H3K56ac | Unknown | LHY, PRR9, CCA1, | [66] | |
| PRR7, TOC1, LUX | ||||
| H3K9/14ac | Unknown | CCA1, LHY, TOC1 | [60] | |
| H3ac | Unknown | CCA1, LHY, TOC1 | [57,61] | |
| H3ac | HAF2 | PRR5, LUX | [62] | |
| Histone Deacetylation | H3K9ac/H4ac | HDA19 | LHY | [63] |
| H3ac | HDA6 | TOC1 | [45] | |
| Histone Methylation | H3K4Me3 | SDG2/ATXR3 | LHY, PRR9, CCA1, | [60,61,66] |
| PRR7, TOC1, LUX | ||||
| H3K36me2 | Unknown | LHY, CCA1, TOC1 | [61] | |
| Histone Demethylation | H3K4Me2 | LDL1, LDL2 | TOC1 | [45] |
| H3K9Me3 | JMJ30/JMJD5 | CCA1, LHY | [67,68,69] | |
| Histone Monoubiquitination | H2BUb | HUB1 | [71,72] | |
| Histone Phosphorylation | H3S28ph | Unknown | CCA1, LHY, TOC1, PRR5, GI | [59] |
| H2AS95ph | MLK4 | GI | [73] |
For example, the expression of CCA1 correlates with methylation levels of CHH (where H = A, T, or C) sites in the promoter region in heterosis [56]. In contrast, the expression of TOC1 is accompanied by the clock-controlled pattern of histone acetylation [57]. Indeed, at dawn, TOC1 expression is repressed by CCA1 via the binding of CCA1 to the TOC1 promoter, which depends on a repressive chromatin environment promoted by histone H3 deacetylation [57]. Recent studies indicated that HDA6 is involved in this process [45]. Unlike CCA1, which favors histone hypoacetylation, RVE8/LCL5 leads to H3 hyperacetylation at the TOC1 promoter, since overexpression of RVE8/LCL5 results in a rising phase of TOC1 expression, which in turn coincides with increased H3 acetylation [58].
In addition to TOC1, the regulation of other oscillators by oscillating histone marks is also investigated. For instance, histone acetylation (H3ac, H3K9/H3K27ac, H3K56ac, and H3K9/ 14ac) is closely correlated with the rhythmic expression of LHY, CCA1, and TOC1 [57,59,60,61], as well as PRR5, PRR9, PRR7, GI, and LUX [59,62]. Recent studies showed that the H3ac level of PRR5 and LUX loci is regulated by HAF2, a histone acetyltransferase belonging to the TAFII250 family [62]. Interestingly, a correlation of increased H3K9ac and H4ac levels with the up-regulation of LHY was observed in hda19 mutants [63]. Furthermore, the histone deacetylase HDA6 represses the expression of TOC1 by histone deacetylation [45]. These studies indicate that both HDA6 and HDA19 are involved in the regulation of circadian functions.
The molecular components responsible for the histone methylation at the core of the plant clock are just beginning to emerge. The H3K4me3 accumulation at the promoters of LHY, CCA1, and TOC1 is positively correlated with the rhythmic transcript levels of these genes, regulated by the histone methyltransferase SDG2/ATXR3 (set domain group 2/Arabidopsis trithorax-related) [60,64,65,66]. In contrast, the histone H3K36me2 levels and transcription levels of these genes show a negative correlation [61]. More recently, it was found that CCA1 and LHY recruit the histone modification complex containing LDL1/2 and HDA6 to the TOC1 locus to reduce H3ac and H3K4Me2 levels, resulting the repression of TOC1 [45]. The Jumonji C domain–containing histone demethylase, JMJ30/JMJD5, is also involved in circadian control. As a demethylase of H3K9me3 [67], JMJ30/JMJD5 is directly repressed by the core oscillators CCA1 and LHY [68]. In turn, JMJ30/JMJD5 also promotes expression of CCA1 and LHY, presumably through modulating the H3K9me3 levels at the promoter of these genes [67,68]. Interestingly, the Arabidopsis JMJD5/JMJ30 can rescue the circadian phenotypes of the mammalian cells that are deficient for the human JMJD5/JMJ30, suggesting a conserved function of JMJ30/JMJD5 in plants and animals [69].
