Abstract
Genome-wide association studies (GWAS) have identified copy number variants (CNVs) associated with obesity in chromosomal regions 1p31.1, 10q11.22, 11q11, 16p12.3, and recently 1p21.1, which contains the salivary amylase gene (AMY1). Recent evidence suggests this enzyme may influence gut microbiota composition through carbohydrate (mainly starch) degradation. The role of these CNVs in obesity has been scarcely explored in the Latino population, and thus the aim of our study was to evaluate the association of 1p31.1, 10q11.22, 11q11, 16p12.3 and 1p21.1 CNVs with obesity in 921 Mexican children, to replicate significant associations in 920 Mexican adults, and to analyze the association of AMY1 copy number with gut microbiota in 75 children and 45 adults. Of the five CNVs analyzed, 1q11 CNV was significantly associated with obesity in children, but not in adults. Only AMY1 CNV was significantly associated with obesity in both age groups. Moreover, gut microbiota analyses revealed a positive correlation between AMY1 copy number and Prevotella abundance. This genus has enzymes and gene clusters essential for complex polysaccharide degradation and utilization. To our knowledge, this is the first study to analyze the association of these five CNVs in the Mexican population and to report a correlation between AMY1 CN and gut microbiota in humans.
Keywords: CNVs, obesity, Mexican, AMY1, Prevotella
1. Introduction
Obesity represents a major global health problem [1]. In Mexico, according to the National Health and Nutrition Survey, the prevalence of overweight and obesity is 33.2% in children and 72.5% in adults [2]. Childhood obesity is the main predictor of adulthood obesity, which is associated with a number of metabolic and cardiovascular risk factors including type 2 diabetes, dyslipidemia, and hypertension [3,4].
Although the high prevalence of obesity has been attributed to an obesogenic environment, obesity has high heritability estimates ranging from 40 to 80% [5]. To date, more than 90 single nucleotide polymorphism (SNPs) have been associated with body mass index (BMI) mainly in Caucasian populations [6,7,8,9,10,11], some of which were also associated with BMI in Mexican children and adults [12]. Altogether, these SNPs account for less of 5% of the variability of this trait [11,12]. It has been suggested that copy number variation (CNV) might contribute to explain the missing heritability of obesity [13]. In this regard, genome-wide association studies (GWAS) have revealed certain CNV regions associated with obesity in European and Asian populations, including chromosomal regions 1p31.1, 10q11.22 11q11, 16p12.3, and more recently 1p21.1 which includes the salivary amylase gene AMY1 [9,10,14,15,16]. Interestingly, Falchi et al. reported that the effect size of the AMY1 CNV on obesity risk was considerably higher than that of other genetic variants such as fat mass and obesity-associated (FTO) gene polymorphisms [16]. However, subsequent studies evaluating the association of AMY1 with obesity have reported conflicting results [17,18].
Because AMY1 CNV has been associated with obesity and is considered one of the strongest signals of recent selection on human populations [19], there is great interest in unraveling the role of AMY1 in human obesity. AMY1 copy number (CN) correlates positively with salivary amylase amount and activity [19,20,21], and could likely influence gut microbiota composition through dietary carbohydrate processing. In this regard, the Prevotella-driven enterotype appears to be predominant in subjects consuming high proportions of dietary carbohydrate and fiber [22,23], however, its relationship with AMY1 CN has not been studied. Moreover, a study in mice reported an association of the AMY1 locus with both weight gain and increased Enterobacteria relative abundance in the gut [24].
The role of CNVs in obesity has been scarcely explored in the Mexican population, and thus the aim of our study was to evaluate the association of five candidate copy number variants (1p31.1, 10q11.22, 11q11, 16p12.3 and 1p21.1) with obesity in Mexican children, to replicate significant associations in Mexican adults, and to analyze the association of AMY1 copy number with gut genera and species belonging to Enterobacteriaceae and Prevotellaceae families and their role in obesity in Mexican children and adults.
2. Materials and Methods
2.1. Case-Control Studies in Children and Adults
A total of 921 unrelated Mexican mestizo children aged 6–12 years (485 normal weight controls and 436 obesity cases) were recruited from a summer camp for children of employees of the Mexican Health Ministry and Hospital Infantil de México. The adult cohort included 920 unrelated Mexican-Mestizo adults aged 18–75 years (536 with normal weight and 384 with obesity). Obese and normal-weight adults were recruited from several health institutions and public universities in Mexico City. Recruitment and inclusion criteria for children and adults have been described elsewhere [12,25]. All participants answered a detailed questionnaire providing demographic and lifestyle information. The study protocol was performed in accordance with the Declaration of Helsinki and was approved by the Ethic Committees of participant institutions. All adult participants and parents or legal guardians of children provided informed consent, and all children assented to participate.
2.2. Anthropometric and Biochemical Parameters
Anthropometric measurements including weight, height, waist, and hip circumference were determined following the procedures recommended by Lohman et al. [26]. BMI was calculated as body weight divided by height squared (kg/m2). For children, BMI percentile was calculated using age and sex-specific BMI reference data as recommended by the Centers for Disease Control and Prevention, and obesity was defined as BMI ≥95th percentile [27]. In adults, obesity status was determined according to World Health Organization (WHO) criteria [28]. Body composition was measured by bioelectrical impedance using a BIA (BIA 101 RJL System) body composition analyzer (RJL Systems, Clinton Township, MI, USA) only in children. Biochemical parameters including fasting total cholesterol (TC), triglyceride (TG), HDL-C (High Density Lipoprotein Cholesterol), glucose and insulin levels were performed using standardized procedures as previously described [12].
