Abstract
Purpose
The purpose of this study was to characterize the palmitoyl-proteome in lens fiber cells. S-palmitoylation is the most common form of protein S-acylation and the reversible nature of this modification functions as a molecular switch to regulate many biological processes. This modification could play important roles in regulating protein functions and protein–protein interactions in the lens.
Methods
The palmitoyl-proteome of bovine lens fiber cells was investigated by combining acyl-biotin exchange (ABE) chemistry and mass-spectrometry analysis. Due to the possibility of false-positive results from ABE experiment, a method was also developed for direct detection of palmitoylated peptides by mass spectrometry for validating palmitoylation of lens proteins MP20 and AQP5. Palmitoylation levels on AQP5 in different regions of the lens were quantified after iodoacetamide (IAA)-palmitate exchange.
Results
The ABE experiment identified 174 potential palmitoylated proteins. These proteins include 39 well-characterized palmitoylated proteins, 92 previously reported palmitoylated proteins in other tissues, and 43 newly identified potential palmitoylated proteins including two important transmembrane proteins in the lens, AQP5 and MP20. Further analysis by direct detection of palmitoylated peptides confirmed palmitoylation of AQP5 on C6 and palmitoylation of MP20 on C159. Palmitoylation of AQP5 was found to only occur in a narrow region of the inner lens cortex and does not occur in the lens epithelium, in the lens outer cortex, or in the lens nucleus.
Conclusions
AQP5 and MP20 are among 174 palmitoylated proteins found in bovine lens fiber cells. This modification to AQP5 and MP20 may play a role in their translocation from the cytoplasm to cell membranes during fiber cell differentiation.
Keywords: S-palmitoylation, palmitoyl proteome, acyl-biotin exchange, AQP5 and MP20
Protein S-palmitoylation is an important reversible lipid modification produced by the covalent attachment of a 16-carbon palmitic acid to cysteine residues via a thioester linkage. Like other lipid modifications, the attached palmitate serves as a hydrophobic anchor for membrane targeting.1,2 Palmitoylation has also been found to regulate protein trafficking and subcellular localization3–5 and the reversible nature of this modification allows proteins to rapidly shuttle between intracellular membrane compartments.6,7 Palmitoylation also regulates protein stability,8,9 impacts the lateral distribution of proteins on the plasma membrane by targeting them to lipid rafts,10,11 as well as regulates the dynamic assembly of many neuronal proteins.12,13
Detection of protein palmitoylation in the past has been hindered due to lack of specific antibodies, the hydrophobic nature of the modification, and the labile thioester linkage.14 In early days, metabolic labeling was used for detection of palmitoylation15 and mutagenesis was used for deducing palmitoylation sites.16,17 Over the past decade, large-scale S-palmitoylation profiling using the acyl biotin exchange (ABE) method18 and fatty acid chemical reporters19 has enabled more rapid and highly sensitive detection of palmitoylated proteins. Based on these methods, a protein palmitoylation database was established suggesting that 10% or more of the human proteome is susceptible to S-palmitoylation.20
S-palmitoylation, the most common acylation of proteins, could play important roles in regulating protein functions and protein–protein interactions in the lens. To date, only a few studies have examined protein palmitoylation in the lens. In 1990, Cenedella21 reported palmitoylation of a few lens proteins by radiolabeling with [3H] palmitate, but the identities of palmitoylated proteins were not determined. Manenti et al.22 reported palmitoylation of AQP0 and alphaA-crystallin, but AQP0 was found to not be palmitoylated by Cenedella.21 Radiolabeling does not distinguish the nature of the chemical linkage of palmitate to the proteins and N-palmitoylation via an amide linkage, reported previously,23 may explain the radioactive signal detected by Manenti et al.22 Due to detection limits of the methodologies used, only a few proteins were detected to be palmitoylated in these two studies.21,22 We anticipated that many more proteins are palmitoylated in the lens.
Lens fiber cell differentiation is accompanied by extensive fiber cell elongation, programmed elimination of cytoplasmic organelles and nuclei, membrane remodeling, and expression of fiber cell-specific proteins.24–26 During these processes, several important lens proteins undergo modifications or change their cellular localization. For example, MP20 and AQP5 are both found in the cytoplasm of peripheral fiber cells and translocated to the cell plasma membrane during fiber cell differentiation.27–29 The mechanisms driving protein trafficking in lens fiber cells are not entirely elucidated. One of the important functions of palmitoylation is to target proteins to cell membranes1,3; therefore, palmitoylation could be another potential mechanism that is involved in inducing AQP5 trafficking. In this study, we applied the ABE method to screen potential palmitoylated proteins in ocular lens tissue and subsequently confirmed palmitoylation of MP20 and AQP5 using direct detection of palmitoylated peptides by mass spectrometry.
Material and Methods
Acyl-Biotin Exchange (ABE)
The ABE experiment was carried out as previously described.18 Using this method, free sulfhydryl groups after reduction are alkylated by N-ethylmaleimide (NEM). Hydroxylamine is then used to cleave the thioester bonds of palmitoylated cysteine residues, thereby generating new free sulfhydryl groups. A biotinylated thiol–reactive reagent is then added to react with free-cysteine residues and streptavidin beads are used to pulldown biotinylated proteins and to enrich for palmitoylated proteins. Finally, the biotin groups are removed and sulfhydryl groups are alkylated by iodoacetamide.