Histone monoubiquitination and phosphorylation were also found to be involved in regulating circadian function. Histone monoubiquitination 1 (HUB1), an E3 ligase, controls H2B monoubiquitination (H2BUb) [70]. H2BUb is associated with H3K4me3 accumulation at several clock-controlling genes and alters their amplitude [71,72], indicating that this chromatin regulator participates in circadian rhythmicity. However, the H2BUb levels of oscillator genes have not been investigated. Moreover, chromatin immunoprecipitation-sequencing data shows that core oscillators like CCA1 and LHY have increased levels of histone H3 phosphorylation on serine 28 (H3S28ph) at the end of the night, while TOC1, PRR5, and GI have high levels of H3S28ph at the end of the day [59]. A recent report demonstrated that MUT9P-like-kinase (MLK4), which phosphorylates histone H2A on serine 95 (H2AS95ph), interacts with CCA1, allowing MLK4 to bind to the GI promoter [73]. Meanwhile, CCA1 also interacts with YAF9a, a co-subunit of the Swi2/Snf2-related ATPase (SWR1) and NuA4 complexes, which themselves are responsible for incorporating the histone variant H2A.Z into chromatin and possess histone H4 acetylase activity [74,75]. Thus, decreased H2AS95ph, along with the attenuated accumulation of H2A.Z and the acetylation of H4, leads to reduced GI expression [73].
Interestingly, a previous study showed that TOC1 circadian induction is accompanied by clock-controlled cycles of histone acetylation that favor transcriptionally permissive chromatin structures at the TOC1 locus [60]. Further data shows that H3 activating marks, such as H3K9/K14Ac and H3K4Me3, associating with the transcriptional start sites (TSSs) of CCA1/LHY and TOC1 are also circadian regulated [59]. These results seem to indicate that circadian-regulated histone activating marks lead to the circadian expression profiles of oscillators. In agreement with a recent study, the expression of some chromatin remodeling factor genes, such as HAG3, HDA2/6, SUVH3, and JMJ28 displayed circadian oscillation [76]. However, in the CCA1-overexpression plants, the H3K9/14Ac and H3K4Me3 levels associated with CCA1, LHY, and TOC1 TSSs were reduced [59], suggesting that the circadian oscillators may also affect histone modifications. Our recent results showed that CCA1/LHY recruits the LDL1/LDL2-HDA6 histone modification complex, reducing the H3Ac and H3K4Me levels in the promoter of TOC1 [45] and indicating that circadian oscillators are key factors in histone modifications regulation. Nevertheless, considering the complexity of the circadian regulatory network, further studies are required to deepen our mechanistic understanding of circadian regulation and histone modification.
5. Concluding Remarks
As an integral part of plant biology, the circadian system coordinates external stimuli and an internal timing mechanism, in order to optimize growth and development. However, despite emerging evidence on new components involved in alternative splicing, regulation of protein stability, and histone modifications responsible for regulation of the core clock components, many questions still persist over the mechanism of circadian regulation. The repressive mechanism of the core components CCA1/LHY and TOC1 is still poorly understood. CCA1/LHY have been recently shown to recruit a histone modification complex containing LDL1/2 and HDA6 to their target loci (such as TOC1) to repress gene expression by histone deacetylation and H3K4 demethylation [45]. These findings indicate a link between chromatin modifications and the repressive mechanisms of a plant circadian rhythm’s core components. Nevertheless, transcriptional regulation associated with chromatin modifications is only one level nestled within a multi-layered regulatory network. Post-transcriptional mechanisms, protein–protein interactions, and protein degradation all have their own essential roles in aiding rhythm generation [76,77,78,79]. It is only through the integration of all of these layers of activity that plants generate a comprehensive circadian network sustaining robust rhythms. Thus, further investigation into the cross-talk between multi-layered regulatory networks will help us to understand how plants thrive in an unpredictable and changing environment.