2.3. CNV Quantification
Genomic DNA was isolated from peripheral leukocytes using the QIAmp DNA Blood Mini Kit (QIAmp 96 DNA Blood Kit, Quiagen, Hilden, Germany). Copy number quantification was estimated by duplex quantitative real-time PCR (qPCR) with two TaqMan assays (Table S1), one for the target CNV and the other for the reference gene (RNaseP) (Life Technologies, Pleasanton, CA, USA). Assays were performed in a ViiA7 Real-Time PCR instrument (Thermo Fisher, Waltham, MA, USA). Relative copy number values were calculated by the ΔΔCT method using Copy Caller software (v.2.0, Applied Biosystems, Foster City, CA, USA), and results were validated only when calling confidence was >80% and ΔCq standard deviation between replicates was <0.20. All quantification assays were performed in triplicate. AMY1 CNV quantification was verified by droplet digital PCR (ddPCR System, Bio-Rad). Reactions were performed following the manufacturer’s recommendations using ddPCR™ Supermix and Taqman probes. Droplets were generated in a QX100 droplet device and amplification was performed by a C1000 Touch PCR thermal cycler (BioRad, Hercules, CA, USA). ddPCR data were analyzed using QuantaSoft software version 1.3.1.0. Four control samples obtained from the Coriell Institute for Medical Research were previously quantified and validated by Fiber-FISH [NA11930 (2 AMY1 copies), NA10852 (6 AMY1 copies), NA11993 (10 AMY1 copies) and NA18972 (18 AMY1 copies)] [29] were included as references to quantify AMY1 copy number. We further validated AMY1 CNs of these samples by digital PCR.
2.4. Gut Microbiota Analyses
Fecal samples were obtained from 75 children and 45 adult participants and DNA was extracted from these samples as previously described [30]. The V4 hypervariable region was amplified using 515F and 806R primers [31] and sequenced in an Illumina MiSeq 2 × 250 device. Sequences were analyzed using QIIME 1.9.1 [32], and phylogenetic distances were calculated by the UniFrac method [33]. A full description of 16S rRNA sequencing analyses has been described elsewhere [30]. Abundances were normalized using an arcsin transformation.
2.5. Dietary Assessment
A semi-quantitative food frequency questionnaire previously validated for the Mexican population [34] was applied to children and adults with available fecal samples. Average daily energy and nutrient intake was computed through the Evaluation System of Nutritional Habits and Nutrient Intake Software, and was expressed both as grams and percentage of total energy.
2.6. Statistical Analyses
Statistical analyses were performed with SPSS software version 18.0. Anthropometric and biochemical parameters in obese and normal-weight subjects were compared using the Student’s t-test or Mann-Whitney U-test. Associations of 1p31.1, 10q11.22, 11q11 and 16p12.3 CNVs with obesity were tested comparing the frequency of deletions (<2 copies) and duplications (>2 copies) in normal-weight and obese children. Given the high range of AMY1 copy numbers (2–19), two different cutoff points were used to compare the copy number in normal and obese subjects: The AMY1 CN median (CN < 6 vs. CN > 6), and the cutoff point used by Falchi et al. (CN ≤ 4 or low vs. CN ≥ 10 or high) [16]. All associations were tested by logistic regression, adjusting for age and sex. In addition, lineal regression models were used to test associations of CNVs with anthropometric and metabolic parameters adjusting for age, sex, and BMI as appropriate. Correlations between microbial relative abundance (genera and species belonging to Enterobacteriaceae and Prevotellaceae families) and AMY1 CN or diet were evaluated using Spearman’s tests. Hochberg false discovery rate (FDR)-adjusted q-values < 0.05 were considered significant for the entire microbiota analysis [35]. Relative abundance of genera and species belonging to Enterobacteriaceae and Prevotellaceae families in low and high AMY1 CN carriers was compared using the Mann-Whitney U-test.
3. Results
3.1. Clinical Characteristics of Case-Control Study
Clinical and biochemical characteristics of 921 children and 920 adults stratified by nutritional status are shown in Table S2. As expected, normal-weight children and adults had significantly lower obesity-related anthropometric measurements and lower biochemical measurements (insulin, HOMA-IR (Homeostatic Model Assessment for Insulin Resistance), triglycerides and total cholesterol levels) as compared to children and adults with obesity (P ≤ 0.01).
