Abstract
Multidrug efflux pumps constitute a category of antibiotic resistance determinants that are a part of the core bacterial genomes. Given their conservation, it is conceivable that they present functions beyond the extrusion of antibiotics currently used for therapy. Pseudomonas aeruginosa stands as a relevant respiratory pathogen, with a high prevalence at hospitals and in cystic fibrosis patients. Part of its success relies on its low susceptibility to antibiotics and on the production of virulence factors, whose expression is regulated in several cases by quorum sensing (QS). We found that overexpression of the MexCD-OprJ multidrug efflux pump shuts down the P. aeruginosa QS response. Our results support that MexCD-OprJ extrudes kynurenine, a precursor of the alkyl-quinolone signal (AQS) molecules. Anthranilate and octanoate, also AQS precursors, do not seem to be extruded by MexCD-OprJ. Kynurenine extrusion is not sufficient to reduce the QS response in a mutant overexpressing this efflux pump. Impaired QS response is mainly due to the extrusion of 4-hydroxy-2-heptylquinoline (HHQ), the precursor of the Pseudomonas Quinolone Signal (PQS), leading to low PQS intracellular levels and reduced production of QS signal molecules. As the consequence, the expression of QS-regulated genes is impaired and the production of QS-regulated virulence factors strongly decreases in a MexCD-OprN P. aeruginosa overexpressing mutant. Previous work showed that MexEF-OprJ, another P. aeruginosa efflux pump, is also able of extruding kynurenine and HHQ. However, opposite to our findings, the QS defect in a MexEF-OprN overproducer is due to kynurenine extrusion. These results indicate that, although efflux pumps can share some substrates, the affinity for each of them can be different. Although the QS response is triggered by population density, information on additional elements able of modulating such response is still scarce. This is particularly important in the case of P. aeruginosa lung chronic infections, a situation in which QS-defective mutants are accumulated. If MexCD-OprJ overexpression alleviates the cost associated to triggering the QS response when un-needed, it could be possible that MexCD-OprJ antibiotic resistant overproducer strains might be selected even in the absence of antibiotic selective pressure, acting as antibiotic resistant cheaters in heterogeneous P. aeruginosa populations.
Keywords: Pseudomonas aeruginosa, quorum sensing, antibiotic resistance, multidrug efflux pump, MexCD-OprJ
Introduction
Pseudomonas aeruginosa is a free-living microorganism able to survive in different environments that not only plays an ecological role in natural ecosystems (Green et al., 1974; Romling et al., 1994; Morales et al., 2004; Walker et al., 2004), but it is also an important causative agent of infections in patients with underlying diseases (Obritsch et al., 2005; Driscoll et al., 2007; Martinez-Solano et al., 2008; Talwalkar and Murray, 2016). The characteristic low susceptibility to antibiotics of this organism relies on several factors (Fajardo et al., 2008). Particularly relevant is the activity of chromosomally-encoded multidrug resistance (MDR) efflux pumps (Vila and Martínez, 2008; Hernando-Amado et al., 2016). Further, the acquisition of mutation-driven resistance is common in this opportunistic pathogen, particularly along chronic infections (Oliver et al., 2015; Palmer and Whiteley, 2015; Lopez-Causape et al., 2018), where the constitutive overexpression of MDR efflux pumps is one of the biggest problems to eradicate these infections (Vila and Martínez, 2008; Vila et al., 2011; Hernando-Amado et al., 2016). Efflux pumps exhibit different functions, with physiological and ecological significances that go beyond their activity as antibiotic resistance elements (Piddock, 2006; Martinez et al., 2009; Alcalde-Rico et al., 2016; Hernando-Amado et al., 2016). In the case of P. aeruginosa, an opportunistic pathogen not fully adapted to human hosts (Alonso et al., 1999; Morales et al., 2004), these functions should be of relevance for the success of P. aeruginosa as a respiratory infectious pathogen.
Pseudomonas aeruginosa harbors several efflux systems that belong to different families (Stover et al., 2000). The most studied because of their clinical relevance are MexAB-OprM (Li et al., 1995), MexCD-OprJ (Poole et al., 1996; Kohler et al., 1997), MexEF-OprN (Kohler et al., 1997), and MexXY (Aires et al., 1999; Mine et al., 1999). They all belong to the Resistance-Nodulation-Division (RND) family of MDR systems (Hernando-Amado et al., 2016). Mutants that exhibit constitutive overexpression of each of these efflux pumps are selected upon treatment with antibiotics; the mutations are frequently located in the regulatory elements adjacent to the respective operon encoding for these MDR systems (Llanes et al., 2004; Sobel et al., 2005; Jeannot et al., 2008; Zaborskyte et al., 2017).
The unregulated overexpression of an efflux system not only contributes to antibiotic resistance but may also have pleiotropic effects in the bacterial physiology. We have recently reported that overexpression of RND systems in P. aeruginosa leads to an excessive internalization of protons that acidify the cytoplasm, which causes a biological cost in absence of oxygen or nitrate, since both are necessary to compensate for the intracellular H+ accumulation (Olivares et al., 2014; Olivares Pacheco et al., 2017). In addition to these non-specific effects, other effects might be due to the unregulated extrusion of intracellular compounds, some of which may be relevant for the ecological behavior of P. aeruginosa (Blanco et al., 2016). Indeed, different studies have shown that overexpression of MDR efflux pumps may challenge the P. aeruginosa quorum sensing (QS) response (Evans et al., 1998; Kohler et al., 2001; Linares et al., 2005; Olivares et al., 2012), which is in turn determinant for modulating several physiological processes, including bacterial pathogenicity, in response to population density (Williams et al., 2007).
In P. aeruginosa, the QS-signaling network consists of three main interconnected regulatory systems: Las, Rhl, and Pqs, which synthetize and respond to the autoinducers N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL), N-butanoyl-L-homoserine lactone (C4-HSL), and the 2-alkyl-4(1H)-quinolones (AQs) Pseudomonas Quinolone Signal (PQS, or its immediate precursor 2-heptyl-4-hydroxyquinoline, HHQ), respectively (Williams and Camara, 2009). These autoinducers are able to bind to their respective transcriptional regulators, namely LasR, RhlR and PqsR, thus controlling the expression of a large number of genes including those responsible for their own synthesis: lasI, rhlI, and pqsABCDE, respectively.
Some P. aeruginosa RND systems have been associated with QS. MexAB-OprM is induced by C4-HSL (Maseda et al., 2004) and has been proposed to extrude 3-oxo-C12-HSL and other 3-oxo-HSL related compounds (Evans et al., 1998; Pearson et al., 1999; Minagawa et al., 2012). MexEF-OprN is able to efflux HHQ (Lamarche and Déziel, 2011) and kynurenine (Olivares et al., 2012), both precursors of the PQS autoinducer signal (Farrow and Pesci, 2007; Palmer et al., 2013). In agreement with these findings, the antibiotic resistant mutants that overproduce MexAB-OprM or MexEF-OprN have been associated with a low production of QS-controlled virulence factors (Evans et al., 1998; Pearson et al., 1999; Sanchez et al., 2002; Olivares et al., 2012). Some studies have demonstrated that acquisition of antibiotic resistance due to constitutive overexpression of mexCD-oprJ correlates with a decrease in the production of several virulence factors, some of them controlled by QS (Sanchez et al., 2002; Linares et al., 2005; Jeannot et al., 2008; Stickland et al., 2010). However, the underlying reasons for this correlation remain to be elucidated. In this work, we analyzed in depth the production of each QS signal molecule (QSSM) and the expression levels of the genes controlled by these regulation systems in order to understand how overexpression of MexCD-OprJ could be affecting the P. aeruginosa QS response, and consequently the behavior of this bacterial pathogen in the infected patient.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Culture Conditions
The Escherichia coli and P. aeruginosa strains and the plasmids used in this work, are listed in the Table 1. The primers used are listed in the Table 2.
Table 1.
Bacterial strains and plasmids used in the present work.
| Bacterial strain/plasmids | Description | Reference/origin |
|---|---|---|
| Escherichia coli | ||
| One Shot OmniMaxTM 2 T1 | Host strain used for the maintenance of cloning plasmids: F′ proAB lacIq lacZΔM15 Tn10(TetR) Δ(ccdAB) mcrA, Δ(mrr,hsdRMS-mcrBC) ϕ80(lacZ)ΔM15 Δ(lacZYA-argF)U169 endA1 recA1 supE44 thi-1 gyrA96 relA1 tonA panD | Invitrogen |
| S17-1λ pir | Conjugative donor strain used for transferring plasmids to P. aeruginosa acceptor strains by conjugation assays: F− thi pro hsdR hsdM+ recA RP42-Tc::Mu-Km::Tn7 | Simon et al., 1986 |
| S17 miniCTX::PpqsA-lux | S17-1λ pir strain containing the miniCTX::PpqsA-lux plasmid | Fletcher et al., 2007a,b |
| JM109-pSB1142 (LasR-based Biosensor) | Biosensor strain used for detecting the QS signal, 3-oxo-C12-HSL, produced by P. aeruginosa strains | Winson et al., 1998 |
| JM109-pSB536 (RhlR-based Biosensor) | Biosensor strain used for detecting the QS signal, C4-HSL, produced by P. aeruginosa strains | Swift et al., 1997 |
| Pseudomonas aeruginosa | ||
| PAO1 | Wild-type PAO1-V clinic strain given from the lab of V. de Lorenzo | Linares et al., 2005 |
| PAO1 miniCTX::PpqsA-lux (PAO1_PpqsA) | PAO1-V strain with the reporter construction PpqsA-luxCDABE inserted in the specific attB site of the chromosome | Present work |
| JFL28 (nfxB∗) | Spontaneous resistant mutant obtained from PAO1-V strain, which overproduces the MexCD-OprJ efflux system by punctual inactivating mutation in nfxB gene | Linares et al., 2005 |
| JFL28 miniCTX::PpqsA-lux (nfxB∗_PpqsA) | JFL28 strain with the reporter construction PpqsA-luxCDABE inserted in the specific attB site of the chromosome | Present work |
| nfxB∗ΔmexD | JFL28 strain with an inactive MexCD-OprJ efflux system by partial deletion of the mexD gene | Present work |
| PAO1 CTX::PpqsA-lux::pqsA (PqsR-based Biosensor) | PAO1-ΔpqsA strain with the reporter construction PpqsA-luxCDABE inserted in the specific attB site of the chromosome. Used for detecting the AQs produced by other P. aeruginosa strains | Fletcher et al., 2007a,b |
| Plasmid | ||
| pGEM-T Easy | Commercial plasmid used for cloning optimization of PCR products (AmpR) | Promega |
| pGEM-T-ΔmexD | pGEM-T Easy vector with the flanking DNA sequences of a 2058 bp inner region of mexD gene (AmpR) | Present work |
| pEX18Ap | Plasmid with conjugative properties used for deleting genes in P. aeruginosa by homolog recombination. AmpR | Hoang et al., 1998 |
| pEX18Ap-ΔmexD | pEX18Ap vector with the flanking DNA sequences of a 2058 bp inner region of mexD gene used for deleting mexD gene in P. aeruginosa strains. AmpR | Present work |
| Mini-CTX-lux-PpqsA | Plasmid derived from mini-CTX-lux Becher and Schweizer (2000) in which the expression of the luxCDABE operon is under the transcriptional control of the pqsABCDE promoter region of P. aeruginosa. TcR | Fletcher et al., 2007a,b |
| pSB1142 | Plasmid carried by the LasR-Bioreporter strain necessary for detecting 3-oxo-C12-HSL. TcR | Winson et al., 1998 |
| pSB536 | Plasmid carried by the RhlR-Bioreporter strain necessary for detecting C4-HSL. AmpR | Swift et al., 1997 |
Table 2.