Specific experimental details for our study are as follows: a frozen 1-year-old bovine lens (Pel-Freez Biologicals, Rogers, AK, USA) was decapsulated and the outer 2-mm thick tissue was isolated from the surface of the lens. Tissue was homogenized in homogenizing buffer (HM buffer) containing 25 mM Tris, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 150 mM NaCl, and 50 mM NEM, pH 7.4. The sample was centrifuged at 33,000g for 20 minutes and the supernatant was discarded. The pellets were washed twice with HM buffer to generate the water-insoluble fraction (WIF). The WIF (1.5 mg) was dissolved in 1 mL of 4% SDS in 25 mM Tris, 5 mM EDTA, 1 mM PMSF, 10 mM tris(2-carboxyethyl)phosphine (TCEP), and 150 mM NaCl and incubated at room temperature for 45 minutes. Then the sample was alkylated by 50 mM NEM at 4°C overnight followed by two sequential chloroform/methanol precipitations to remove excess NEM. Precipitated proteins were solubilized in 800 μL of 25 mM Tris, 5 mM EDTA, 1 mM PMSF, and 150 mM NaCl, 2M urea, and 4% SDS. Two hundred μL of 10 mM N-[6-(Biotinamido) hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP; ThermoFisher Scientific, Rockford, IL, USA) was added. The sample was then divided into two portions and one was mixed with 500 μL of 1 M hydroxylamine in 0.5 M Tris (pH 7.2) and the other was mixed with 500 μL of 0.5 M Tris buffer (pH 7.2). The samples were incubated at room temperature for 1 hour. The proteins were precipitated twice as described above and the pellets were dissolved in 50 μL of 4% SDS in HM buffer and then diluted by 1950 μL of HM buffer containing of 0.2% Triton X-100. The samples were centrifuged to remove particulates and added to 200 μL of streptavidin-agarose beads. The beads were incubated at room temperature for 90 minutes and washed eight times with HM buffer containing 0.1% SDS and 0.2% Triton X-100. The bound proteins were eluted by incubation with 300 μL of elution buffer (25 mM Tris, 5 mM EDTA, 1 mM PMSF, and 150 mM NaCl, 0.1% SDS, 0.2% Triton X-100 and 10 mM dithiothreitol) at 56°C for 10 minutes. Beads were then further washed with 300 μL elution buffer. The eluant and wash were pooled together and iodoacetamide was added to 100 mM. The samples were incubated at 25°C in the dark for 1 hour and speedvac concentrated to 100 μL. The proteins were precipitated as described above and suspended in 100 μL of 10% acetonitrile (ACN) in 50 mM Tris, pH 8.0. Trypsin (1 μg; ThermoFisher Scientific) was added and the sample was incubated at 37°C for 18 hours. The samples were then dried in a speedvac and reconstituted in 0.1% formic acid prior to LC-MS/MS analysis. This experiment was repeated three times using three different lenses.
Gel Electrophoresis, Immunoblotting, and In-Gel Digestion
The WIF from a bovine lens cortex was prepared as described for the ABE experiment. The WIF was washed with 8 M urea once. The sample was reduced at 25°C for 1 hour in 100 μL 8 M urea containing 20 mM TCEP and then alkylated by 50 mM NEM at 25°C for 1 hour. The sample was diluted to 1 mL with 8 M urea and centrifuged at 33,000g for 20 minutes to obtain the urea-insoluble fraction (UIF). The UIF was also prepared as above using two 32-year-old human lens cortex samples (1-mm thick tissue from the outer lens surface) from different donors (NDRI, Philadelphia, PA, USA). The pellets were then dissolved in 5% SDS and mixed with lithium dodecyl sulfate sample buffer and loaded onto a 4-12% NuPAGE Novex Bis-Tris gel (ThermoFisher Scientific). MOPS running buffer was used for separation. The sample was either run through a full SDS-PAGE gel or run into the stacking gel for only 1 cm. After SDS-PAGE, gels were stained with GelCode Blue Safe Protein Stain (ThermoFisher Scientific). Western blots for both AQP5 and AQP0 were done to confirm the position of AQP5 in the gel relative to AQP0. For Western blots, the proteins in the gel were electrophoretically transferred to a nitrocellulose membrane. The membrane was blocked with 5% non-fat milk in TBST (25 mM Tris-buffered saline, 0.1% Tween 20, pH 8.0) for 1 hour. The AQP0 Western blot was done using goat anti-AQP0 extracellular domain primary antibody (Santa Cruz Biotechnology, Dallas, TX, USA) and IRDye 800CW donkey anti-goat IgG (LI-COR Biosciences, Lincoln, NE, USA) as the secondary antibody. After washing away the donkey anti-goat secondary antibody, the AQP5 Western blot was done on the same membrane using rabbit anti-AQP5 primary antibody (Abcam, Cambridge, MA, USA) and goat anti-rabbit secondary antibody labeled with Dylight 680 (ThermoFisher Scientific). The results were read using an Odyssey Infrared Imager (LI-COR Biosciences).