3.2. Association of 11q11and 1p21.1 (AMY1) CNVs with Obesity in Mexican Children
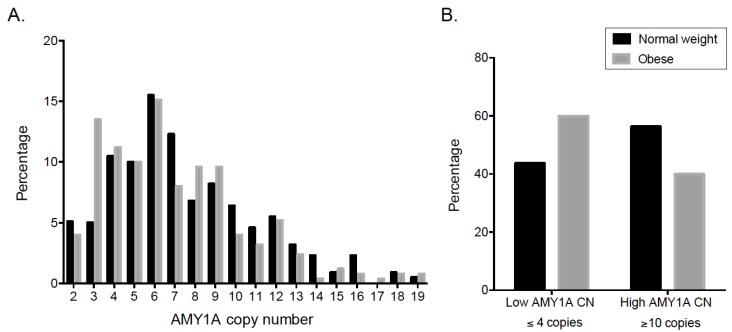
The associations of the five CNV loci with obesity in children are shown in Table 1. Copy numbers in loci 16p12.3, 1p31.1 and 10q11.22 ranged from 1 to 4 in Mexican children. Deletions within these loci were infrequent (≤5%), and were not significantly associated with obesity (P ≥ 0.1). In contrast, 11q11/OR4P4/OR4S2/OR4C6 (olfactory receptors family) and 1p21.1/AMY1 CNVs ranged from 0 to 8 and from 2 to 19, respectively. Heterozygous or homozygous deletions of OR4P4/OR4S2/OR4C6 CNV were significantly associated with lower obesity risk (OR = 0.774; 95% CI = 0.634–0.945; P = 0.047). The distribution of AMY1 copy numbers in obese and normal weight children is shown in Figure 1A (median CN = 6). Children with <6 AMY1 copies had a borderline significantly higher risk of obesity as compared to those with ≥6 AMY1 CNs (OR = 1.323; 95% CI = 0.984–1.780, P = 0.064) (Table 1). Moreover, using the previously reported cutoff values of ≤4 AMY1 copies (low) and ≥10 AMY1 copies (high), low AMY1 copy numbers were significantly associated with obesity (OR = 1.530; 95% CI = 1.030–2.273, P = 0.035; after adjusting for age and gender) (Figure 1B). No significant associations between 11q11/OR4P4/OR4S2/OR4C6 or 1p21.1/AMY1 CNVs and biochemical traits were observed in children (Table S3).
Table 1.
Association of five copy number variants with obesity risk in Mexican children (n = 921).
| Locus | Gene | CN Range | Classification | Normal Weight | Obese | Association | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | OR | 95% CI | P-value | ||||
| 11q11 | OR4P4, OR4S2 | 0–8 | Deletion | 199 | 41.0 | 151 | 34.6 | 0.774 | 0.634–0.945 | 0.047 |
| OR4C6 | ≥2 copies | 286 | 59.0 | 285 | 65.4 | |||||
| 1p21.1 | AMY1 | 2–19 | Less 6 copies | 116 | 23.9 | 128 | 29.4 | 1.323 | 0.984–1.780 | 0.064 |
| ≥6 copies | 369 | 76.1 | 308 | 70.6 | ||||||
| 16p12.3 | GPRC5B | 1–4 | Deletion | 11 | 2.3 | 13 | 2.9 | 1.307 | 0.472–3.622 | 0.606 |
| ≥2 copies | 474 | 97.7 | 423 | 97.1 | ||||||
| 1p31.1 | NEGR1 | 1–4 | Deletion | 25 | 5.2 | 23 | 5.3 | 1.078 | 0.425–3.622 | 0.867 |
| ≥2 copies | 460 | 94.8 | 413 | 94.7 | ||||||
| 10q11.22 | NPY4R | 1–4 | Deletion | 21 | 4.3 | 19 | 4.4 | 1.064 | 0.411–2.758 | 0.898 |
| ≥2 copies | 464 | 95.7 | 417 | 95.6 | ||||||
OR, odds ratio; CI, confidence interval; OR4P4, olfactory receptor family 4 subfamily P member 4 gene; OR4S2, olfactory receptor family 4 subfamily S member 2 gene; OR4C6, Olfactory receptor family 4 subfamily C member 6 gene; AMY1, salivary amylase gene; GPRC5B, G protein-coupled receptor class C group 5 member B gene; NEGR1, neuronal growth regulator 1 gene; NPY4R, neuropeptide Y receptor Y4 gene. Associations were tested by logistic regression and adjusted by sex and age.
Figure 1.
Distribution of AMY1 copy number in normal weight and obese children (n = 921). (A) AMY1 copy numbers ranged from 2 to 19. Normal-weight children are represented by gray bars and obese children by black bars; (B) Distribution of low vs high AMY1 copy numbers in normal weight and obese children. Low AMY1 copy numbers were significantly more frequent in obese than in normal weight children (P = 0.035).
3.3. Association of AMY1 Copy Number with Obesity in Mexican Adults
We then sought whether the associations found in children were also observed in Mexican adults. The 11q11 CNV was not associated with obesity in adults (P = 0.537). However, the presence of less than six AMY1 copies showed a borderline significant association with obesity (OR = 1.521; 95% CI = 0.928–2.495; P = 0.096), and individuals with a low number of AMY1 copies (≤4) had a significantly higher risk of obesity than those with a high number (≥10) of AMY1 copies (OR = 1.536; 95% CI = 1.019–2.313, P = 0.040) (Table 2). As observed in children, AMY1 CNV showed no significant associations with biochemical parameters (Table S4).
Table 2.
Association of 11q11 and 1p21.1 copy number variants with obesity risk in Mexican adults (n = 920).
| Locus | Gene | CN Range | Classification | Normal Weight | Obese | Association | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | OR | 95% CI | P-value | ||||
| 11q11 | OR4P4, OR4S2 | 0–8 | Deletion | 124 | 32.3 | 186 | 34.7 | 1.054 | 0.877–1.266 | 0.537 |
| OR4C6 | ≥2 copies | 260 | 67.7 | 350 | 65.3 | |||||
| 1p21.1 | AMY1 | 2–19 | Less 6 copies | 94 | 24.5 | 165 | 30.8 | 1.521 | 0.928–2.495 | 0.096 |
| ≥6 copies | 290 | 75.5 | 371 | 69.2 | ||||||
OR, odds ratio; CI, confidence interval; OR4P4, olfactory receptor family 4 subfamily P member 4 gene; OR4S2, olfactory receptor family 4 subfamily S member 2 gene; OR4C6, Olfactory receptor family 4 subfamily C member 6 gene; AMY1, salivary amylase gene. Associations were tested by logistic regression and adjusted by sex and age.