Collection of primers used in the present work.
| Name | Sequence | Description |
|---|---|---|
| HindIII_mexD_Fw mexD_int_Rev | 5′-CCCAAGCTTCGAGGTGCGCGCGCGGGTGGCCGGC-3′ | Amplification of the DNA flanking region “Up” for deleting mexD gene |
| 5′-GCGAGCCTGCAGCAGCGCTTATTCGGACATCGGλATCC-3′ | ||
| mexD_int_Fw HindIII_mexD_Rev | 5′-GGATTTTCCGATGTCCGAATAAGCGCTGCTGCAGGCTCGC-3′ | Amplification of the DNA flanking region “Down” for deleting mexD gene |
| 5″-CCCAAGCTTCAGACGλCAGATAGGTACGAACA-3′ | ||
| ΔmexD_check_Fw ΔmexD_check_Rev | 5′-GGTGAAGATCGTGCCGAAG-3′ | To check the deletion of the mexD gene |
| 5′-ATTGGTGAAGTCGTTGATCA-3′ | ||
| M13_Fw M13_Rev | 5′-CACGACGTTGTλACGAC-3′ | To check the insertion of cloning DNA fragment into pGEM-t Easy vector |
| 5′-GGATAACAATTTCACACAGG-3′ | ||
| rplU_Fwd rplU_Rev | 5′-CGCAGTGATTGTTACCGGTG-3′ | To check DNA contamination of RNA samples |
| 5′-AGGCCTGAATGCCGGTGATC-3′ | ||
| rpsL_Fwd rpsL_Rev | 5′-GCAAGCGCATGGTCGACAAGA-3′ | Real-time RT-PCR (Housekeeping) |
| 5′-CGCTGTGCTCTTGCAGGTTGTGA-3′ | ||
| lasA_Fwd lasA_Rev | 5′-ATGGACCAGATCCAGGTGAG-3′ | Real-time RT-PCR |
| 5′-CGTTGTCGTAGTTGCTGGTG-3′ | ||
| lasB_Fwd lasB_Rev | 5′-ATCGGCAAGTACACCTACGG-3′ | Real-time RT-PCR |
| 5′-ACCAGTCCCGGTACAGTTTG-3′ | ||
| rhlA_Fwd rhlA_Rev | 5′-CGAGGTCAATCACCTGGTCT-3′ | Real-time RT-PCR |
| 5′-GACGGTCTCGTTGAGCAGAT-3′ | ||
| rhlB_Fwd rhlB_Rev | 5′-GAGCGACGAACTGACCTACC-3′ | Real-time RT-PCR |
| 5′-GGGAATCCCGTACTTCTCGT-3′ | ||
| lecA_Fwd lecA_Rev | 5′-ATAACGAAGCAGGGCAGGTA-3′ | Real-time RT-PCR |
| 5′-TTGCCAATCTTCATGACCAG-3′ | ||
| phzB1_Fwd phzB1_Rev | 5′-AACGAACTTCGCGλAGAA-3′ | Real-time RT-PCR |
| 5′-TTTGTCTTTGCCACGAATGA-3′ | ||
| phzB2_Fwd phzB2_Rev | 5′-GCGAGACGGTGGTCAAGTAT-3′ | Real-time RT-PCR |
| 5′-AATCCGGGAAGCATTTCAG-3′ | ||
| phzS_Fwd phzS_Rev | 5′-CAAGTCGCTGGTGAACTGG-3′ | Real-time RT-PCR |
| 5′-CGGGTACTGCAGGATCAACT-3′ | ||
| mexG_Fwd mexG_Rev | 5′-GGCGAAGCTGTTCGACTATC-3′ | Real-time RT-PCR |
| 5′-AGAAGGTGTGGACGATGAGG-3′ | ||
| lasI_Fwd lasI_Rev | 5′-CTACAGCCTGCAGAACGACA-3′ | Real-time RT-PCR |
| 5′-ATCTGGGTCTTGGCATTGAG-3′ | ||
| rhlI_Fwd rhlI_Rev | 5′-CTCTCTGAATCGCTGGAAGG-3′ | Real-time RT-PCR |
| 5′-GACGTCCTTGAGCAGGTAGG-3′ | ||
| pqsA_Fwd pqsA_Rev | 5′-CAATACACCTCGGGTTCCAC-3′ | Real-time RT-PCR |
| 5′-TGAACCAGGGλGAACAGG-3′ | ||
| pqsD_Fwd pqsD_Rev | 5′-CATGTGATCTGCCATCAACC-3′ | Real-time RT-PCR |
| 5′-AGCCGTAGGTCAGGACCAG-3′ | ||
| pqsE_Fwd pqsE_Rev | 5′-GACATGGAGGCTTACCTGGA-3′ | Real-time RT-PCR |
| 5′-CTCAGTTCGTCGAGGGATTC-3′ | ||
| phnB_Fwd phnB_Rev | 5′-CACTCGCTGGTGGTCAGTC-3′ | Real-time RT-PCR |
| 5′-AGAGTAGAGCGTTCTCCAGCA-3′ | ||
| pqsH_Fwd pqsH_Rev | 5′-ATGTCTACGCGACCCTGAAG-3′ | Real-time RT-PCR |
| 5′-AACTCCTCGAGGTCGTTGTG-3′ |
Unless other conditions are specified, experiments were carried out at 37°C in 100 ml flasks containing 25 ml of LB broth (Lennox). The E. coli strains carrying plasmids with ampicillin (AmpR) or tetracycline (TcR) resistance genes were grown in LB medium with 100 μg/ml of ampicillin or 10 μg/ml of tetracycline, respectively. For determining the effect of different carbon sources on P. aeruginosa growth, overnight cultures were washed with M63 medium containing MgSO4 1 mM and diluted to an OD600 = 0.01 in clear bottom 96-well plates containing 150 μl/well of M63 with the corresponding carbon source at a final concentration of 10 mM. The growth of each strain was measured at 37°C using a multi-plate reader.
Whole Genome Sequence of the nfxB∗ Strain and Generation of a nfxB∗ΔmexD Mutant
The nfxB∗ mutant was fully sequenced at Parque Científico de Madrid using Illumina technology as described (Garcia-Leon et al., 2014). Two ≈1000 bp DNA regions adjacent to the fragment of mexD to be deleted were amplified by PCR using the primers listed in Table 2. The amplicons were purified and used together for a nested PCR reaction in which a recombinant 2058 bp DNA was generated and cloned into pGEM-t Easy (pGEM-T-ΔmexD). E. coli OmniMaxTM cells were transformed with this plasmid and the sequence of the construction was verified by Sanger sequencing. The fragment was excised using HindIII and subcloned into pEX18Ap. The resulting pEX18Ap-ΔmexD construction was incorporated into E. coli S17-1λ pir by transformation. Introduction of the deleted allele into P. aeruginosa nfxB∗ was performed by conjugation using S17-1λ pir (pEX18Ap-ΔmexD) as donor strain as described (Hoang et al., 1998). mexD deletion was confirmed by PCR using the primers described in Table 2.
Analysis of the Production of QS-Regulated Virulence Factors
The secretion of elastase and protease IV was measured after 20 h of incubation of the bacterial cultures in LB at 37°C following the methods described in Kessler and Safrin (2014). Rhamnolipids detection was carried out as described (Wittgens et al., 2011). Pyocyanin was determined as detailed (Essar et al., 1990). For the swarming motility assay, O/N cultures were washed with sterile 0.85% NaCl and diluted to an OD600 = 1.0. Five-microliters drops were poured on the center of Petri dishes containing 25 ml of a defined medium (0.5% casamino acids, 0.5% bacto agar, 0.5% glucose, 3.3 mM K2HPO4, and 3 mM MgSO4), which were incubated 16 h at 37°C.
RNA Extraction and Real-Time RT-PCR
RNA was obtained using the RNeasy mini kit (QIAGEN) as described (Olivares et al., 2012). After treatment with DNase (Olivares et al., 2012), the presence of DNA contamination was checked by PCR using rplU primers. Real-time RT-PCR was performed as described in Olivares et al. (2012) using the primers listed in Table 2. The experiments were carried out in triplicate. The 2−ΔΔCt method (Livak and Schmittgen, 2001) was used for quantifying the results, normalizing the results to the housekeeping gene, rpsL.
Thin Layer Chromatography (TLC) and Time Course Monitoring of QSSMs Accumulation
Bacterial O/N cultures were washed with fresh LB medium and diluted to an OD600 = 0.01 for subsequent growth. For TLC assays, the QSSMs extractions were carried out as described (Fletcher et al., 2007a). For time course assays, this protocol was optimized to simultaneous monitoring QSSMs accumulation and cell density. For each extraction time, 1.8 ml aliquots from cultures were centrifuged (7,000 × g, 10 min at 4°C). The supernatants were filtered through 0.22 μm pore size membrane and the cellular pellets were resuspended in 1.8 ml of methanol HPLC grade to extract the QSSMs. 900 μl of cell-free supernatants were used to extract the QSSMs by adding 600 μl of acidified ethyl acetate twice. The resulting acidified ethyl acetate extracts were dried and subsequently dissolved in 900 μl of methanol HPLC grade.