Abbreviations
| AS | Alternative splicing | BOA | Brother of LUX ARRHYTHMO |
| CCA1 | Circadian clock associated 1 | CHE | CCA1 hiking expedition |
| COP1 | E3-ubiquitin ligase, Constitutive photomorphogenic 1 | DET1 | De-etiolated 1 |
| EE | Evening element | ELF3 | Early flowering 3 |
| FKF1 | Flavin binding, KELCH repeat, F-BOX | GI | GIGANTEA |
| HDA6 | Histone deacetylase 6 | HUB1 | Histone monoubiquitination 1 |
| HAG3 | Histone acetyltransferase 8 | HDA3 | Histone deacetylase 3 |
| SUVH3 | SU(VAR)3-9 HOMOLOG 3 | JMJ28 | Jumonji C domain–containing histone demethylase 28 |
| JMJ3 | Jumonji C domain–containing histone demethylase 3 | LHY | Late elongated hypocotyl |
| LDL1 | LSD1-Like 1 | LDL2 | LSD1-Like 2 |
| LUX | LUX ARRHYTHMO | LSD1 | Lysine-Specific Demethylase 1 |
| LNK | Night light-inducible and clock-regulated gene 1 | LKP2 | LOV KELCH protein 2 |
| LOV | Light, Oxygen, and Voltage | MLK4 | MUT9P-like-kinase |
| PRR5 | Pseudo-response regulators 5 | PRMT5 | Protein arginine methyltransferase 5 |
| RVE8/LCL5 | CCA1-like 5 | SKIP | SNW/Ski-interacting protein |
| STIPL1 | Spliceosomal timekeeper locus 1 | SDG2/ATXR3 | Set domain group 1/Arabidopsis trithorax-related |
| SWR1 | Swi2/Snf2-related ATPase | TOC1 (PRR1) | Timing of CAB expression 1 |
| YAF9 | a co-subunit of the SWR1 | ZTL | ZEITLUPE |
Funding
This work was supported by the Guangdong Natural Science Funds for Distinguished Young Scholars (2016A030306047); the Youth Innovation Promotion Association, CAS (2017398), and the National Natural Science Foundation of China (No. 31771423). This work was also supported by the Ministry of Science and Technology of Taiwan (105-2311-B-002-012-MY3 and 107-2811-B-002-001-) and National Taiwan University (107L893101).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wijnen H., Young M.W. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 2.Harmer S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 3.Gould P.D., Ugarte N., Domijan M., Costa M., Foreman J., Macgregor D., Rose K., Griffiths J., Millar A.J., Finkenstadt B., et al. Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol. 2013;9:650. doi: 10.1038/msb.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClung C.R. The genetics of plant clocks. Adv. Genet. 2011;74:105–139. doi: 10.1016/B978-0-12-387690-4.00004-0. [DOI] [PubMed] [Google Scholar]
- 5.Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carre I.A., Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/S0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z.Y., Tobin E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/S0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 7.Matsushika A., Makino S., Kojima M., Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- 8.Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Mas P., Panda S., Kreps J.A., Kay S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 9.Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yakir E., Hilman D., Kron I., Hassidim M., Melamed-Book N., Green R.M. Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 2009;150:844–857. doi: 10.1104/pp.109.137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 12.Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2015;112:E4802–E4810. doi: 10.1073/pnas.1513609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis Circadian Clock. Plant Cell. 2016;28:696–711. doi: 10.1105/tpc.15.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alabadi D., Yanovsky M.J., Mas P., Harmer S.L., Kay S.A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 2002;12:757–761. doi: 10.1016/S0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- 15.Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen M.D., Thomashow M.F. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–339. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- 17.Adams S., Manfield I., Stockley P., Carre I.A. Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS ONE. 2015;10:e0143943. doi: 10.1371/journal.pone.0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai S.H., Wei X.P., Pei L.P., Thompson R.L., Liu Y., Heard J.E., Ruff T.G., Beachy R.N. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell. 2011;23:961–972. doi: 10.1105/tpc.111.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S.X., Webb C.J., Knowles S.M., Kim S.H.J., Wang Z.Y., Tobin E.M. CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagel D.H., Kay S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012;22:R648–R657. doi: 10.1016/j.cub.