3.4. Association of AMY1 Copy Number with Prevotella Abundance
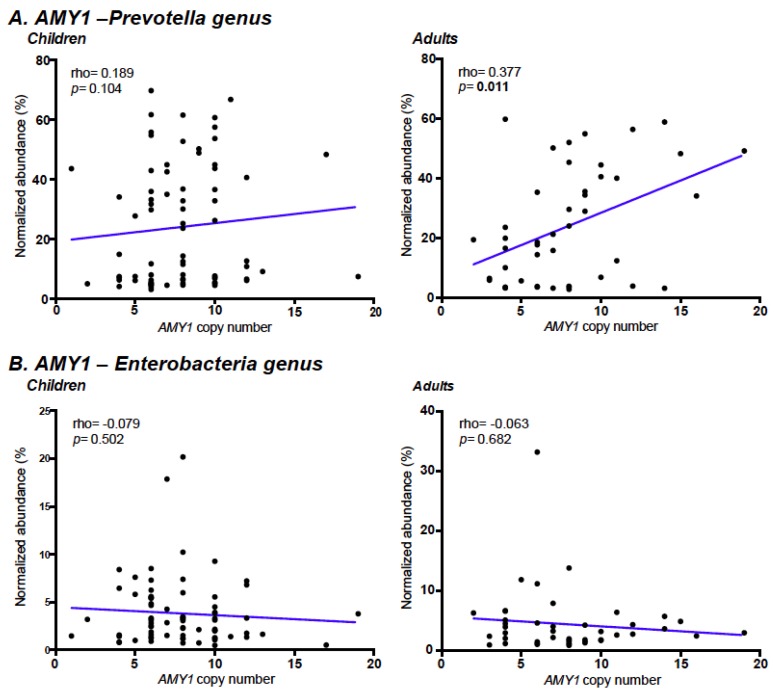
Correlations between AMY1 copies and the relative abundance of Prevotella and Enterobacteria genera in both children and adults are shown in Figure 2. No significant correlations between Enterobacteria and AMY1 copy number were observed in any age group (Figure 2B). However, a positive and significant correlation between AMY1 copy number and Prevotella abundance was observed in adults (r = 0.377; P = 0.011), while a positive correlation that did not reach statistical significance was observed in children (r = 0.189; P = 0.104) (Figure 2A). We then explored correlations between AMY1 CN and all other identified gut microbiota genera. In addition to the relative abundance of Prevotella that showed a positive and significant correlation in adults, no other taxa were significantly associated with AMY1 CN (Table S5).
Figure 2.
Correlations of AMY1 CN with relative abundance of Prevotella and Enterobacteria genera in Mexican individuals. Bacterial abundances of Prevotella (A) and Enterobacteria (B) were normalized using arcsin sqrt transformation, and Spearman’s correlation coefficients were estimated.
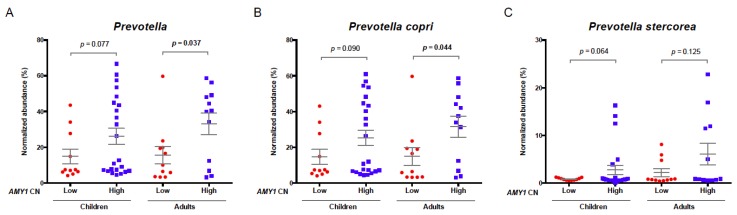
Moreover, the relative abundance of Prevotella, specifically of Prevotella copri, was twofold higher in adults with a high number of AMY1 copies (≥10) as compared to those with a low AMY1 copy number (≤4). These comparisons were similar in children, without reaching statistical significance (P = 0.077 and 0.090, respectively; adjusted by age, gender and BMI percentile) (Figure 3; Table S6).
Figure 3.
Comparison of Prevotella relative abundance in Mexican individuals with low vs. high AMY1 CNs. Average abundances of Prevotella (A), Prevotella copri (B) and Prevotella stercorea (C) were higher in both children and adults with high (≥10) as compared to low (≤4 copies) AMY1 copy numbers. Differences reached statistical significance only for Prevotella genus and Prevotella copri in adults.
We then compared dietary carbohydrate intake in individuals with low and high AMY1 CN, observing no differences in the consumption of any carbohydrate (Table S7). Moreover, no significant correlations were observed between Prevotella genus or species abundance with dietary carbohydrates, fiber or starch intake (P > 0.05) (Figure S1).