Alkyl-quinolone signal (AQs) were detected by TLC as described (Fletcher et al., 2007a) using the PAO1 CTX::PpqsA-lux biosensor strain. C4-HSL and 3-oxo-C12-HSL were analyzed using the JM109-pSB536 (RhlR- based biosensor) and JM109-pSB1142 (LasR-based biosensor) biosensor strains, respectively (Yates et al., 2002). The image processing software “ImageJ” was used for densitometry analysis of the light spots.
For time course accumulation assays, flat white 96-well plates with optical bottom were filled with a mix containing 5 μl of sample and 195 μl of a 1/100 dilution of the corresponding O/N biosensor cultures. The experiments were carried out on a multi-plate luminometer/spectrophotometer reader. The highest relative light units (RLU = luminescence/OD600 ratio) obtained for each biosensor strain and the OD600 in which the samples were taken from P. aeruginosa cultures were represented.
Analysis by HPLC-MS of Kynurenine and Anthranilate Accumulation in Cell-Free Supernatants
Bacterial strains were grown in M63 containing succinate (10 mM) and tryptophan (10 mM). After 24 h at 37°C, the supernatants were filtered through a 0.22 μm pore size membrane and lyophilized. 100 mg of each sample were resuspended in 2 ml of 3 mM ammonium acetate and dissolved in H2O/methanol (50/50). The amounts of anthranilate and kynurenine were determined by HPLC-MS at Laboratorio de Cromatografía-SIdI from the Universidad Autónoma de Madrid.
Insertion of the Reporter Construction, miniCTX::PpqsA-lux, in the Chromosome of P. aeruginosa and Analysis of pqsABCDE Expression
The insertion of the miniCTX::PpqsA-lux reporter into the chromosomes of the different P. aeruginosa strains was carried out by conjugation as described (Hoang et al., 1998) using E. coli S17-1λ pir containing miniCTX::PpqsA-lux (Fletcher et al., 2007b) as donor strain. The resulting P. aeruginosa reporter strains were inoculated in flat white 96-well plates with optical bottom containing 200 μL of LB with or without 4 mM anthranilate at an initial OD600 = 0.01. The growth (OD600) and the bioluminescence emitted by the PpqsA-luxCDABE construction was monitored using a multi-plate reader.
Statistical Analysis
At least three biological replicates were analyzed in each experiment. Statistical significance was evaluated by using a Student’s two-tailed test with a confidence interval of 95%. The differences were considered significant for P-values < 0.05 (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). The quantification of the areas under the curves was carried out using the GraphPad Prim software and the mean of each biological replicates were used to calculate statistical significance.
Results
Increased expression of efflux pumps due to mutations in their regulators can produce different changes in bacterial physiology. In most cases, the phenotypes observed in this kind of mutants are attributed to the activity of the overexpressed efflux pump. However, in other instances, the mutations in the local regulator itself might have effects on the bacterial physiology, which affect bacterial virulence and are independent of the activity of the efflux pump (Tian et al., 2009a,b). To address this possibility, we used a previously described mutant that overexpresses MexCD-OprJ (Linares et al., 2005). To discard the possibility that other mutations besides those in the mexCD-oprJ repressors might have been selected in this strain during its stay in the laboratory, the genome of the mutant was fully sequenced. Only the already described nfxB mutation (Linares et al., 2005) was found. From this mutant, an nfxB∗ΔmexD strain, which keeps the nfxB mutation in addition to a partial deletion of the mexD gene, was generated. By comparing nfxB∗ and nfxB∗ΔmexD strains, we were able to define more precisely which phenotypes depend on the activity of the efflux pump and which are solely due to the inactivation of the NfxB repressor, independently of the activity of the efflux pump.
Overexpression of MexCD-OprJ Results in a Decrease in the Production of QS-Controlled Virulence Factors in P. aeruginosa
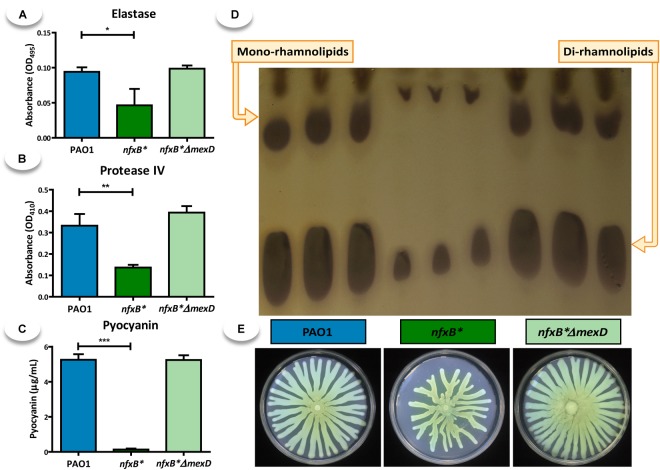
Swarming motility and the production of elastase, proteinase IV, pyocyanin, and, rhamnolipids were analyzed to establish whether or not MexCD-OprJ affects the production of P. aeruginosa QS-regulated virulence elements. As Figure 1 shows and in agreement with previous studies (Sanchez et al., 2002; Stickland et al., 2010), the nfxB∗ strain exhibits a decrease in swarming motility and in the production of all analyzed virulence factors in comparison with the wild-type PAO1 strain. The fact that the deletion of mexD fully restores the production of QS-regulated virulence factors in an nfxB∗ background, indicates that the observed impairment is solely due to the activity of MexCD-OprJ, independently of the potential activity of the NfxB regulator protein.
FIGURE 1.
Overexpression of the MexCD-OprJ efflux pump results in a decrease in the production of different virulence factors regulated by the QS system. The elastase (A), protease IV (B), pyocyanin (C), rhamnolipids, (D) and swarming (E) assays were conducted as described in Methods using cultures of the PAO1, nfxB∗ and nfxB∗ΔmexD strains. Statistical significances were evaluated by using a Student’s two-tailed test and considered significant if P < 0.05, with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). nfxB∗ presented a lower production of all tested virulence factors that the parental wild-type PAO1. The deletion of mexD in strain nfxB∗mexD restores the phenotypes to the levels of the wild-type strain, indicating that the defects in the expression of virulence factors were solely due to the activity of the mexCD-oprJ efflux pump.
Overproduction of the MexCD-OprJ Efflux System Results in a Lower Expression of QS- Regulated Genes
Expression of a set of QS-regulated genes (Pearson et al., 1997; Deziel et al., 2003, 2005; Dietrich et al., 2006; Rampioni et al., 2016) was analyzed to determine if a low production of virulence factors in the nfxB∗ mutant correlates with a deregulated expression of QS-regulated genes. LasB controls elastase production (Pearson et al., 1997; Kessler and Safrin, 2014). RhlA and RhlB are implicated in rhamnolipids biosynthesis (Pearson et al., 1997; Deziel et al., 2003), which in turn is important for swarming motility (Deziel et al., 2003). PhzB1, PhzB2, and PhzS are implicated in pyocyanin biosynthesis and the MexGHI-OpmD efflux pump has been described to be regulated by this phenazine (Dietrich et al., 2006). As shown in Figure 2A, the expression levels of the tested genes are lower in the nfxB∗ strain than in PAO1. In addition, the expression of these genes is restored to PAO1 levels, even overcoming them, upon mexD deletion in the nfxB∗ strain, further confirming that mexCD-oprJ overexpression is what causes an impaired QS response in the nfxB∗ mutant. These results are in agreement with the lower production of virulence factors observed in nfxB∗ (Figure 1).
FIGURE 2.
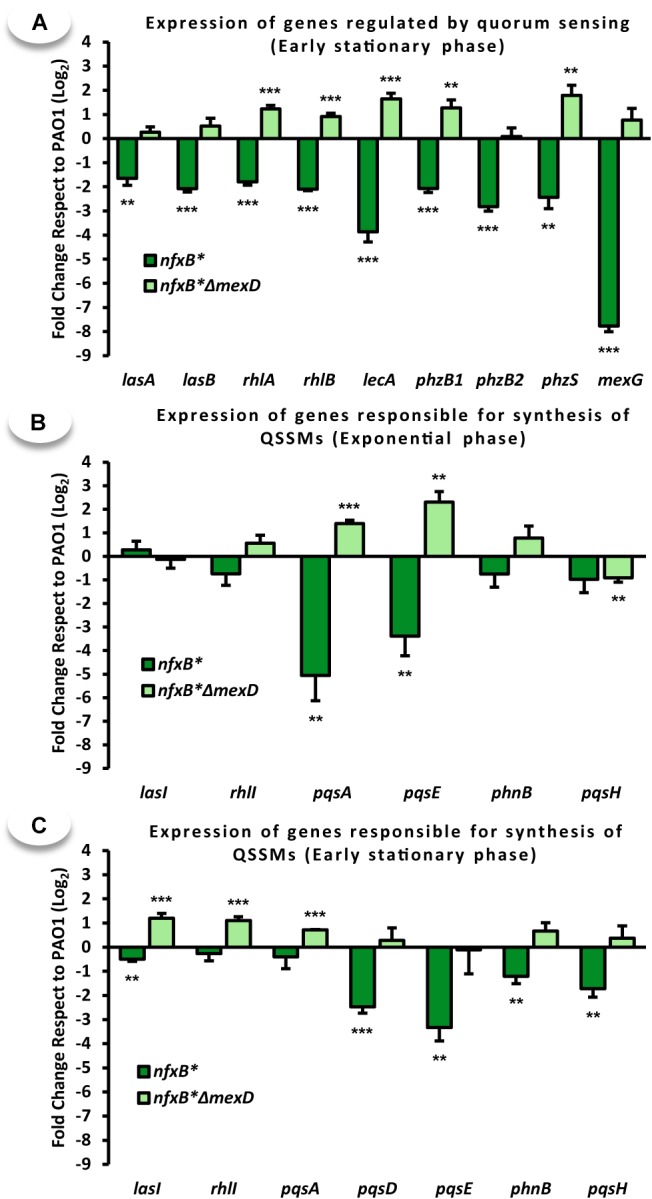
Overexpression of the MexCD-OprJ efflux system affects the expression levels of QS-regulated genes. Transcriptional analysis by real-time RT-PCR of (A) genes regulated by quorum sensing (QS) response (lasA, lasB, rhlA, rhlB, lecA, phzB1, phzB2, phzS, and mexG) and (B,C) genes responsible for QSSMs production (lasI, rhlI, pqsA, pqsD, pqsE, phnB, and pqsH) from samples obtained in (B) exponential (OD600 = 0.6) and (A,C) early stationary phase of growth (OD600 = 2.5) in PAO1, nfxB∗ and nfxB∗ΔmexD strains grown in LB medium. Statistical significances were evaluated by using a Student’s two-tailed test and considered significant if P < 0.05, with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). The results showed that the genes implicated in the synthesis of PQS pqsA and pqsE were expressed at lower level in the nfxB∗ strain than in the wild-type PAO1 strain at exponential phase of growth (B). In early stationary growth phase (A,C), the expression levels of the PQS-biosynthesis genes (pqsD, pqsE, phnB, and pqsH), as well as all of the analyzed QS-regulated genes, were significantly lower in the nfxB∗ strain than in the PAO1 wild-type. The deletion of mexD in strain nfxB∗mexD restores or even increase the levels of expression to those of the wild-type strain, indicating that these defects were solely due to the activity of the MexCD-OprJ efflux pump.