To identify proteins in the gel, gel bands, or the whole stained area of the short stack gel were excised. Proteins in the gel were digested by either endoproteinase Lys-C or trypsin (ThermoFisher Scientific) and the peptides were extracted from the gel as previously described.30 The peptide extraction procedure was modified for direct detection of palmitoylated peptides as follows: peptides were extracted using 0.1% formic acid twice and 5% ACN (0.1% formic acid) twice. The remaining gel pieces were extracted with 15% ACN (0.1% formic acid) three times, 25% ACN (0.1% formic acid) three times, and 50% ACN (0.1% formic acid) three times. Each extraction was done for 30 minutes. To identify palmitoylated AQP5 peptides, 25% and 50% ACN extracts were pooled and dried in a speedvac and reconstituted in 50% ACN (0.1% formic acid). The extract was diluted to 20% ACN (0.1% formic acid) and loaded on a C8 trap column for LC-MS/MS analysis. To identify the palmitoylated MP20 peptides, 50% ACN extracts were pooled and dried in a speedvac and reconstituted in 50% ACN (0.1% formic acid) and directly loaded onto a C4 trap column for LC-MS/MS analysis.
Iodoacetamide (IAA)-Palmitate Exchange and Quantification of Palmitoylation Levels in Different Lens Regions
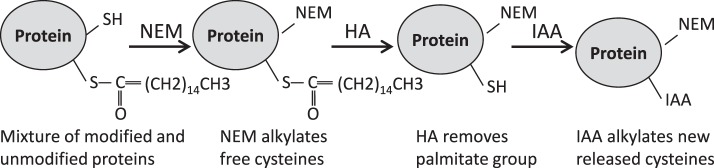
IAA-palmitate exchange was done as shown in Figure 1 to alkylate nonmodified cysteines by NEM and to alkylate palmitoylated cysteines by IAA after hydroxylamine removal of the palmitate moiety from palmitoylated proteins. This method was applied for both quantification of palmitoylation levels of AQP5 in different lens regions and for the study of potential palmitoylation sites in MP20. To quantify the levels of palmitoylation in the different lens regions, a frozen bovine lens was decapsulated and different lens regions were isolated as described previously30 by vortexing the lens in 25 mM Tris, 5 mM EDTA, 150 mM NaCl, 50 mM NEM, 1 mM PMSF, pH 7.4 for 5 seconds. Normalized lens distances were defined as previously described.30 The resulting samples were vortexed and centrifuged at 200,000g for 20 minutes and the supernatants were discarded. The pellets were washed with HM buffer once and reduced at 25°C for 1 hour in 100 μL of 8 M urea containing 20 mM TCEP and alkylated by 100 mM NEM at 25°C for 1 hour. The samples were diluted with 1 mL of 4 M urea and centrifuged at 200,000g for 20 minutes. The pellets were washed with water three times and suspended in 1 M hydroxylamine (HA) in 1M Tris buffer and incubated at 25°C for 1 hour. The samples were centrifuged to remove HA and washed with water twice. The pellets were then suspended in 100 μL of 8 M urea containing 55 mM IAA at 25°C in the dark for 45 minutes. The samples were then diluted with water to 4 M urea and centrifuged at 200,000g for 20 minutes to remove excess reagents to obtain UIF. The urea-insoluble pellets were dissolved in 5% SDS for SDS-PAGE analysis. For the IAA-palmitate exchange experiment to detect MP20 palmitoylation, the lens cortical UIF was prepared and separated by SDS-PAGE and the experiment for removal of the palmitate moiety was done in the gel. Briefly the corresponding gel bands were washed with 50 mM ammonium bicarbonate buffer twice and incubated with 100 μL of 1M hydroxylamine (HA) in 1M Tris buffer (pH7.2) at 25°C for 1 hour. The samples were then washed with 50 mM ammonium bicarbonate buffer twice and incubated with 100 μL 100 mM IAA at 25°C in the dark for 1 hour. The gel bands were then processed for in-gel digestion. One set of gel bands was digested by trypsin as described above and the other set was digested with chymotrypsin (Promega, Madison, WI, USA) at 37°C for 1 hour. The peptides were extracted as described in the Gel Electrophoresis, Immunoblotting, and In-Gel Digestion section and analyzed by LC-MS/MS.
Figure 1.
IAA-palmitate exchange. IAA-palmitate exchange was performed following the steps in this flow chart. The nonpalmitoylated cysteines in the sample were alkylated with NEM. The cysteines that are blocked by palmitate were not alkylated by NEM. After NEM alkylation, the excess NEM in the samples was removed and hydroxylamine was then used to remove palmitate from palmitoylated proteins and to expose new cysteine thiol groups that were then alkylated with IAA. Therefore, if all alkylation steps go to completion, all NEM alkylated cysteines were not palmitoylated and all IAA alkylated cysteines were from the palmitoylated cysteines (ignoring other thioester linkage modifications on cysteines).