4. Discussion
Association studies of copy number variants with obesity are limited and have reported inconsistent results [9,10,14,15,16,17,18]. In the present study, we sought associations of five obesity-related CNV loci [1p31.1/NEGR1 (Neuronal growth regulator 1), 10q11.22/GPRC5B (G Protein-Coupled Receptor Class C Group 5 Member B), 11q11/OR4P4/OR4S2/OR4C6, 16p12.3/NPY4R (Neuropeptide Y Receptor Y4) and 1p21.1/AMY1] with obesity risk in the Mexican population. In contrast with previous findings, NEGR1, GPRC5 and NPY4R CNVs were not associated with obesity in Mexican children. As with all association studies, this could be due to an array of factors including ethnic, study design and methodological differences, or gene-environment interactions, among others [36,37]. The lack of association of the 1p31.1/NEGR1 CNV is in agreement with previous reports in Mexican children and adults where rs2815752 (in high LD with 1p31.1/NEGR1 CNV) was not associated with obesity [12,38], but in disagreement with the findings of Antúnez-Ortiz et al. who found a significant association of another NEGR1 CNV in a group of Mexican children [39]. Regarding the GPCR5 CNV, it has been previously suggested that the effect of the deletion of CNV is ethnic-specific, as it was significantly associated with obesity in Europeans, who have a higher CNV deletion frequency (0.27); but not in the Chinese population who have a lower CNV deletion frequency (0.008) [40]. The GPCR5 CNV deletion frequency was 0.025 in the Mexican population, and was not associated with obesity, which is consistent with the previous findings in Chinese individuals. Finally, previous studies associating the NPY4R CNV with obesity risk have reported contradictory findings, as the CNV deletion allele has been associated both with higher and lower obesity risk [14,15,41,42]. Our findings are consistent with the report of Sun et al., who found no association of this CNV deletion with obesity in the Chinese population [43].
Interestingly, 11q11/OR4P4/OR4S2/OR4C6 and 1p21.1/AMY1 CNVs were significantly associated with obesity in Mexican children. The association of the OR4P4/OR4S2/OR4C6 CNV is consistent with the previously reported role of olfactory receptor gene polymorphisms in obesity [44], and the role of the olfactory system in thermogenesis and energy homeostasis in the murine model [45]. However, while the 11q11 CNV deletion was associated with a decreased risk of obesity in Mexican children, previous studies have reported this CNV deletion is associated with increased obesity risk [15,46]. Moreover, the 11q11 CNV deletion was not associated with obesity in Mexican adults. These age-dependent association differences could be due to higher BMI heritability in childhood [47], higher effect sizes of obesity-associated loci in childhood, or to age-dependent gene–environment interactions [48,49].
The role of AMY1 copy number variation in obesity risk has been widely studied [16,50,51,52], with inconsistent results [17,18]. It has been suggested that AMY1 CN variation among human populations may result from natural selection; however, starch digestion does not seem to be the major selective force [53]. In this study, we did not observe AMY1 CN differences between Mexican Mestizo and Indigenous subjects (mean 7.0 ± 3.2 in Mexican-Mestizos and 7.2 ± 3.7 in 130 Mexican indigenous; P = 0.511). This is in agreement with a recent report suggesting that a high number of AMY1 copies were fixed in modern human populations, after the separation of humans and Neanderthals [54].
Interestingly, AMY1 was the only loci associated with obesity in both children and adults. Falchi et al. first reported a remarkably strong association of low AMY1 copy numbers with increased obesity risk in Europeans [16]. Further studies replicated this association in other populations and in different age groups, although with considerably lower effect sizes [50,51,52]. A previous study in Mexican children reported an association of AMY1 copy numbers with obesity [55], although Usher et al. [17] suggested that this study described an outlier set of control samples with unusually high AMY1 copy number measurements, which is not necessarily a replication of earlier findings. In the present study, we observed the initially reported shifted distribution of AMY1 copy number between individuals with normal weight and obesity. Altogether, these findings confirm the role of this CNV in obesity in Mexican children and adults.
The mechanism by which AMY1 copy number plays a role in obesity remains unclear. Our study evaluated the AMY1 CN-gut microbiota-obesity association in a subgroup of Mexican children and adults. We did not observe significant correlations between AMY1 copy number and Enterobacteriaceae family (Table S6), as previously reported in the murine model [24]. However, the gut microbiome of children and adults with high AMY1 copy numbers was enriched in Prevotella genus and species, but not with any other gut microbiota genera, stressing the importance of further studies on the possible role of AMY1 function on this bacterial genus. Prevotella is one of the most abundant enterotypes in the intestinal microbiome and has enzymes and gene clusters essential for fermentation and utilization of complex polysaccharides [22,23,56]. This is consistent with results of several studies showing that increased Prevotella is metabolically favorable [23], and that a high Prevotella/Bacteroides ratio favors weight loss in response to certain dietary interventions [57,58]. However, while we observed significant associations of high AMY1 CN with both normal weight and increased Prevotella abundance (specifically Prevotella copri), Prevotella abundance was not significantly associated with nutritional status or with total dietary carbohydrates (including fiber and starch). Further studies using larger sample sizes will define whether this lack of association is due to insufficient statistical power.
This study has certain limitations that should be acknowledged. Firstly, the sample size was insufficient to detect associations of obesity with low frequency alleles (1p31.1/NEGR1, 10q11.22/GPRC5B, and 16p12.3/NPY4R), although it was sufficient to find associations with 11q11/OR4P4/OR4S2/OR4C6, and 1p21.1/AMY1 CNs. Secondly, although amylase activity is clearly associated with obesity [19,20,21], this parameter was not measured in the present study. Finally, intestinal microbiome analysis was performed in a small number of individuals, limiting statistical power to associate the entire intestinal microbiome with AMY1 CNs. However, the sample size was sufficient to find a positive correlation between Prevotella abundance and AMY1 CNs.
5. Conclusions
In conclusion, of the five CNVs previously associated with obesity, only AMY1 CNV was significantly associated with obesity in Mexican children and adults. Moreover, gut microbiota analyses identified a positive correlation between AMY1 copy number and Prevotella abundance, which highlights the role of genetics in the modulation of intestinal microbiota. To our knowledge, this is the first study to report the association of AMY1 CN and gut microbiota in humans. Future studies are required to identify mechanisms explaining these associations.