To gain more insights on the reasons for this impaired QS-response, we analyzed the expression of genes responsible for the production of both families of autoinducers AHLs (lasI and rhlI) (Pesci et al., 1997) and AQs (pqsABCDE-phnAB and pqsH) (Gallagher et al., 2002). This was performed along the exponential growth phase when expression of these QS biosynthesis genes starts, and in early stationary phase, when the Pqs-system is fully active (Lepine et al., 2003). As shown in Figures 2B,C, expression of the genes responsible for the synthesis of PQS and HHQ exhibit a marked decrease in the nfxB∗ strain at both time points. These changes were restored to wild-type levels upon MexCD-OprJ inactivation in an nfxB∗ background. pqsA, from the pqsABCDE operon responsible for the biosynthesis of AQs (Gallagher et al., 2002), exhibits the sharpest decrease in expression during exponential growth phase (Figure 2B). Expression of phnB, implicated in the synthesis of anthranilate through the chorismic acid pathway (Farrow and Pesci, 2007; Palmer et al., 2013), as well as pqsH, which codify the enzyme responsible for the conversion of HHQ into PQS (Gallagher et al., 2002), decreases more in early stationary phase (Figures 2B,C).
In contrast to the strong variations in expression of PQS-related genes, the activity of MexCD-OprJ had a minor impact on the expression of AHLs-related genes in both exponential and stationary growth phases. The nfxB∗ strain did not present alterations in rhlI expression, the gene responsible for the synthesis of C4-HSL, neither in exponential (Figure 2B) nor in stationary phase of growth (Figure 2C). A similar behavior was observed for lasI, the gene responsible for the synthesis of 3-oxo-C12-HSL, detecting just a slight decreased expression in the nfxB∗ strain during early stationary growth phase (Figures 2B,C).
MexCD-OprJ Overexpression Entails a Decrease in the Production and Accumulation of AQs Due to Their Extrusion Through This Efflux Pump
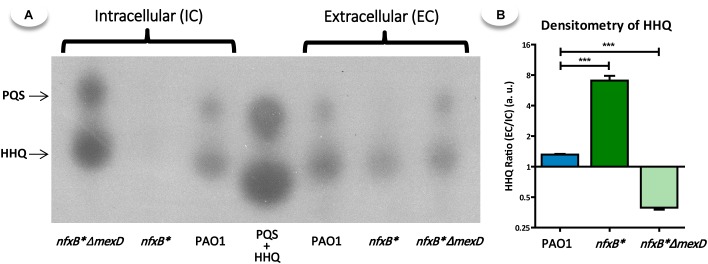
The production and accumulation of PQS and HHQ in both supernatant and cellular extracts decreased in the nfxB∗ mutant (Figure 3A). This effect is directly dependent on MexCD-OprJ activity, since PQS/HHQ accumulation increased in the nfxB∗ΔmexD strain, even overcoming the wild-type levels in cell extracts (Figure 3A). Interestingly, the proportion of HHQ present in the supernatants with respect to cell-extracts is different among the three strains. As Figure 3B shows, the nfxB∗ mutant has a higher supernatant/cell extract HHQ ratio than PAO1. Further, the deletion of mexD in the nfxB∗ strain produced the opposite effect, decreasing the HHQ ratio to lower values than those of the wild-type strain, suggesting that MexCD-OprJ may be extruding HHQ, affecting the progressive intracellular accumulation of this signal. Since the expression of the pqsABCDE-phnAB operon, responsible of AQs biosynthesis (Gallagher et al., 2002), is activated in presence of PQS/HHQ (Wade et al., 2005; Hazan et al., 2010; Rampioni et al., 2016), we postulate that HHQ extrusion by MexCD-OprJ could be the main cause for the lower production of AQs observed in the nfxB∗ strain, ultimately resulting in a defective QS-system.
FIGURE 3.
PQS and HHQ production is impaired in the strain that overproduces the MexCD-OprJ efflux pump. (A) To determine the accumulation levels of the autoinducers synthesized by Pseudomonas aeruginosa, a technique based on TLC coupled with a PqsR-based biosensor was used. The samples were extracted from cultures in early stationary phase (OD600 = 2.5). (B) The TLC-spots corresponding to HHQ were quantified by densitometry and the ratio between the HHQ present in the supernatant respect to cell extract was calculated and represented. Statistical significances were evaluated by using a Student’s two-tailed test and considered significant if P < 0.05, with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). As shown, overexpression of the MexCD-OprJ efflux system in nfxB∗ strongly reduces the production of PQS and HHQ as compared with PAO1 and nfxB∗ΔmexD strains. Furthermore, the analysis by densitometry of the HHQ ratio shows that this defect in AQs production is likely caused by an excessive extrusion of HHQ through MexCD-OprJ.
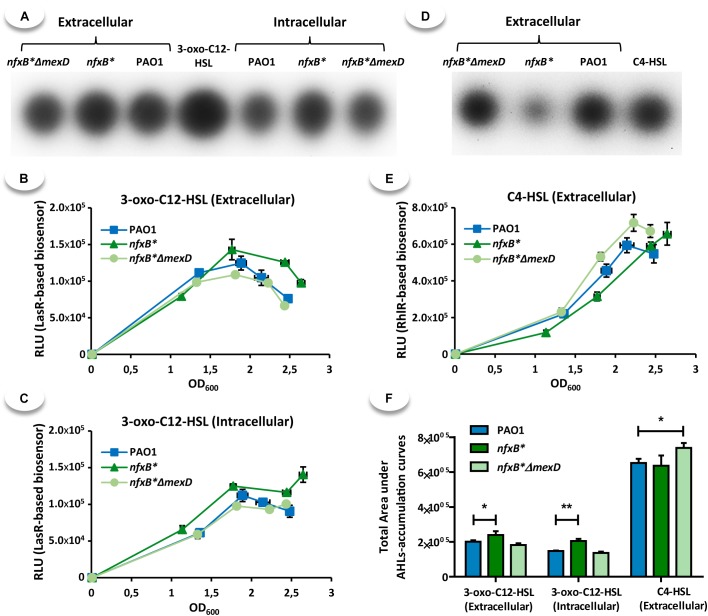
Overexpression of MexCD-OprJ Produces Minor Effects in the Synthesis of 3-oxo-C12-HSL and C4-HSL Autoinducers
Since the Las, Rhl, and Pqs regulation systems are highly interconnected (McKnight et al., 2000; Diggle et al., 2003; Kasper et al., 2016), we wanted to know whether or not the excessive HHQ extrusion through MexCD-OprJ in the nfxB∗ mutant could be also affecting the production of the QS signals, 3-oxo-C12-HSL (autoinducer signal for Las system) and C4-HSL (autoinducer signal for Rhl system). As shown in Figures 4A–C,F, both intracellular and extracellular amounts of 3-oxo-C12-HSL are slightly higher in nfxB∗ cultures than in either the wild-type PAO1 strain or the nfxB∗ΔmexD mutant. The opposite effect was observed for C4-HSL; the nfxB∗ mutant accumulates slightly lower extracellular levels of this QS signal during late exponential phase (Figure 4D), reaching the levels of extracellular accumulation observed in both PAO1 and nfxB∗ΔmexD in early stationary phase (Figures 4E,F). This variation may also exist inside the cell due to the ability of C4-HSL to freely diffuse through cytoplasmic membrane (Pearson et al., 1999). Altogether, these results indicate that overexpression of MexCD-OprJ leads to minor alterations of AHLs production. These changes might be due to the strongly impaired production of PQS and HHQ.
FIGURE 4.
The nfxB∗ mutant displays minor alterations in the kinetic of accumulation of 3-oxo-C12-HSL autoinducer. TLCs (A,D) and time course accumulation assays (B,C,E) were used to determine the accumulation of either 3-oxo-C12-HSL or C4-HSL autoinducer compounds. The samples for the TLC assays were extracted from cultures in late exponential phase (OD600 = 1.7) and the samples for the time course assay were taken at different time along the cell cycle (4, 5, 6, and 7 h post-inoculation). The area under the curve of each time course assay was calculated (F) and statistical significances were evaluated by using a Student’s two-tailed test with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). As shown, the overexpression of MexCD-OprJ has a slightly but significant effect on 3-oxo-C12-HSL accumulation. The nfxB∗ strain presented higher levels of 3-oxo-C12-HSL than PAO1 and nfxB∗ΔmexD both outside (A,B,F) and inside the cells (A,C,F). In contrast, the supernatant accumulation of C4-HSL in late exponential phase was lower in the MexCD-OprJ overexpressing mutant as compared with PAO1 and nfxB∗ΔmexD strains (D,E). However, the quantification of total area under the curves (F) only showed a significant increase in nfxB∗ΔmexD strain respect to both PAO1 and nfxB∗.
MexCD-OprJ Is Able to Extrude Kynurenine but Not Anthranilate, Both Precursors of AQs Signals
Our results indicate that the impaired QS response associated to the overexpression of the MexCD-OprJ efflux pump is mainly caused by a decreased production of PQS and HHQ, likely due to an excessive HHQ extrusion through this efflux system. The MexEF-OprN efflux pump is able to extrude both HHQ and its precursor kynurenine (Lamarche and Déziel, 2011; Olivares et al., 2012); extrusion of the latter is the main cause for the impaired QS response observed in MexEF-OprN overproducer strains (Olivares et al., 2012). A similar situation might also apply to MexCD-OprJ.