LC-MS/MS Analysis
Samples from the ABE experiment were analyzed by MudPIT as previously described.31 A six-step salt pulse gradient (50, 100, 200, and 500 mM and 1 and 2 M ammonium acetate) was used. For direct detection of palmitoylated AQP5 peptides, samples were pressure loaded on a C8 trap column (50 mm × 100 μm) packed with Phenomenex Jupiter resin (5-μm mean particle size, 300 Å pore size) and separated on a one-dimensional fused silica capillary column (250 mm × 100 μm) packed with Phenomenex Jupiter resin (C8, 5-μm mean particle size, 300 Å pore size). The LC gradient was run as follows: 0 to 25 minutes: 2% to 30% ACN (0.1% formic acid), 25 to 55 minutes: 35% to 98% ACN (0.1% formic acid), 55 to 80 minutes: 98% ACN (0.1% formic acid) balanced with 0.1% formic acid. For direct detection of palmitoylated MP20 peptides, samples were pressure loaded on a C4 trap column (50 mm × 100 μm) packed with Phenomenex Jupiter resin (5-μm mean particle size, 300 Å pore size) and separated on a one-dimensional fused silica capillary column (250 mm × 100 μm) packed with Phenomenex Jupiter resin (C4, 5-μm mean particle size, 300 Å pore size). The LC gradient was run as follows: 0 to 2 minutes: 30% ACN (0.1% formic acid), 2 to 55 minutes: 30% to 98% ACN (0.1% formic acid), 55 to 80 minutes: 98% ACN (0.1% formic acid) balanced with 0.1% formic acid. For all other samples, samples were pressure loaded onto a C18 trap column (50 mm × 100 μm) packed with Phenomenex Jupiter resin (5-μm mean particle size, 300 Å pore size) and separated on a one-dimensional fused silica capillary column (250 mm × 100 μm) packed with Phenomenex Jupiter resin (C18, 3-μm mean particle size, 300 Å pore size) using the following gradient at a flow rate of 0.5 μL/min: 0 to 10 minutes: 2% ACN (0.1% formic acid), 10 to 50 minutes: 2% to 35% ACN (0.1% formic acid), 50 to 60 minutes: 35% to 90% ACN (0.1% formic acid) balanced with 0.1% formic acid. For all experiments, the eluate from analytical column was directly infused into a Velos Pro mass spectrometer (ThermoFisher Scientific) equipped with a nanoelectrospray source. The instrument was operated in data dependent mode with one precursor scan event (mass-to-charge ratio [m/z] 300–2000) to identify the top 15 most abundant ions for fragmentation by collision-induced dissociation with normalized collision energy of 35. The automatic gain control (AGC) was 3e4 for MS1 and 1e4 for MS2. Dynamic exclusion (exclude after 1 spectrum, release after 15 seconds, and exclusion list size of 300) was enabled. Singly charged ions were not excluded. To quantify the level of palmitoylation, the instrument was operated in a targeted mode only for nine AQP5 peptides, including NEM or IAA alkylated AQP5 peptide 4-12. To further confirm palmitoylation on MP20, the sample was run on a Q Exactive Instrument (ThermoFisher Scientific) with the same column and LC conditions described above. The data-dependent instrument method consisted of MS1 acquisition (m/z: 350–1800, R = 70,000), using an MS AGC target value of 1e6, followed by up to 15 MS/MS scans (R = 17,500) of the most abundant ions detected in the preceding MS scan. The MS2 AGC target value was set to 2e5 ions, with a maximum ion time of 200 ms, and a 5% underfill ratio, and intensity threshold of 5e4. Higher energy collisional dissociation was set to 27, dynamic exclusion was set to 5 seconds, and peptide match and isotope exclusion were enabled. Singly charged ions were not excluded.
Data Analysis
Protein database searching was done as previously described32 against a concatenated forward and reversed bovine uniprot database (September 2016, 32,523 entries in the forward database). Other than modifications described previously,32 variable modifications of cysteines by NEM and IAA were used. The MVH and Xcorr values were used for score optimization for ABE experiments and results were filtered to a 1% peptide false-discovery rate (FDR) with the requirement for a minimum of two peptides per reported protein using IDPicker.33 For palmitoylated peptide identification, one peptide was required to identify a protein because nonmodified peptides were removed from the sample, but the data were manually verified. For quantifying the level of palmitoylation, the AQP5 peptide 4-12 with either NEM or IAA alkylation on C6 was targeted for fragmentation. The resulting raw files were imported into Skyline34 for peak-picking and quantitation. Twelve product ions (y2+, y3+, y4+, y5+, y6+, y7+, y72+, b2+, b5+*, b6+*, b7+*, b8+*) from the AQP5 peptide 4-12 were used. Asterisks indicate b-ions that lose a water molecule. The total peak areas (A) from these product ions for AQP5 peptide 4-12 with either NEM or IAA alkylation on C6 were obtained. The level of palmitoylation was defined as AIAA/(AIAA+ANEM).
For data from ABE experiments, the spectral counts for HA-treated samples were normalized by the corresponding control based on the spectral counts of αB-crystallin because αB-crystallin is a major lens protein and that does not contain cysteine residues. After normalization, the spectral counts for all three biological replicates were averaged. Statistical analysis was then performed as previously described35 to obtain a threshold ratio for high-confidence identification of palmitoylated proteins. Because quantification for proteins with low-spectral counts is not accurate, statistical analysis was only performed on the proteins with average spectral counts of at least three in the HA-treated group. As reported previously,35 all zeros in the control group were replaced with 1 to avoid division by zero. Ratios (HA+/HA−) of spectral counts were calculated and the protein-wise Log2 of spectral count ratios were clustered using the Gaussian mixture model by XLSTAT software in Excel (Microsoft, Redmond, WA, USA). The expectation-maximization (EM) algorithm was chosen for modeling the data with the Bayesian information criterion option selected.