Acknowledgments
We thank Luz E. Guillén and Alfredo Mendoza-Vargas for her technical assistance.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6643/10/11/1607/s1, Figure S1: Correlations of Prevotella and Enterobacteria abundances with AMY1 CN, metabolic parameters and dietary carbohydrate intake. The correlation matrix depicts positive correlations in red and negative correlations in blue. The color scale indicates estimated Spearman’s correlation coefficient values. Statistical significance of each correlation is proportional to the size of the circle, Table S1: CNVs and assay designs used to test associations, Table S2: Clinical, anthropometric and biochemical parameters of the study population, Table S3: Association of 11q11 (OR4P4/OR4S2/OR4C6) and 1p21.1 (AMY1) copy number with biochemical parameters in Mexican children stratified by nutritional status, Table S4: Association of 11q11 and 1p21.1 copy number with biochemical parameters in Mexican adults stratified by nutritional status, Table S5: Correlations of AMY1 CN with relative abundances of gut microbiota at the genus level in Mexican children and adults, Table S6: Comparison of normalized Prevotellaceae and Enterobacteriaceae abundances in Mexican individuals with low and high AMY1 copy numbers, Table S7: Comparison of energy and dietary carbohydrate intake in Mexican individuals with low and high AMY1 copy numbers.
Author Contributions
P.L.-M. and S.C.-Q. designed the study. V.A.-A., B.E.d.R.-N., J.S., R.V.-C., T.V.-M. and C.A.A.-S. contributed reagents/materials. P.L.-M., H.V.-R., B.E.L.-C. and S.M.-R. performed the experiments. P.L.-M., S.M.-R. and L.R.M.-K. analyzed data. P.L.-M., T.V.-M. and S.C.-Q. wrote the paper, incorporating input from authors. All authors read and approved the final content of the manuscript.
Funding
This study was supported by PAPIIT/UNAM, Universidad Nacional Autónoma de México grant number IA202413. PL-M is in the Ciencias Bioquímicas PhD program from Universidad Nacional Autónoma de México (UNAM), and is supported by a PhD fellowship from CONACyT, México.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Arroyo-Johnson C., Mincey K.D. Obesity Epidemiology Worldwide. Gastroenterol. Clin. N. Am. 2016;45:571–579. doi: 10.1016/j.gtc.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shamah-Levy T., Rivera-Dommarco J., Hernández-Ávila M. Encuesta Nacional de Salud y Nutrición de Medio Camino. Instituto Nacional de Salud Pública; Cuernavaca, Mexico: 2016. [Google Scholar]
- 3.Freedman D.S., Khan L.K., Serdula M.K., Dietz W.H., Srinivasan S.R., Berenson G.S. The relation of childhood BMI to adult adiposity: The Bogalusa Heart Study. Pediatrics. 2005;115:22–27. doi: 10.1542/peds.2004-0220. [DOI] [PubMed] [Google Scholar]
- 4.Juonala M., Magnussen C.G., Berenson G.S., Venn A., Burns T.L., Sabin M.A., Srinivasan S.R., Daniels S.R., Davis P.H., Chen W., et al. Childhood adiposity, adult adiposity, and cardiovascular risk factors. N. Engl. J. Med. 2011;365:1876–1885. doi: 10.1056/NEJMoa1010112. [DOI] [PubMed] [Google Scholar]
- 5.Elks C.E., den Hoed M., Zhao J.H., Sharp S.J., Wareham N.J., Loos R.J., Ong K.K. Variability in the heritability of body mass index: A systematic review and meta-regression. Front. Endocrinol. 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 9.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leon-Mimila P., Villamil-Ramirez H., Villalobos-Comparan M., Villarreal-Molina T., Romero-Hidalgo S., Lopez-Contreras B., Gutierrez-Vidal R., Vega-Badillo J., Jacobo-Albavera L., Posadas-Romeros C., et al. Contribution of common genetic variants to obesity and obesity-related traits in mexican children and adults. PLoS ONE. 2013;8:e70640. doi: 10.1371/journal.pone.0070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sha B.Y., Yang T.L., Zhao L.J., Chen X.D., Guo Y., Chen Y., Pan F., Zhang Z.X., Dong S.S., Xu X.H., et al. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J. Hum. Genet. 2009;54:199–202. doi: 10.1038/jhg.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarick I., Vogel C.I., Scherag S., Schafer H., Hebebrand J., Hinney A., Scherag A. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum. Mol. Genet. 2011;20:840–852. doi: 10.1093/hmg/ddq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falchi M., El-Sayed Moustafa J.S., Takousis P., Pesce F., Bonnefond A., Andersson-Assarsson J.C., Sudmant P.H., Dorajoo R., Al-Shafai M.N., Bottolo L., et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat. Genet. 2014;46:492–497. doi: 10.1038/ng.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usher C.L., Handsaker R.E., Esko T., Tuke M.A., Weedon M.N., Hastie A.R., Cao H., Moon J.E., Kashin S., Fuchsberger C., et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat. Genet. 2015;47:921–925. doi: 10.1038/ng.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yong R.Y., Mustaffa S.B., Wasan P.S., Sheng L., Marshall C.R., Scherer S.W., Teo Y.Y., Yap E.P. Complex Copy Number Variation of AMY1 does not Associate with Obesity in two East Asian Cohorts. Hum. Mutat. 2016;37:669–678. doi: 10.1002/humu.22996. [DOI] [PubMed] [Google Scholar]