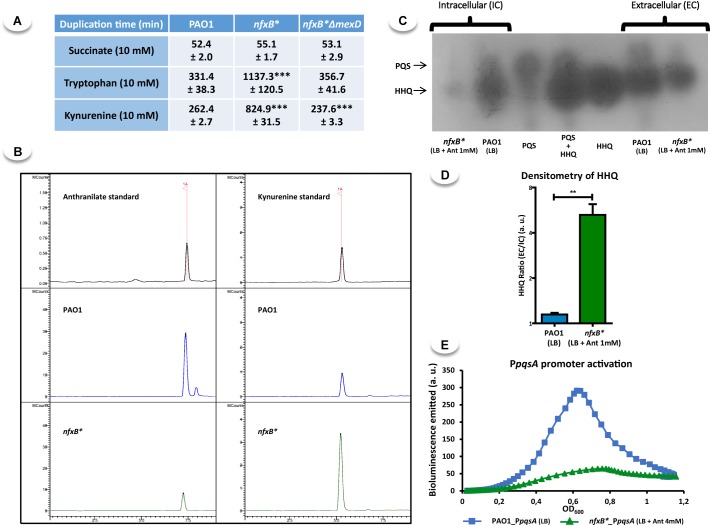
One of the immediate precursors of AQs in P. aeruginosa is anthranilate, which can < be synthetized either by PhnAB from chorismic acid or by kynurenine pathway when tryptophan is present in the medium (Deziel et al., 2004; Farrow and Pesci, 2007). Since the kynurenine pathway is the main source of anthranilate for AQs production when bacteria grow in rich LB medium (Farrow and Pesci, 2007), it could be possible that extrusion of some of the biosynthetic intermediates through MexCD-OprJ might affect the AQs production in nfxB∗. To test this hypothesis, we first analyzed the growth kinetic of the strains PAO1, nfxB∗, and nfxB∗ΔmexD in minimal medium containing tryptophan, kynurenine, or succinate as the sole carbon source. As shown in Figure 5A, the nfxB∗ mutant presents a growth defect in both tryptophan or kynurenine as the sole carbon source when compared to PAO1. These results strongly suggest extrusion of one or more intermediates of the kynurenine pathway through the MexCD-OprJ efflux system. As shown (Figure 5A) deletion of mexD in this mutant was enough to restore the wild-type growth rate, indicating that the observed growth defects, and the potential extrusion of these intermediates was solely due to the activity of MexCD-OprJ.
FIGURE 5.
Impaired intracellular accumulation of anthranilate produced by an excessive kynurenine extrusion through MexCD-OprJ is not the cause for lower AQs production of the nfxB∗ mutant. Statistical significances were evaluated by using a Student’s two-tailed test and considered significant if P < 0.05, with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). (A) The duplication time of PAO1, nfxB∗, and nfxB∗ΔmexD strains growing in minimal medium with succinate (control), tryptophan or kynurenine (both anthranilate precursors) as sole carbon sources was determined. As shown, the nfxB∗ mutant presents an impaired growth in both tryptophan and kynurenine, and deletion of mexD gene restored the growth rate in nfxB∗ mutant, suggesting these compounds might be substrates of MexCD-OprJ. (B) Anthranilate and kynurenine accumulation in cell-free supernatants was quantified by HPLC-MS from PAO1 and nfxB∗ cultures grown along 24 h in M63 minimal medium with succinate (10 mM) and tryptophan (10 mM) as sole carbon sources. Left panels anthranilate, right panels kynurenine. As shown, the supernatants from nfxB∗ cultures contained more kynurenine and less anthranilate than those from the wild-type PAO1 strain, indicating that kynurenine is a substrate of MexCD-OprJ and anthranilate is not extruded by this efflux pump. (C) The production of AQs in PAO1 and nfxB∗ strains growing in LB medium supplemented with anthranilate 1 mM was analyzed in early stationary phase (OD600 = 2.5) by TLC. (D) The extracellular vs. intracellular HHQ ratios were calculated measuring each one of the HHQ spots obtained in the TLC-assays by densitometry. (E) Real-time pqsABCDE expression was analyzed in both PAO1 and nfxB∗ strains growing in LB medium and LB supplemented with anthranilate 4 mM respectively, using a chromosomal insertion of the reporter construction PpqsA::luxCDABE. The results show that anthranilate supplementation of LB medium does not restore the AQs production in the nfxB∗ strain (C,E), reinforcing our hypothesis that HHQ extrusion (D) rather than kynurenine extrusion through MexCD-OprJ is the main cause for the QS-defective response of the nfxB∗ strain.
To analyze this possibility, we looked for the presence of kynurenine and anthranilate in the supernatants of PAO1 and nfxB∗ cultures. We observed a lower amount of anthranilate and a higher accumulation of kynurenine in the supernatants of nfxB∗ cultures (Figure 5B). Altogether, these results show that MexCD-OprJ is able to extrude kynurenine, but not anthranilate.
The Low Levels of PQS and HHQ Observed in the nfxB∗ Strain Is Not Just Due to Kynurenine Extrusion
Having established that the constitutive overexpression of MexCD-OprJ efflux pump leads to a decrease in the extracellular accumulation of anthranilate, we wondered whether a low intracellular availability of anthranilate could be the cause of the impaired PQS and HHQ production observed in the nfxB∗ strain. To address this possibility, we grew PAO1 and nfxB∗ in LB medium supplemented with 1 mM anthranilate and analyzed the production and accumulation of these two signals. As shown in Figure 5C, anthranilate supplementation does not restore PQS/HHQ production to wild-type levels in the nfxB∗ strain. In addition, our results indicate that the nfxB∗ strain continues to extrude HHQ at higher levels than those observed in the wild-type strain under these conditions (Figure 5D).
We entertained the possibility that a higher anthranilate concentration was needed to restore AQs production to wild-type levels in nfxB∗. To this end, we supplemented LB medium with up to 4 mM anthranilate and analyzed the activation of the pqsABCDE promoter in real-time in both PAO1 and nfxB∗. As shown in Figure 5E, a higher concentration of anthranilate did not restore the activation of the pqsABCDE promoter in the nfxB∗ strain. These results indicate that a low anthranilate concentration caused by kynurenine extrusion is not the main underlying cause for the impaired PQS and HHQ production observed in this strain. These results further support the notion that an excessive, non-physiological, extrusion of HHQ caused by the overexpression of MexCD-OprJ is likely the main cause for the lower accumulation and production of HHQ and PQS in the multidrug resistant nfxB∗ mutants.
The Low Production of AQs Associated to MexCD-OprJ Overexpression Is Not Caused by an Impaired Intracellular Accumulation of Octanoate
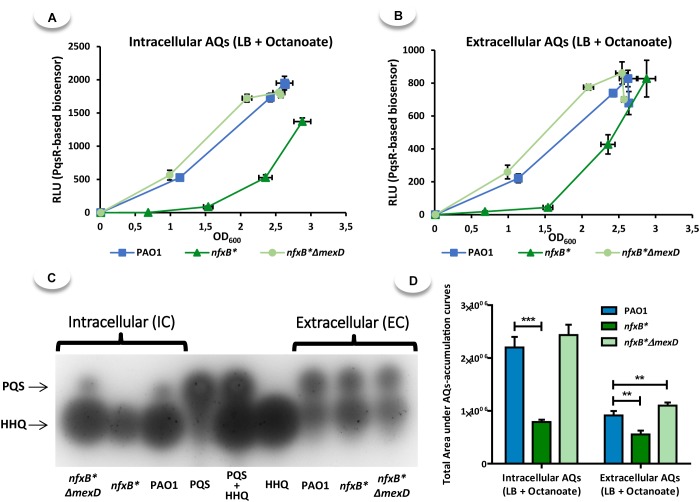
Octanoate is the other direct precursor of PQS and HHQ (Dulcey et al., 2013). Once we established that anthranilate synthesis is not the limiting step in the production of AQs by the nfxB∗ strain, we wondered whether a hypothetical low production or intracellular accumulation of octanoate might be affecting the AQs production in this strain. For that purpose, we measured the progressive accumulation of AQs in both cell-free supernatants and cellular extracts from PAO1, nfxB∗, and nfxB∗ΔmexD cultures grown in LB supplemented with 5 mM octanoate.
In agreement with previous findings (Dulcey et al., 2013), we found that the intracellular accumulation of AQs (Figure 6A) and pyocyanin production (Figure 7) increase when octanoate is added. However, these increases were similar in all strains, and both the pyocyanin production and the absolute AQs levels reached inside cells were still lower in nfxB∗ than in PAO1 or in nfxB∗ΔmexD. These results indicate that a lower availability of octanoate is not the cause of the impaired QS response displayed by the nfxB∗ strain.
FIGURE 6.
LB supplementation with octanoate increased the AQs production in PAO1, nfxB∗, and nfxB∗ΔmexD strains but, in the case of nfxB∗, but the accumulation levels remain being lower in nfxB∗ than observed in the other strains. To determine the time course production of AQs in PAO1, nfxB∗, and nfxB∗ΔmexD strains, we extracted these compounds from both the cells (A) and the cell-free supernatants (B) at different times along the cell cycle (4, 5, 6, and 7 h post-inoculation). Additionally, the last points of time course extractions were analyzed by TLC (C) in order to know the proportion of PQS and HHQ present on each AQs-extracts. The total area under each time course accumulation curve was quantified (D) and statistical significances were evaluated by using a Student’s two-tailed test and considered significant if P < 0.05, with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). The results show that supplementation of LB with 5 mM octanoate, even allowing nfxB∗ strain to accumulate levels of AQs out of the cells near to those in PAO1 and nfxB∗ΔmexD (B–D), was insufficient to restore the intracellular accumulation of PQS and HHQ (A,C,D). Furthermore, the fact that in TLC assay (C), the spot corresponding with HHQ present in nfxB∗ supernatant is slightly higher than that in PAO1 and nfxB∗ΔmexD, together with the evident low intracellular accumulation of HHQ in the nfxB∗ strain, confirm our hypothesis that MexCD-OprJ is able to extrude HHQ.
FIGURE 7.