Results
The Palmitoyl-Proteome of Lens Fiber Cells From ABE Experiments
Palmitoylated proteins in bovine lens fiber cells were enriched in all three biological replicates by ABE experiments. After removing all keratins, 1386 proteins were detected in three samples with at least two distinct peptides. Four hundred ninety proteins had at least an average spectral count of three in the HA-treated group and these proteins were clustered based on the spectral count ratio in HA-treated and nontreated samples resulting in two Gaussian components shown in Figure 2A. The left and right Gaussian components represent the Log2-ratio distributions of contaminating protein-dominant and palmitoylated protein-dominant protein groups, respectively. The cutoff ratio (HA+/HA−) for putative palmitoylated proteins was calculated to be 4.2 based on the left Gaussian component using a significance level of 0.05. The spectral count distribution for 490 proteins in both control and HA-treated sample can be found in Figure 2B. Highly confident palmitoylated proteins appear close to the x-axis and have ratio (HA+/HA−) greater than 4.2. Previously identified palmitoylated proteins are highlighted in color. Synaptobrevin homolog (YKT6) is the only well-known palmitoylated protein that was detected in the sample, but failed to pass the cutoff criteria. YKT6 also failed to be detected in a previous study,18 probably due to low level of palmitoylation.
Figure 2.
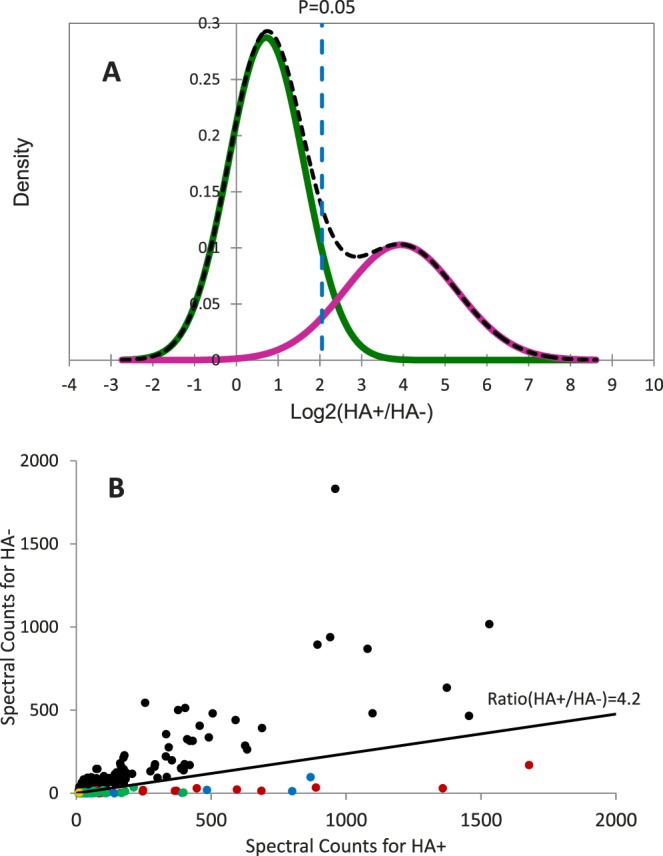
Global analysis of palmitoylated proteins in lens fiber cells membranes. Palmitoylated proteins were ABE-enriched and analyzed by mass spectrometry. The data were clustered based on a Bayesian information criterion-based Gaussian mixture model and resulted in two Gaussian distributions (A). The left and right Gaussian components represent the Log2-ratio distributions of contaminating protein-dominant and palmitoylated protein-dominant protein groups, respectively. Proteins with Log2 (HA+/HA−) > 2.07 (corresponding to HA+/HA− > 4.2, P < 0.05) were treated as palmitoylated proteins with high confidence. The spectral count distribution of 490 proteins that was used for statistical analysis is also plotted (B). Proteins that were not identified as palmitoylated proteins are shown in black and newly identified palmitoylated proteins are shown in green. Palmitoylated proteins that are reported in the Swiss Protein database are shown in red and potential palmitoylated proteins that reported in the SwissPalm database are shown in blue. The only known palmitoyated protein YKT6 was detected (shown in orange), but its HA+/HA− ratio did not reach the cutoff threshold to be considered as a palmitoylated protein.
Using a significance level of 0.05, we report 174 proteins identified as palmitoylated proteins with high confidence (Supplementary Table S1). Among these proteins, 39 have been reported as S-palmitoylated proteins in the uniprot protein database and their sites of palmitoylation have been identified or predicted. An additional 92 proteins have been previously reported as potential palmitoylated proteins.20 The remaining 43 proteins have not been reported as palmitoylated proteins. Potential palmitoylation sites in these 43 proteins were analyzed by CSS-PALM 3.036 and 33 proteins were predicted to have at least one palmitoylation site based on three types of palmitoylation (-CC- pattern, -CXXC- pattern, and others). Thus, these 33 proteins are very likely to be palmitoylated in the lens (Table 1). Additionally, Table 2 lists top 25 proteins ranked by spectral counts and excluding proteins listed in Table 1 that have been previously reported as palmitoylated proteins. Some of these proteins are important and highly abundant in the lens fiber cells. Others are not abundant lens proteins and the high spectral count detected in the sample suggests these proteins undergo significant palmitoylation in lens fiber cells.