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W., Perez-Garcia P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 23.Pokhilko A., Fernandez A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farre E.M., Liu T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 2013;16:621–629. doi: 10.1016/j.pbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 26.Pruneda-Paz J.L., Breton G., Para A., Kay S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farre E.M., Kay S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrero E., Kolmos E., Bujdoso N., Yuan Y., Wang M.M., Berns M.C., Uhlworm H., Coupland G., Saini R., Jaskolski M., et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., Sakakibara H., Mizuno T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawat R., Takahashi N., Hsu P.Y., Jones M.A., Schwartz J., Salemi M.R., Phinney B.S., Harmer S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu P.Y., Devisetty U.K., Harmer S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife. 2013;2:e00473. doi: 10.7554/eLife.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James A.B., Syed N.H., Bordage S., Marshall J., Nimmo G.A., Jenkins G.I., Herzyk P., Brown J.W.S., Nimmo H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X.X., Wu F.M., Xie Q.G., Wang H.M., Wang Y., Yue Y.L., Gahura O., Ma S.S., Liu L., Cao Y., et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 2012;24:3278–3295. doi: 10.1105/tpc.112.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon Y.J., Park M.J., Kim S.G., Baldwin I.T., Park C.M. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 2014;14:136. doi: 10.1186/1471-2229-14-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo P.J., Park M.J., Lim M.H., Kim S.G., Lee M., Baldwin I.T., Park C.M. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–2442. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mancini E., Sanchez S.E., Romanowski A., Schlaen R.G., Sanchez-Lamas M., Cerdan P.D., Yanovsky M.J. Acute effects of light on alternative splicing in light-grown plants. Photochem. Photobiol. 2016;92:126–133. doi: 10.1111/php.12550. [DOI] [PubMed] [Google Scholar]
- 39.Hong S., Song H.R., Lutz K., Kerstetter R.A., Michael T.P., McClung C.R. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez S.E., Petrillo E., Beckwith E.J., Zhang X., Rugnone M.L., Hernando C.E., Cuevas J.C., Herz M.A.G., Depetris-Chauvin A., Simpson C.G., et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Santangelo S., Mancini E., Francey L.J., Schlaen R.G., Chernomoretz A., Hogenesch J.B., Yanovsky M.J. Role for LSM genes in the regulation of circadian rhythms. Proc. Natl. Acad. Sci. USA. 2014;111:15166–15171. doi: 10.1073/pnas.1409791111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones M.A., Williams B.A., McNicol J., Simpson C.G., Brown J.W.S., Harmer S.L. Mutation of Arabidopsis SPLICEOSOMAL TIMEKEEPER LOCUS1 causes circadian clock defects. Plant Cell. 2012;24:4066–4082. doi: 10.1105/tpc.112.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau O.S., Huang X., Charron J.B., Lee J.H., Li G., Deng X.W. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Kim J., Somers D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA. 2013;110:761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung F.Y., Chen F.F., Li C., Chen C., Lai Y.C., Chen J.H., Cui Y., Wu K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Q.G., Wang P., Liu X., Yuan L., Wang L.B., Zhang C.G., Li Y., Xing H.Y., Zhi L.Y., Yue Z.L., et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell. 2014;26:2843–2857. doi: 10.1105/tpc.114.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Garcia P., Ma Y., Yanovsky M.J., Mas P. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. USA. 2015;112:5249–5253. doi: 10.1073/pnas.1420792112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baudry A., Ito S., Song Y.H., Strait A.A., Kiba T., Lu S., Henriques R., Pruneda-Paz J.L., Chua N.H., Tobin E.M., et al. F-Box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mas P., Kim W.Y., Somers D.E., Kay S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 50.Wang L., Fujiwara S., Somers D.E. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. Embo J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J., Geng R.S., Gallenstein R.A., Somers D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development. 2013;140:4060–4069. doi: 10.1242/dev.096651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J.W., Rubio V., Lee N.Y., Bai S.L., Lee S.Y., Kim S.S., Liu L.J., Zhang Y.Y., Irigoyen M.L., Sullivan J.A., et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujiwara S., Wang L., Han L.Q., Suh S.S., Salome P.A., McClung C.R., Somers D.E. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 54.Pfluger J., Wagner D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007;10:645–652. doi: 10.1016/j.pbi.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibney E.R., Nolan C.M. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 56.Ng D.W.K., Miller M., Yu H.H., Huang T.Y., Kim E.D., Lu J., Xie Q.G., McClung C.R., Chen Z.J. A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell. 2014;26:2430–2440. doi: 10.1105/tpc.113.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perales M., Mas P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farinas B., Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 59.Baerenfaller K., Shu H., Hirsch-Hoffmann M., Futterer J., Opitz L., Rehrauer H., Hennig L., Gruissem W. Diurnal changes in the histone H3 signature H3K9ac|H3K27ac|H3S28p are associated with diurnal gene expression in Arabidopsis. Plant Cell Environ. 2016;39:2557–2569. doi: 10.1111/pce.12811. [DOI] [PubMed] [Google Scholar]
- 60.Hemmes H., Henriques R., Jang I.C., Kim S., Chua N.H. Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 2012;53:2016–2029. doi: 10.1093/pcp/pcs148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song H.R., Noh Y.S. Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells. 2012;34:279–287. doi: 10.1007/s10059-012-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee K., Seo P.J. The HAF2 protein shapes histone acetylation levels of PRR5 and LUX loci in Arabidopsis. Planta. 2018;248:513–518. doi: 10.1007/s00425-018-2921-y. [DOI] [PubMed] [Google Scholar]
- 63.Tian L., Fong M.P., Wang J.Y.J., Wei N.E., Jiang H.M., Doerge R.W., Chen Z.J. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics. 2005;169:337–345. doi: 10.1534/genetics.104.033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berr A., McCallum E.J., Menard R., Meyer D., Fuchs J., Dong A., Shen W.H. Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 2010;22:3232–3248. doi: 10.1105/tpc.110.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo L., Yu Y., Law J.A., Zhang X. SET DOMAIN GROUP2 is the major histone H3 lysine [corrected] 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2010;107:18557–18562. doi: 10.1073/pnas.1010478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malapeira J., Khaitova L.C., Mas P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA. 2012;109:21540–21545. doi: 10.1073/pnas.1217022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee K., Park O.S., Seo P.J. JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 2018;95:961–975. doi: 10.1111/tpj.14002. [DOI] [PubMed] [Google Scholar]
- 68.Lu S.X., Knowles S.M., Webb C.J., Celaya R.B., Cha C., Siu J.P., Tobin E.M. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu X.F., Jiang D.H., Wang Y.Q., Bachmair A., He Y.H. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 71.Bourbousse C., Ahmed I., Roudier F., Zabulon G., Blondet E., Balzergue S., Colot V., Bowler C., Barneche F. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 2012;8:e1002825. doi: 10.1371/journal.pgen.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Himanen K., Woloszynska M., Boccardi T.M., Groeve S., Nelissen H., Bruno L., Vuylsteke M., Lijsebettens M. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 2012;72:249–260. doi: 10.1111/j.1365-313X.2012.05071.x. [DOI] [PubMed] [Google Scholar]
- 73.Su Y.H., Wang S.L., Zhang F., Zheng H., Liu Y.N., Huang T.T., Ding Y. Phosphorylation of histone H2A at serine 95: A plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell. 2017;29:2197–2213. doi: 10.1105/tpc.17.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C.Y., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altaf M., Auger A., Monnet-Saksouk J., Brodeur J., Piquet S., Cramet M., Bouchard N., Lacoste N., Utley R.T., Gaudreau L., et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee H.G., Lee K., Jang K., Seo P.J. Circadian expression profiles of chromatin remodeling factor genes in Arabidopsis. J. Plant Res. 2015;128:187–199. doi: 10.1007/s10265-014-0665-8. [DOI] [PubMed] [Google Scholar]
- 77.Hsu P.Y., Harmer S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014;19:240–249. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seo P.J., Mas P. Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell. 2014;26:79–87. doi: 10.1105/tpc.113.119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nohales M.A., Kay S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016;23:1061–1069. doi: 10.1038/nsmb.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]