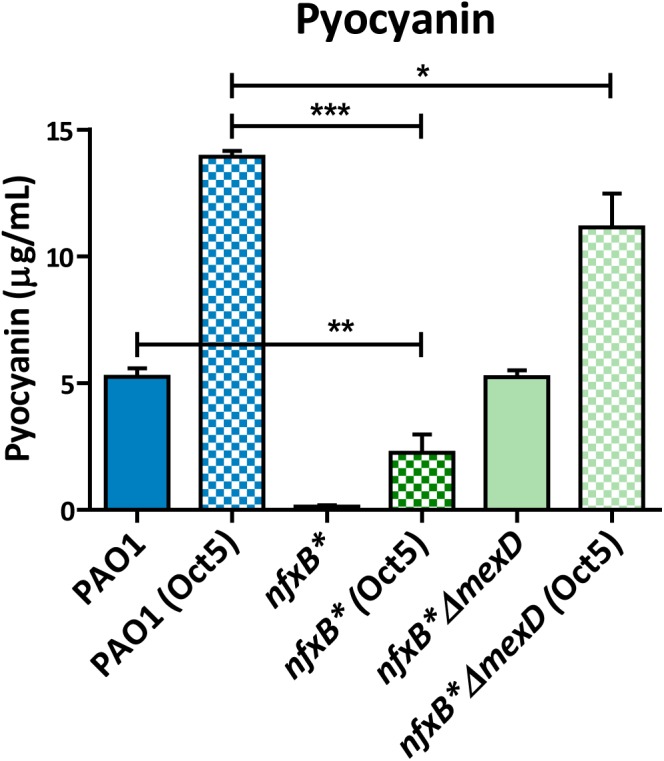
LB supplementation with octanoate increased the pyocyanin production in all analyzed strains, but nfxB∗ remained producing lower levels than those observed in PAO1 when grew in LB medium. For pyocyanin assay, the strains were grown in LB medium with or without octanoate (5 mM) along a time lapse of 20 h and the pyocyanin was extracted with chloroform-based protocol as is described in Methods. Statistical significances were evaluated by using a Student’s two-tailed test and considered significant if P < 0.05, with a confidence interval of 95% (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). The results show that supplementation of LB with 5 mM octanoate, even allowing nfxB∗ strain increase the pyocyanin production, was insufficient to reach the levels produced by PAO1 in LB without octanoate. This indicate that a hypothetical low intracellular accumulation of octanoate is not the main cause for defective pyocyanin production observed in nfxB∗ strain.
It is worth mentioning that, although the nfxB∗ supernatants exhibit a delay in AQs accumulation in the presence of 5 mM octanoate, the supernatants from all three strains exhibit similar levels when the cultures reach high cell densities (OD600 > 2.5) (Figure 6B). In contrast, the intracellular AQs accumulation remains lower in the nfxB∗ strain (Figure 6A). In the same way, the quantification of the areas under the curve for AQs-accumulation shows that the differences observed in these strains are lower in the supernatants when compared to cell extracts (Figure 6D). Further, the analysis by TLC of AQs extracted from the last point of the time course assay showed that, while the intracellular accumulation of PQS and HHQ remained being lower in the case of nfxB∗, the extracellular accumulation of these two AQs were similar among PAO1, nfxB∗ and nfxB∗ΔmexD cultures (Figure 6C). These results further reinforce the hypothesis that MexCD-OprJ is able to extrude HHQ (and likely PQS as well), and confirm that overexpression of this system adversely affects the intracellular accumulation of AQs. This extrusion can be considered the bottleneck that precludes a proficient PQS production as well as the onset of a proper QS response in nfxB∗-type mutants.
Discussion
In the current work, we demonstrate that a P. aeruginosa nfxB∗ mutant, which overexpresses the MexCD-OprJ efflux pump, exhibits an impaired QS response due the extrusion of HHQ. Specifically, this non-physiological extrusion leads to a decrease in the expression of the pqsABCDE operon responsible for AQs synthesis, which affects AQs-dependent and the PqsE-dependent regulons that comprise the genes involved in swarming motility, and in the production of pyocyanin, rhamnolipids, and proteases among others (Hazan et al., 2010; Rampioni et al., 2010, 2016).
The QS response in P. aeruginosa consists mainly on the Las, Rhl, and Pqs systems which are dependent on the 3-oxo-C12-HSL, C4-HSL, and PQS/HHQ autoinducers, respectively (Williams and Camara, 2009). The cross-regulation between these QS-systems is hierarchically understood, with the Las system located at the top, activating the other two QS systems, and followed by the Pqs-dependent activation of the Rhl-system, and by the Rhl-dependent repression of the Pqs-system (Dubern and Diggle, 2008; Williams and Camara, 2009). However, evidence exists that the hierarchy and the relationship between these QS-systems may be modulated depending on environmental conditions and the activity of global regulators as MvaT or RsmA among others (Choi et al., 2011; Mellbye and Schuster, 2014; Hammond et al., 2016; Maura et al., 2016; Welsh and Blackwell, 2016). In addition, recent studies have highlighted the relevant role of the feedback-regulation between Las, Rhl, and Pqs systems as well as the relevance of the PqsE and RhlR regulators (Deziel et al., 2005; Farrow et al., 2008; Dekimpe and Deziel, 2009; Hazan et al., 2010; Rampioni et al., 2010, 2016). Indeed, it has been demonstrated that the expression of approximately 90% of the genes in the AQs-regulon may be regulated through pqsE induction (Hazan et al., 2010). Likewise, the non-virulent phenotype prompted by the absence of AQs synthesis may be by-passed through pqsE induction, restoring the full P. aeruginosa virulence (Deziel et al., 2005; Hazan et al., 2010; Rampioni et al., 2010). Further, the regulation of several QS-dependent factors could be redundant. In such a way, the production of elastase, rhamnolipids, or pyocyanin, which are mainly under the control of Las, Rhl, and Pqs systems, respectively, are also regulated by PqsE independently of AQs production (Deziel et al., 2005; Hazan et al., 2010; Rampioni et al., 2010, 2016). In addition, expression of rhlR increases upon pqsE induction at the same time that some functions of PqsE as a QS-regulator are dependent on RhlR and C4-HSL production, thus establishing a complex feedback regulation loop (Hazan et al., 2010). Even more, exogenous addition of C4-HSL to the cultures may partially complement some of the phenotypes impaired in a pqsE mutant, such as pyocyanin production (Farrow et al., 2008; Hazan et al., 2010; Rampioni et al., 2010). Recent work indicates that PqsE is an alternative ligand synthase and PqsE and RhlR function as a QS-autoinducer synthase–receptor pair able of regulate the P. aeruginosa QS response independently of RhlI (Mukherjee et al., 2018).
Given this role of PqsE as one of the main elements in the QS regulatory network, we postulate that a reduced production of PQS and HHQ, together with a decreased pqsE expression, are the main causes for the lack of QS-response associated to the constitutive overexpression of the MexCD-OprJ efflux system. In this work, we show that expression of QS-regulated genes decreases in an nfxB∗ antibiotic resistant mutant and that inactivation of the MexCD-OprJ efflux pump in this background restores expression of these genes to wild-type levels, being even higher in some cases (Figure 2). Similar results were obtained with some QS-regulated phenotypes such as the production of elastase, protease IV, pyocyanin, rhamnolipids, and swarming motility (Figure 1), indicating that the alterations in the QS-response displayed by the nfxB∗ mutant are directly caused by the increased expression and activity of the MexCD-OprJ efflux system. We also demonstrated that loss of function of NfxB leads to an excessive extrusion of HHQ through the overexpressed MexCD-OprJ efflux pump, resulting in a low intracellular accumulation. Expression of pqsABCDE during exponential and early stationary growth phases is subjected to a positive feed-back transcriptional regulation under the control of the PqsR-(PQS/HHQ) complex (Rampioni et al., 2016). Therefore, the non-physiological HHQ extrusion through MexCD-OprJ may abrogate this positive feed-back regulation and directly cause the decrease in pqsABCDE-phnAB expression (Figure 2) and the AQs synthesis impairment (Figure 3) observed in the nfxB∗ mutant. We also showed that this defective AQs accumulation could not be restored by adding either anthranilate or octanoate, the two PQS/HHQ main precursors (Figures 5, 6), reinforcing the concept that the main cause for the defective QS-response associated to nfxB mutations is an excessive extrusion of HHQ through MexCD-OprJ, and not of metabolic precursors as kynurenine, also extruded by MexCD-OprJ. Additionally, the presence of similar levels of PQS in the supernatants of PAO1, nfxB∗ and nfxB∗ΔmexD growing in presence of octanoate, together with the absence of this autoinducer signal in the cell-extracts of nfxB∗ (Figure 6C) suggests that PQS could also be a MexCD-OprJ substrate.
To sum up, here we show that the AQs production is affected by the increased efflux of HHQ by the MexCD-OprJ RND system overexpressed in the nfxB∗ ciprofloxacin-resistant mutants. To note here that this type of antibiotic resistant mutants that are isolated in vivo from ciprofloxacin-treated patients (Jeannot et al., 2008). As a consequence, expression of the Pqs-regulon, which also comprises those PqsE-regulated genes in a PQS-independent way (Rampioni et al., 2010, 2016), is strongly altered in an nfxB∗ mutant. This alteration may have minor collateral effects on the AHLs-dependent QS systems and is likely the main cause for the low virulence profile observed in antibiotic resistant mutants overproducing MexCD-OprJ.
Previous work has shown that the P. aeruginosa MDR efflux pump MexEF-OprN is able of extruding kynurenine and HHQ as well (Lamarche and Déziel, 2011; Olivares et al., 2012). However, different to the situation with MexCD-OprJ, the reason for the impairment in the QS response of a MexEF-OprN overexpressing mutant was mainly the extrusion of kynurenine (Olivares et al., 2012), the HHQ precursor. Similarly, both efflux pumps can accommodate the same antibiotics, although the affinity for each of them can be different. Indeed, overexpression of whatever these two efflux systems increases ciprofloxacin and chloramphenicol resistance but at different levels, being MexCD-OprJ more efficient extruding ciprofloxacin and MexEF-OprN extruding chloramphenicol (Linares et al., 2005). It has been shown that, in addition to contributing to a coordinated response of the bacterial population, several QS signal molecules (Williams, 2007; Pacheco and Sperandio, 2009; Martinez, 2014) are also involved in inter-specific communication. For example, it has been shown that AQs may function as antimicrobial compounds against Staphylococcus aureus, a bacterial species commonly detected together P. aeruginosa in polymicrobial infections (Mashburn et al., 2005; Nguyen et al., 2015; Nguyen and Oglesby-Sherrouse, 2016). Further, HHQ also is able to induce apoptosis in human mesenchymal stem cells (Holban et al., 2014), and to impair the production of several factors implicated in the innate immune response affecting the binding of the nuclear factor-κβ to its targets (Kim et al., 2010). The fact that in this work we demonstrate that MexCD-OprJ is able to extrude HHQ, altering the accumulation level of the autoinducer signals produced by P. aeruginosa, opens a new perspective over the potential functions of this RND efflux system in the interactions between this opportunistic pathogen and other co-existing species (including the human host); a hypothesis that remain to be explored.