Table 1.
Newly Identified Potential Palmitoylated Proteins in the Lens Fiber Cells
Table 2.
Top 25 Palmitoylated Proteins Detected in Bovine Lenses*†
Among the 43 new potential palmitoylated proteins, two important transmembrane proteins in the lens fiber cells appear, AQP5 and lens fiber membrane intrinsic protein (MP20) also known as LIM2.37 Considering the possibility of false positive identification using the ABE method, palmitoylation of AQP5 and MP20 was further confirmed by direct detection of palmitoylated peptides using mass spectrometry.
Direct Detection of Palmitoylated AQP5 Peptides
Direct detection of palmitoylated peptides by mass spectrometry is a reliable way to confirm protein palmitoylation and to identify the site(s) of palmitoylation. However, direct detection of protein palmitoylation is challenging due to the hydrophobic nature of the modification. Furthermore, detection of the palmitoylated AQP5 peptides in the lens is expected to be more difficult due to interference from highly abundant AQP0 signals. To reduce interference from AQP0, the lens membrane fraction was separated by SDS-PAGE and Western blots for both AQP5 and AQP0 were performed to locate AQP5 in the gel relative to AQP0 (Fig. 3). Although both proteins have similar molecular weights, SDS-PAGE separates AQP5 from the majority of AQP0; however, AQP5 co-localizes with the truncated AQP0 band. Additional steps to reduce interference from AQP0 and other nonmodified peptides include digestion by Lys-C and separation of hydrophobic fatty acylated peptides from abundant hydrophilic peptides using solubility differences. Lysine residues are only found in the C-terminal region of AQP0; therefore, Lys-C digestion only provides hydrophilic AQP0 C-terminal peptides that can be removed from the gel by hydrophilic solvent extraction. Hydrophobic fatty acylated peptides together with other hydrophobic peptides remain in the gel and can then be extracted with more hydrophobic solvents. After optimization of the sample preparation and analysis methods, palmitoylated AQP5 peptide 4-12 was detected in bovine lenses (Fig. 4, top panel). Figure 4 shows signals matching both the parent ion and a fragment ion. Palmitoylation on C6 is confirmed by the tandem mass spectrum in Figure 5A. For comparison, a tandem mass spectrum for nonmodified AQP5 peptide 4-12 with C6 alkylated by NEM is shown in Figure 5B. Similar fragment ions can be detected when comparing the tandem mass spectrum of the nonmodified and modified peptide. The tandem mass spectrum of the palmitoylated peptide contains a fragment corresponding to a neutral loss of 238 Da (C16H30O), a characteristic of S-palmitoylated peptides.38 Additional loss of water may be due to facile water loss from N-terminal glutamic acid as has been reported previously.39 B-ions of the AQP5 4-12 peptide also show significant water loss that can be seen in tandem mass spectra of both the nonmodified and modified peptides.
Figure 3.

Separation of AQP0 and AQP5 by SDS-PAGE. The lens membrane fraction was separated by SDS-PAGE and stained by Coomassie blue (A). Western blots for AQP0 and AQP5 were done to show the position of AQP5 in the gel relative to AQP0 band. (B) Western blot for AQP5 (green); (C) Western blot for both AQP0 (red) and AQP5 co-localized with truncated AQP0 (yellow).
Figure 4.

Detection of palmitoylated AQP5 peptide 4-12 in both bovine and human lenses. Hydrophobic Lys-C peptides from AQP5 gel bands were analyzed by LC-MS/MS. Extracted ion chromatograms corresponding to a parent ion (m/z: 617.38) and one fragment ion (m/z: 478.5) were plotted. Asterisks indicate the peaks representing the palmitoylated AQP5 peptide 4-12 confirmed by tandem mass spectra at the corresponding retention time. The experiments were repeated in both bovine (A) and human lenses (B). There are several unidentified peaks in human samples; however, the tandem mass spectra at the corresponding retention time indicate those peaks are not from palmitoylated AQP5. (C) Protein alignment for the N-terminus of AQP5 in different species.
Figure 5.
Tandem mass spectra for AQP5 peptide 4-12 with or without a palmitoyl group. Tandem mass spectra of palmitoylated (A) and nonpalmitoylated (B) AQP5 4-12 are shown. The b- and y- ions are labeled. Asterisks indicate ions that lost a water molecule. #Indicates modification with palmitate (ΔM = 238.2297 Da); $Indicates modification with NEM (ΔM = 125.0477 Da).
The N-terminal sequence of AQP5 is highly conserved between species (Fig. 4C); therefore, palmitoylation on C6 of AQP5 was expected to be present in lens tissue from other species. For example, human AQP5 has the identical N-terminal sequence as bovine AQP5. Palmitoylated AQP5 4-12 was detected in both human lenses and the signal from one lens can be seen in Figure 4B and the modification was confirmed by tandem mass spectrometry (data not shown). Palmitoylation on other cysteine residues in AQP5 was not detected.