- 19.Perry G.H., Dominy N.J., Claw K.G., Lee A.S., Fiegler H., Redon R., Werner J., Villanea F.A., Mountain J.L., Misra R., et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandel A.L., Peyrot des Gachons C., Plank K.L., Alarcon S., Breslin P.A. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS ONE. 2010;5:e13352. doi: 10.1371/journal.pone.0013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z.M., Lin J., Chen L.H., Zhang M., Chen W.W., Yang X.R. The roles of AMY1 copies and protein expression in human salivary alpha-amylase activity. Physiol. Behav. 2015;138:173–178. doi: 10.1016/j.physbeh.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 22.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovatcheva-Datchary P., Nilsson A., Akrami R., Lee Y.S., De Vadder F., Arora T., Hallen A., Martens E., Bjorck I., Backhed F. Dietary Fiber-Induced Improvement in Glucose Metabolism is Associated with Increased Abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Parks B.W., Nam E., Org E., Kostem E., Norheim F., Hui S.T., Pan C., Civelek M., Rau C.D., Bennett B.J., et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villalobos-Comparan M., Teresa Flores-Dorantes M., Teresa Villarreal-Molina M., Rodriguez-Cruz M., Garcia-Ulloa A.C., Robles L., Huertas-Vazquez A., Saucedo-Villarreal N., Lopez-Alarcon M., Sanchez-Munoz F., et al. The FTO gene is associated with adulthood obesity in the Mexican population. Obesity. 2008;16:2296–2301. doi: 10.1038/oby.2008.367. [DOI] [PubMed] [Google Scholar]
- 26.Lohman T.G., Roche A.F., Martorell R. Anthropometric Standardization Reference Manual Abridged Edition. Human Kinetics Books; Champaign, IL, USA: 1991. 90p. p. vi. [Google Scholar]
- 27.Kuczmarski R.J., Ogden C.L., Guo S.S., Grummer-Strawn L.M., Flegal K.M., Mei Z., Wei R., Curtin L.R., Roche A.F., Johnson C.L. 2000 CDC Growth Charts for the United States: Methods and development. Vital Health Stat. 11. 2002;246:1–190. [PubMed] [Google Scholar]
- 28.World Health Organization . Obesity: Preventing and Managing the Global Epidemic. Report of a WHO Consultation. WHO; Geneva, Switzerland: 2000. pp. 1–253. (Technical Report Series). [PubMed] [Google Scholar]
- 29.Carpenter D., Dhar S., Mitchell L.M., Fu B., Tyson J., Shwan N.A., Yang F., Thomas M.G., Armour J.A. Obesity, starch digestion and amylase: Association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Hum. Mol. Genet. 2015;24:3472–3480. doi: 10.1093/hmg/ddv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Contreras B.E., Moran-Ramos S., Villarruel-Vazquez R., Macias-Kauffer L., Villamil-Ramirez H., Leon-Mimila P., Vega-Badillo J., Sanchez-Munoz F., Llanos-Moreno L.E., Canizalez-Roman A., et al. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr. Obes. 2018;13:381–388. doi: 10.1111/ijpo.12262. [DOI] [PubMed] [Google Scholar]
- 31.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C.A., Stombaugh J., Gonzalez A., Ackermann G., Wendel D., Vazquez-Baeza Y., Jansson J.K., Gordon J.I., Knight R. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez-Avila M., Romieu I., Parra S., Hernandez-Avila J., Madrigal H., Willett W. Validity and reproducibility of a food frequency questionnaire to assess dietary intake of women living in Mexico City. Salud Publica Mex. 1998;40:133–140. doi: 10.1590/S0036-36341998000200005. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 36.Stryjecki C., Alyass A., Meyre D. Ethnic and population differences in the genetic predisposition to human obesity. Obes. Rev. 2018;19:62–80. doi: 10.1111/obr.12604. [DOI] [PubMed] [Google Scholar]
- 37.Huang T., Hu F.B. Gene-environment interactions and obesity: Recent developments and future directions. BMC Med. Genom. 2015;8(Suppl. 1):S2. doi: 10.1186/1755-8794-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abadi A., Peralta-Romero J., Suarez F., Gomez-Zamudio J., Burguete-Garcia A.I., Cruz M., Meyre D. Assessing the effects of 35 European-derived BMI-associated SNPs in Mexican children. Obesity. 2016;24:1989–1995. doi: 10.1002/oby.21590. [DOI] [PubMed] [Google Scholar]
- 39.Antunez-Ortiz D.L., Flores-Alfaro E., Burguete-Garcia A.I., Bonnefond A., Peralta-Romero J., Froguel P., Espinoza-Rojo M., Cruz M. Copy Number Variations in Candidate Genes and Intergenic Regions Affect Body Mass Index and Abdominal Obesity in Mexican Children. Biomed. Res. Int. 2017;2017:2432957. doi: 10.1155/2017/2432957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang T.L., Guo Y., Li S.M., Li S.K., Tian Q., Liu Y.J., Deng H.W. Ethnic differentiation of copy number variation on chromosome 16p12.3 for association with obesity phenotypes in European and Chinese populations. Int. J. Obes. 2013;37:188–190. doi: 10.1038/ijo.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aerts E., Beckers S., Zegers D., Van Hoorenbeeck K., Massa G., Verrijken A., Verhulst S.L., Van Gaal L.F., Van Hul W. CNV analysis and mutation screening indicate an important role for the NPY4R gene in human obesity. Obesity. 2016;24:970–976. doi: 10.1002/oby.21435. [DOI] [PubMed] [Google Scholar]