The activation of the QS-response implies an increase in expression of 100s of genes, and it has been estimated that this consumes approximately 10% of P. aeruginosa metabolic resources (Haas, 2006). Under this panorama, QS-defective mutants, which are unable to produce different exoproducts such as siderophores or proteases relevant for nutrients uptake, could be cheaters supported by neighbor bacteria able to produce these QS-dependent factors (Wilder et al., 2011; Pollak et al., 2016). The evolution of P. aeruginosa along the chronic infection of the lungs of cystic fibrosis patients involves a radiative evolution, with different morphotypes, including QS-deficient mutants, co-existing in the lung of each patient (LaFayette et al., 2015). It is conceivable that the mutants lacking a proficient QS response remain in the population because they can obtain the benefits brought about by an appropriate QS response carried out by co-existing bacteria without the cost associated with it. In agreement with this hypothesis, it might be possible that antibiotic resistance mediated by MexCD-OprJ overproduction allows nfxB∗ mutants to function as cheaters in the mixed populations colonizing the lung of the infected patient. This feature may have important implications concerning the persistence of antibiotic resistant mutants even in the absence of selection (Knoppel et al., 2017).
Author Contributions
MA-R performed the experimental work. JO-P and JM designed the study. MA-R, JO-P, MC, CA-O, and JM contributed to the interpretation of the results and in writing the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks are given to Diana Palenzuela for her help in the assays of quorum-sensing regulated phenotypes.
Footnotes
Funding. Work in JM laboratory was supported by grants from the Instituto de Salud Carlos III [Spanish Network for Research on Infectious Diseases (RD16/0016/0011)], from the Spanish Ministry of Economy and Competitivity (BIO2017-83128-R), and from the Autonomous Community of Madrid (B2017/BMD-3691).
References
- Aires J. R., Kohler T., Nikaido H., Plesiat P. (1999). Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43 2624–2628. 10.1128/AAC.43.11.2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalde-Rico M., Hernando-Amado S., Blanco P., Martinez J. L. (2016). Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 7:1483 10.3389/fmicb.2016.01483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Rojo F., Martinez J. L. (1999). Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1 421–430. 10.1046/j.1462-2920.1999.00052.x [DOI] [PubMed] [Google Scholar]
- Becher A., Schweizer H. P. (2000). Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29 948–950. 10.2144/00295bm04 [DOI] [PubMed] [Google Scholar]
- Blanco P., Hernando-Amado S., Reales-Calderon J. A., Corona F., Lira F., Alcalde-Rico M., et al. (2016). Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:E14. 10.3390/microorganisms4010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Park H. Y., Park S. J., Park S. J., Kim S. K., Ha C., et al. (2011). Growth phase-differential quorum sensing regulation of anthranilate metabolism in Pseudomonas aeruginosa. Mol. Cells 32 57–65. 10.1007/s10059-011-2322-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe V., Deziel E. (2009). Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155 712–723. 10.1099/mic.0.022764-0 [DOI] [PubMed] [Google Scholar]
- Deziel E., Gopalan S., Tampakaki A. P., Lepine F., Padfield K. E., Saucier M., et al. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55 998–1014. 10.1111/j.1365-2958.2004.04448.x [DOI] [PubMed] [Google Scholar]
- Deziel E., Lepine F., Milot S., He J., Mindrinos M. N., Tompkins R. G., et al. (2004). Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 101 1339–1344. 10.1073/pnas.0307694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E., Lepine F., Milot S., Villemur R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149 2005–2013. 10.1099/mic.0.26154-0 [DOI] [PubMed] [Google Scholar]
- Dietrich L. E., Price-Whelan A., Petersen A., Whiteley M., Newman D. K. (2006). The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61 1308–1321. 10.1111/j.1365-2958.2006.05306.x [DOI] [PubMed] [Google Scholar]
- Diggle S. P., Winzer K., Chhabra S. R., Worrall K. E., Camara M., Williams P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50 29–43. 10.1046/j.1365-2958.2003.03672.x [DOI] [PubMed] [Google Scholar]
- Driscoll J. A., Brody S. L., Kollef M. H. (2007). The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67 351–368. 10.2165/00003495-200767030-00003 [DOI] [PubMed] [Google Scholar]
- Dubern J. F., Diggle S. P. (2008). Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 4 882–888. 10.1039/b803796p [DOI] [PubMed] [Google Scholar]
- Dulcey C. E., Dekimpe V., Fauvelle D. A., Milot S., Groleau M. C., Doucet N., et al. (2013). The end of an old hypothesis: the pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20 1481–1491. 10.1016/j.chembiol.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essar D. W., Eberly L., Hadero A., Crawford I. P. (1990). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172 884–900. 10.1128/jb.172.2.884-900.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K., Passador L., Srikumar R., Tsang E., Nezezon J., Poole K. (1998). Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180 5443–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo A., Martinez-Martin N., Mercadillo M., Galan J. C., Ghysels B., Matthijs S., et al. (2008). The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619. 10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. M., III, Pesci E. C. (2007). Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J. Bacteriol. 189 3425–3433. 10.1128/JB.00209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. M., III, Sund Z. M., Ellison M. L., Wade D. S., Coleman J. P., Pesci E. C. (2008). PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J. Bacteriol. 190 7043–7051. 10.1128/JB.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M. P., Diggle S. P., Camara M., Williams P. (2007a). Biosensor-based assays for PQS, HHQ and related 2-alkyl-4-quinolone quorum sensing signal molecules. Nat. Protoc. 2 1254–1262. [DOI] [PubMed] [Google Scholar]
- Fletcher M. P., Diggle S. P., Crusz S. A., Chhabra S. R., Camara M., Williams P. (2007b). A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environ. Microbiol. 9 2683–2693. [DOI] [PubMed] [Google Scholar]
- Gallagher L. A., Mcknight S. L., Kuznetsova M. S., Pesci E. C., Manoil C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184 6472–6480. 10.1128/JB.184.23.6472-6480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Leon G., Salgado F., Oliveros J. C., Sanchez M. B., Martinez J. L. (2014). Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ. Microbiol. 16 1282–1296. 10.1111/1462-2920.12408 [DOI] [PubMed] [Google Scholar]
- Green S. K., Schroth M. N., Cho J. J., Kominos S. K., Vitanza-Jack V. B. (1974). Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl. Microbiol. 28 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D. (2006). Cost of cell-cell signalling in Pseudomonas aeruginosa: why it can pay to be signal-blind. Nat. Rev. Microbiol. 4:562; author rely 562. [DOI] [PubMed] [Google Scholar]
- Hammond J. H., Hebert W. P., Naimie A., Ray K., Van Gelder R. D., Digiandomenico A., et al. (2016). Environmentally endemic Pseudomonas aeruginosa strains with mutations in lasR are associated with increased disease severity in corneal ulcers. mSphere 1:e00140-16. 10.1128/mSphere.00140-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R., He J., Xiao G., Dekimpe V., Apidianakis Y., Lesic B., et al. (2010). Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. PLoS Pathog. 6:e1000810. 10.1371/journal.ppat.1000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Amado S., Blanco P., Alcalde-Rico M., Corona F., Reales-Calderon J. A., Sanchez M. B., et al. (2016). Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist. Updat. 28 13–27. 10.1016/j.drup.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Holban A. M., Bleotu C., Chifiriuc M. C., Bezirtzoglou E., Lazar V. (2014). Role of Pseudomonas aeruginosa quorum sensing (QS) molecules on the viability and cytokine profile of human mesenchymal stem cells. Virulence 5 303–310. 10.4161/viru.27571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannot K., Elsen S., Kohler T., Attree I., Van Delden C., Plesiat P. (2008). Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 52 2455–2462. 10.1128/AAC.01107-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper S. H., Bonocora R. P., Wade J. T., Musah R. A., Cady N. C. (2016). Chemical inhibition of kynureninase reduces Pseudomonas aeruginosa quorum sensing and virulence factor expression. ACS Chem. Biol. 11 1106–1117. 10.1021/acschembio.5b01082 [DOI] [PubMed] [Google Scholar]
- Kessler E., Safrin M. (2014). Elastinolytic and proteolytic enzymes. Methods Mol. Biol. 1149 135–169. 10.1007/978-1-4939-0473-0_13 [DOI] [PubMed] [Google Scholar]
- Kim K., Kim Y. U., Koh B. H., Hwang S. S., Kim S. H., Lepine F., et al. (2010). HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-kappaB pathway. Immunology 129 578–588. 10.1111/j.1365-2567.2009.03160.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppel A., Nasvall J., Andersson D. I. (2017). Evolution of antibiotic resistance without antibiotic exposure. Antimicrob. Agents Chemother. 61:e01495-17. 10.1128/AAC.01495-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., Michea-Hamzehpour M., Henze U., Gotoh N., Curty L. K., Pechere J. C. (1997). Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23 345–354. 10.1046/j.1365-2958.1997.2281594.x [DOI] [PubMed] [Google Scholar]
- Kohler T., Van Delden C., Curty L. K., Hamzehpour M. M., Pechere J. C. (2001). Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183 5213–5222. 10.1128/JB.183.18.5213-5222.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFayette S. L., Houle D., Beaudoin T., Wojewodka G., Radzioch D., Hoffman L. R., et al. (2015). Cystic fibrosis-adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci. Adv. 1:e1500199. 10.1126/sciadv.1500199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche M. G., Déziel E. (2011). MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. 10.1371/journal.pone.0024310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepine F., Deziel E., Milot S., Rahme L. G. (2003). A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 1622 36–41. 10.1016/S0304-4165(03)00103-X [DOI] [PubMed] [Google Scholar]
- Li X. Z., Nikaido H., Poole K. (1995). Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39 1948–1953. 10.1128/AAC.39.9.1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares J. F., Lopez J. A., Camafeita E., Albar J. P., Rojo F., Martinez J. L. (2005). Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187 1384–1391. 10.1128/JB.187.4.1384-1391.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ CT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Llanes C., Hocquet D., Vogne C., Benali-Baitich D., Neuwirth C., Plesiat P. (2004). Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48 1797–1802. 10.1128/AAC.48.5.1797-1802.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Causape C., Rubio R., Cabot G., Oliver A. (2018). Evolution of the Pseudomonas aeruginosa aminoglycoside mutational resistome in vitro and in the cystic fibrosis setting. Antimicrob. Agents Chemother. 62 e2583–e2517. 10.1128/AAC.02583-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L. (2014). Interkingdom signaling and its consequences for human health. Virulence 5 243–244. 10.4161/viru.28073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., Sanchez M. B., Martinez-Solano L., Hernandez A., Garmendia L., Fajardo A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33 430–449. 10.1111/j.1574-6976.2008.00157.x [DOI] [PubMed] [Google Scholar]