Direct Detection of Palmitoylated MP20 Peptides
The bovine MP20 sequence contains six cysteines. Considering palmitoylation in transmembrane proteins normally occurs on juxtamembranous cysteine residues,40,41 only C159 and C166 were the selected targets for our palmitoylation analysis. Using a method similar to that used for AQP5 palmitoylation detection, the lens membrane fraction was separated by SDS-PAGE to obtain gel bands corresponding to MP20. Signal from MP20 peptides was detected in two gel bands shown in Figure 6E. A tryptic peptide including C166 was easily detected, but palmitoylation on C166 was not detected. To further confirm that C166 was not palmitoylated, IAA-palmitate exchange was done and the result is shown in Figures 6A and 6B. IAA-alkylated C166 was not detected in either gel bands and C166 was exclusively alkylated with NEM in both gel band 1 (Fig. 6A) and gel band 2 (Fig. 6B). This result further suggested that C166 was not palmitoylated. This experiment was also done for C159. Due to poor signal from the long and hydrophobic C159 containing tryptic peptide, chymotrypsin was then used for digestion. Figures 6C and 6D show that C159 was alkylated by IAA only in band 1 (solid line), but C159 was exclusively alkylated by NEM in band 2 (dotted line). These results suggest that MP20 in band 1 was palmitoylated on C159 and MP20 in band 2 was not palmitoylated.
Figure 6.
IAA-palmitate exchange of MP20. Lens membrane proteins were separated by SDS-PAGE and MP20 signal was detected in gel bands 1 and 2. Alkylation of C166 by IAA was not detected and C166 was exclusively alkylated by NEM in both band 1 (A) and band 2 (B). NEM alkylated C159 was not detected in band 1 ([D] solid line). C159 in band 1 was only alkylated by IAA ([C] solid line) suggesting C159 in this gel band is palmitoylated. C159 in band 2 was exclusively alkylated by NEM ([D] dotted line) and no IAA alkylated C159 was detected in band 2 ([C] dotted line). These results indicate C166 is not palmitoylated, but C159 is partially palmitoylated in bovine lens.
Direct detection of MP20 palmitoylation from in-gel digestion as described above was unsuccessful most likely due to the peptide length and hydrophobic nature of the MP20 tryptic peptide. Therefore, the lens membrane fraction was run into a short-stacking gel (1 cm) with larger pore sizes that assisted with peptide extraction. Using this method, several palmitoylated MP20 peptides were detected (peptide 130-167, 131-168, 136-167) supporting palmitoylation on C159. The sample was further analyzed using a Q Exactive instrument to obtain accurate masses for both parent ions and fragment ions to rule out the possibility of other modifications. The Q Exactive data showed both parent and fragment ions had measured masses falling within expected mass accuracy (1.67 ppm for the parent ion and <10 ppm for all fragment ions). A tandem mass spectrum for the palmitoylated peptide 130-167 is shown in Figure 7. The y2 fragment ion reveals NEM alkylated C166 and indicates palmitoylation on C159.
Figure 7.
Tandem mass spectrum of palmitoylated MP20 peptide 130-167. Tandem mass spectrum of palmitoylated MP20 peptide 130-167 from a Q Exactive instrument is shown. The monoisotopic b- and y- ions are labeled. All fragment ions match their theoretical masses within 5 ppm except y20 (9.6 ppm). The y2 ion indicates NEM alkylation on C166 (2.98 ppm); therefore, the palmitate moiety is on C159.
Quantification of Palmitoylation Levels in Different Lens Regions
Different lens regions were isolated and the relative level of palmitoylated AQP5 in each sample was measured. To obtain enough samples for LC-MS/MS analysis from the lens epithelium as well as from very young fiber cells that were attached to the lens capsule upon removal, cells from three to five capsules were pooled together. The experiments were repeated three times and no palmitoylation was detected in these samples. The level of AQP5 palmitoylation measured in different lens regions can be found in Figure 8, where if the normalized lens distance difference was less than 0.1, the level of palmitoylation was averaged. Even though the absolute level of palmitoylation shows variability among three lenses, which is probably due to the mixing effect of different regions collected in different lenses, the overall trend among three lenses is similar. This result indicated that palmitoylation was lower in the peripheral lens fiber cells and increased significantly around the normalized lens distance of 0.9 and then sharply decreased and disappeared in the nucleus. This result indicates that palmitoylation of AQP5 only occurs in a narrow region of the lens.
Figure 8.

Quantification of the level of AQP5 palmitoylation in the different regions of bovine lenses. The levels of palmitoylation on AQP5 were measured after NEM alkylation of nonmodified cysteines and IAA alkylation of palmitoylated cysteines. The experiments were repeated three times using three different bovine lenses. For the samples collected from regions with similar normalized lens distance (the difference is <0.1), the level of palmitoylation was averaged; otherwise, data are shown from a single experiment. A trend line was obtained by fitting the data using the equation 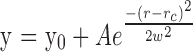 . Origin where y0 represents the baseline, “A” is the maximum level of modification, and rc indicates where the maximum palmitoylation occurs. The results indicate that AQP5 palmitoylation is absent in epithelial cells, very young peripheral fiber cells (data not shown in this figure), and the nucleus, but increases in the cortex. Asterisks indicate that measurements were made in these regions, but no palmitoylation was detected.