- 42.Shebanits K., Andersson-Assarsson J.C., Larsson I., Carlsson L.M.S., Feuk L., Larhammar D. Copy number of pancreatic polypeptide receptor gene NPY4R correlates with body mass index and waist circumference. PLoS ONE. 2018;13:e0194668. doi: 10.1371/journal.pone.0194668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun C., Cao M., Shi J., Li L., Miao L., Hong J., Cui B., Ning G. Copy number variations of obesity relevant loci associated with body mass index in young Chinese. Gene. 2013;516:198–203. doi: 10.1016/j.gene.2012.12.081. [DOI] [PubMed] [Google Scholar]
- 44.Choquette A.C., Bouchard L., Drapeau V., Lemieux S., Tremblay A., Bouchard C., Vohl M.C., Perusse L. Association between olfactory receptor genes, eating behavior traits and adiposity: Results from the Quebec Family Study. Physiol. Behav. 2012;105:772–776. doi: 10.1016/j.physbeh.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Riera C.E., Tsaousidou E., Halloran J., Follett P., Hahn O., Pereira M.M.A., Ruud L.E., Alber J., Tharp K., Anderson C.M., et al. The Sense of Smell Impacts Metabolic Health and Obesity. Cell Metab. 2017;26:198–211. doi: 10.1016/j.cmet.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D., Li Z., Wang H., Yang M., Liang L., Fu J., Wang C., Ling J., Zhang Y., Zhang S., et al. Interactions between obesity-related copy number variants and dietary behaviors in childhood obesity. Nutrients. 2015;7:3054–3066. doi: 10.3390/nu7043054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo G., Liu H., Wang L., Shen H., Hu W. The Genome-Wide Influence on Human BMI Depends on Physical Activity, Life Course, and Historical Period. Demography. 2015;52:1651–1670. doi: 10.1007/s13524-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng X.R., Song J.Y., Ma J., Liu F.H., Shang X.R., Guo X.J., Wang H.J. Association study of childhood obesity with eight genetic variants recently identified by genome-wide association studies. Pediatr. Res. 2014;76:310–315. doi: 10.1038/pr.2014.88. [DOI] [PubMed] [Google Scholar]
- 49.Felix J.F., Bradfield J.P., Monnereau C., van der Valk R.J., Stergiakouli E., Chesi A., Gaillard R., Feenstra B., Thiering E., Kreiner-Moller E., et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum. Mol. Genet. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viljakainen H., Andersson-Assarsson J.C., Armenio M., Pekkinen M., Pettersson M., Valta H., Lipsanen-Nyman M., Makitie O., Lindstrand A. Low Copy Number of the AMY1 Locus Is Associated with Early-Onset Female Obesity in Finland. PLoS ONE. 2015;10:e0131883. doi: 10.1371/journal.pone.0131883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcovecchio M.L., Florio R., Verginelli F., De Lellis L., Capelli C., Verzilli D., Chiarelli F., Mohn A., Cama A. Low AMY1 Gene Copy Number Is Associated with Increased Body Mass Index in Prepubertal Boys. PLoS ONE. 2016;11:e0154961. doi: 10.1371/journal.pone.0154961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnefond A., Yengo L., Dechaume A., Canouil M., Castelain M., Roger E., Allegaert F., Caiazzo R., Raverdy V., Pigeyre M., et al. Relationship between salivary/pancreatic amylase and body mass index: A systems biology approach. BMC Med. 2017;15:37. doi: 10.1186/s12916-017-0784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez C.I., Wiley A.S. Rethinking the starch digestion hypothesis for AMY1 copy number variation in humans. Am. J. Phys. Anthropol. 2017;163:645–657. doi: 10.1002/ajpa.23237. [DOI] [PubMed] [Google Scholar]
- 54.Inchley C.E., Larbey C.D., Shwan N.A., Pagani L., Saag L., Antao T., Jacobs G., Hudjashov G., Metspalu E., Mitt M., et al. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci. Rep. 2016;6:37198. doi: 10.1038/srep37198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mejia-Benitez M.A., Bonnefond A., Yengo L., Huyvaert M., Dechaume A., Peralta-Romero J., Klunder-Klunder M., Garcia Mena J., El-Sayed Moustafa J.S., Falchi M., et al. Beneficial effect of a high number of copies of salivary amylase AMY1 gene on obesity risk in Mexican children. Diabetologia. 2015;58:290–294. doi: 10.1007/s00125-014-3441-3. [DOI] [PubMed] [Google Scholar]
- 56.De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S., Collini S., Pieraccini G., Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hjorth M.F., Roager H.M., Larsen T.M., Poulsen S.K., Licht T.R., Bahl M.I., Zohar Y., Astrup A. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int. J. Obes. 2018;42:284. doi: 10.1038/ijo.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hjorth M.F., Blaedel T., Bendtsen L.Q., Lorenzen J.K., Holm J.B., Kiilerich P., Roager H.M., Kristiansen K., Larsen L.H., Astrup A. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: Results from a post-hoc analysis. Int. J. Obes. 2018 doi: 10.1038/s41366-018-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.