- Martinez-Solano L., Macia M. D., Fajardo A., Oliver A., Martinez J. L. (2008). Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin. Infect. Dis. 47 1526–1533. 10.1086/593186 [DOI] [PubMed] [Google Scholar]
- Maseda H., Sawada I., Saito K., Uchiyama H., Nakae T., Nomura N. (2004). Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48 1320–1328. 10.1128/AAC.48.4.1320-1328.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn L. M., Jett A. M., Akins D. R., Whiteley M. (2005). Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187 554–566. 10.1128/JB.187.2.554-566.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maura D., Hazan R., Kitao T., Ballok A. E., Rahme L. G. (2016). Evidence for direct control of virulence and defense gene circuits by the Pseudomonas aeruginosa quorum sensing regulator. MvfR. Sci. Rep. 6:34083. 10.1038/srep34083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Iglewski B. H., Pesci E. C. (2000). The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182 2702–2708. 10.1128/JB.182.10.2702-2708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellbye B., Schuster M. (2014). Physiological framework for the regulation of quorum sensing-dependent public goods in Pseudomonas aeruginosa. J. Bacteriol. 196 1155–1164. 10.1128/JB.01223-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa S., Inami H., Kato T., Sawada S., Yasuki T., Miyairi S., et al. (2012). RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. 12:70. 10.1186/1471-2180-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine T., Morita Y., Kataoka A., Mizushima T., Tsuchiya T. (1999). Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43 415–417. 10.1128/AAC.43.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales G., Wiehlmann L., Gudowius P., Van Delden C., Tummler B., Martinez J. L., et al. (2004). Structure of Pseudomonas aeruginosa populations analyzed by single nucleotide polymorphism and pulsed-field gel electrophoresis genotyping. J. Bacteriol. 186 4228–4237. 10.1128/JB.186.13.4228-4237.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Moustafa D. A., Stergioula V., Smith C. D., Goldberg J. B., Bassler B. L. (2018). The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115 E9411–E9418. 10.1073/pnas.1814023115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. T., Jones J. W., Ruge M. A., Kane M. A., Oglesby-Sherrouse A. G. (2015). Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J. Bacteriol. 197 2265–2275. 10.1128/JB.00072-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. T., Oglesby-Sherrouse A. G. (2016). Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl. Microbiol. Biotechnol. 100 6141–6148. 10.1007/s00253-016-7596-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obritsch M. D., Fish D. N., Maclaren R., Jung R. (2005). Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy 25 1353–1364. 10.1592/phco.2005.25.10.1353 [DOI] [PubMed] [Google Scholar]
- Olivares J., Alvarez-Ortega C., Linares J. F., Rojo F., Kohler T., Martinez J. L. (2012). Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ. Microbiol. 14 1968–1981. 10.1111/j.1462-2920.2012.02727.x [DOI] [PubMed] [Google Scholar]
- Olivares J., Alvarez-Ortega C., Martinez J. L. (2014). Metabolic compensation of fitness costs associated with overexpression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58 3904–3913. 10.1128/AAC.00121-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares Pacheco J., Alvarez-Ortega C., Alcalde Rico M., Martinez J. L. (2017). Metabolic compensation of fitness costs is a general outcome for antibiotic-resistant Pseudomonas aeruginosa mutants overexpressing efflux pumps. mBio 8:e00500-17. 10.1128/mBio.00500-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Mulet X., Lopez-Causape C., Juan C. (2015). The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 2 41–59. 10.1016/j.drup.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Pacheco A. R., Sperandio V. (2009). Inter-kingdom signaling: chemical language between bacteria and host. Curr. Opin. Microbiol. 12 192–198. 10.1016/j.mib.2009.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G. C., Jorth P. A., Whiteley M. (2013). The role of two Pseudomonas aeruginosa anthranilate synthases in tryptophan and quorum signal production. Microbiology 159 959–969. 10.1099/mic.0.063065-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G. C., Whiteley M. (2015). Metabolism and Pathogenicity of Pseudomonas aeruginosa Infections in the lungs of Individuals with cystic fibrosis. Microbiol Spectr. 3 1–20. 10.1128/microbiolspec.MBP-0003-2014 [DOI] [PubMed] [Google Scholar]
- Pearson J. P., Pesci E. C., Iglewski B. H. (1997). Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179 5756–5767. 10.1128/jb.179.18.5756-5767.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P., Van Delden C., Iglewski B. H. (1999). Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E. C., Pearson J. P., Seed P. C., Iglewski B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179 3127–3132. 10.1128/jb.179.10.3127-3132.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. (2006). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4 629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- Pollak S., Omer-Bendori S., Even-Tov E., Lipsman V., Bareia T., Ben-Zion I., et al. (2016). Facultative cheating supports the coexistence of diverse quorum-sensing alleles. Proc. Natl. Acad. Sci. U.S.A. 113 2152–2157. 10.1073/pnas.1520615113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K., Gotoh N., Tsujimoto H., Zhao Q., Wada A., Yamasaki T., et al. (1996). Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21 713–724. 10.1046/j.1365-2958.1996.281397.x [DOI] [PubMed] [Google Scholar]
- Rampioni G., Falcone M., Heeb S., Frangipani E., Fletcher M. P., Dubern J. F., et al. (2016). Unravelling the genome-wide contributions of specific 2-Alkyl-4-quinolones and PqsE to quorum sensing in Pseudomonas aeruginosa. PLoS Pathog. 12:e1006029. 10.1371/journal.ppat.1006029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G., Pustelny C., Fletcher M. P., Wright V. J., Bruce M., Rumbaugh K. P., et al. (2010). Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ. Microbiol. 12 1659–1673. 10.1111/j.1462-2920.2010.02214.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U., Wingender J., Muller H., Tummler B. (1994). A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60 1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P., Linares J. F., Ruiz-Diez B., Campanario E., Navas A., Baquero F., et al. (2002). Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50 657–664. 10.1093/jac/dkf185 [DOI] [PubMed] [Google Scholar]
- Simon R., O’connell M., Labes M., Puhler A. (1986). Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118 640–659. 10.1016/0076-6879(86)18106-7 [DOI] [PubMed] [Google Scholar]
- Sobel M. L., Neshat S., Poole K. (2005). Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187 1246–1253. 10.1128/JB.187.4.1246-1253.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickland H. G., Davenport P. W., Lilley K. S., Griffin J. L., Welch M. (2010). Mutation of nfxB causes global changes in the physiology and metabolism of Pseudomonas aeruginosa. J. Proteome Res. 9 2957–2967. 10.1021/pr9011415 [DOI] [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406 959–964. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Swift S., Karlyshev A. V., Fish L., Durant E. L., Winson M. K., Chhabra S. R., et al. (1997). Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179 5271–5281. 10.1128/jb.179.17.5271-5281.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwalkar J. S., Murray T. S. (2016). The approach to Pseudomonas aeruginosa in cystic fibrosis. Clin. Chest. Med. 37 69–81. 10.1016/j.ccm.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Tian Z. X., Fargier E., Mac Aogain M., Adams C., Wang Y. P., O’gara F. (2009a). Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res. 37 7546–7559. 10.1093/nar/gkp828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z. X., Mac Aogain M., O’connor H. F., Fargier E., Mooij M. J., Adams C., et al. (2009b). MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb. Pathog. 47 237–241. 10.1016/j.micpath.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Vila J., Fabrega A., Roca I., Hernandez A., Martinez J. L. (2011). Efflux pumps as an important mechanism for quinolone resistance. Adv. Enzymol. Relat. Areas Mol. Biol. 77 167–235. 10.1002/9780470920541.ch5 [DOI] [PubMed] [Google Scholar]
- Vila J., Martínez J. L. (2008). Clinical impact of the over-expression of efflux pump in nonfermentative gram-negative bacilli, development of efflux pump inhibitors. Curr. Drug Targets 9 797–807. 10.2174/138945008785747806 [DOI] [PubMed] [Google Scholar]
- Wade D. S., Calfee M. W., Rocha E. R., Ling E. A., Engstrom E., Coleman J. P., et al. (2005). Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187 4372–4380. 10.1128/JB.187.13.4372-4380.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. S., Bais H. P., Deziel E., Schweizer H. P., Rahme L. G., Fall R., et al. (2004). Pseudomonas aeruginosa-plant root interactions. pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134 320–331. 10.1104/pp.103.027888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. A., Blackwell H. E. (2016). Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem. Biol. 23 361–369. 10.1016/j.chembiol.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder C. N., Diggle S. P., Schuster M. (2011). Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 5 1332–1343. 10.1038/ismej.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153 3923–3938. 10.1099/mic.0.2007/012856-0 [DOI] [PubMed] [Google Scholar]
- Williams P., Camara M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12 182–191. 10.1016/j.mib.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Williams P., Winzer K., Chan W. C., Camara M. (2007). Look who’s talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362 1119–1134. 10.1098/rstb.2007.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson M. K., Swift S., Fish L., Throup J. P., Jorgensen F., Chhabra S. R., et al. (1998). Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163 185–192. 10.1111/j.1574-6968.1998.tb13044.x [DOI] [PubMed] [Google Scholar]
- Wittgens A., Tiso T., Arndt T. T., Wenk P., Hemmerich J., Muller C., et al. (2011). Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Fact 10:80. 10.1186/1475-2859-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates E. A., Philipp B., Buckley C., Atkinson S., Chhabra S. R., Sockett R. E., et al. (2002). N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70 5635–5646. 10.1128/IAI.70.10.5635-5646.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborskyte G., Andersen J. B., Kragh K. N., Ciofu O. (2017). Real-time monitoring of nfxB mutant occurrence and dynamics in Pseudomonas aeruginosa biofilm exposed to subinhibitory concentrations of ciprofloxacin. Antimicrob. Agents Chemother. 61:e02292-16. 10.1128/AAC.02292-16 [DOI] [PMC free article] [PubMed] [Google Scholar]