. Origin where y0 represents the baseline, “A” is the maximum level of modification, and rc indicates where the maximum palmitoylation occurs. The results indicate that AQP5 palmitoylation is absent in epithelial cells, very young peripheral fiber cells (data not shown in this figure), and the nucleus, but increases in the cortex. Asterisks indicate that measurements were made in these regions, but no palmitoylation was detected.
Discussion
ABE together with mass spectrometry techniques have been widely used for global palmitoylproteome studies since it was first introduced in 2006.18 This method avoids traditional [3H] palmitate labeling; a process involving long autoradiographic exposures. Global palmitoylproteome analysis greatly accelerates the identification of new palmitoylated proteins resulting in a rapid expansion in our understanding of the function and mechanism of protein palmitoylation. Previous studies of lens protein palmitoylation have been limited to a small group of lens proteins. The results presented in this paper indicate protein palmitoylation is prevalent in lens fiber cells where a total of 174 palimitoylated proteins were detected. Based on DAVID Bioinformatic Resources,42 several biological processes are enriched in the list of highly confident palmitoylated proteins detected in the lens, including protein folding, cell adhesion, regulation of cell shape, vesicle fusion, and establishment of protein localization to plasma membrane. Considering the many important processes that palmitoylation is involved in, such as protein trafficking, protein aggregation, and membrane-domain association, it is expected that palmitoylation plays important functions in the lens. S-palmitoylation is catalyzed by a family of palmitoyl acyltransferases (PATs).43 The enzymes that are responsible for protein palmitoylation in the lens have not been studied and it will be interesting to know whether there is spatial distribution of these enzymes in the lens. Although direct study of the functional consequences of palmitoylation of lens proteins has not been done, results from previous studies have suggested important roles. For example, paralemmin-1 and -2 were detected with high-spectral counts in our ABE experiment indicating they are highly palmitoylated. The appearance of paralemmins in lens fiber cells has been reported previously to coincide with the appearance of interlocking protrusions, structures that are required for maintaining lens structural order, stability, and transparency.44 Paralemmins have been shown to induce protrusions in a palmitoylation-dependent manner in other tissues.45
One of the major foci of studies on protein palmitoylation is the role in promoting lipid raft association. The palmitoyl moiety could cause protein association with lipid-raft domains through specific affinity of palmitate to other lipids in raft domains.46 Furthermore, palmitoylation of juxtamembranous cysteines affects the conformation of transmembrane domains and affects the effective length of hydrophobic segments, thereby inducing association with lipid-raft domains through membrane thickness.46 Comparing the list of palmitoylated proteins identified with high confidence with proteins detected in lipid rafts of lens fiber cells,32 46% of identified palmitoyl proteins are present in the lipid raft protein list. Therefore, palmitoylation could be important for targeting many proteins to lipid raft domains in lens fiber cell membranes.
AQP5 is an important water channel protein that is primarily localized to secretory glands and is involved in fluid secretion.47 AQP5 is also expressed in several nonglandular tissues, such as cornea and lens.48,49 In lens fiber cells, AQP5 is localized in the cytoplasm in young fiber cells and is translocated to cell membranes in older cells.27,29 The signaling pathways that trigger this translocation in the lens remain unclear; however, translocation of AQP5 to the cell membrane in older fiber cells where AQP0 is truncated could compensate for any water permeability changes induced by AQP0 C-terminal truncation.27,29 Quantification of AQP5 palmitoylation indicates that palmitoylation does not occur in lens epithelial cells or in very young fiber cells, but occurs in a narrow region of the lens cortex and could correlate with AQP5 translocation to the cell membrane. Therefore, AQP5 palmitoylation could be a one of the potential mechanisms for inducing AQP5 trafficking. Similar to AQP5, MP20 also undergoes membrane trafficking in the lens28 and the mechanism of MP20 trafficking in fiber cells has not been studied. Palmitoylation could also play important role for inducing MP20 trafficking. Interestingly, both AQP5 and MP20 were detected in the lipid raft domains of lens fiber cells. Thus, palmitoylation could contribute to the association of these proteins with lipid rafts. Further study is needed to confirm the function of AQP5 and MP20 palmitoylation. It will be interesting to know if palmitoylation of AQP5 also occurs in other tissues.
Direct detection of lipidated peptides by mass spectrometry is challenging due to the typical low stoichiometry and highly hydrophobic nature of such a modification. The method applied in this paper enriches lipidated peptides by removing hydrophilic nonlipidated peptides based on solubility difference. Hydrophobic-lipid contaminants in the sample can be easily removed by using in-gel digestion or chloroform-methanol precipitation. Many myristoylated, palmitoylated, and geranyl-geranylated peptides were detected in the samples (data not shown). Thus, this method appears to be useful as a sample preparation method for direct detection of lipidated peptides from a complex mixture of proteins. Under this enrichment method, S-palmitoylation of AQP0 was not detected; a result that agrees with our ABE result. In addition, CSS-PALM36 does not predict the presence of S-palmitoylation sites in AQP0. Therefore, our results strongly suggest that the major lens membrane protein AQP0 is not palmitoylated on cysteine residues. Because AQP0 is highly palmitoylated or acylated by other fatty acids through an amide linkage,23 metabolic labelling for detection of palmitoylation could result in a positive result as shown in previous study.22
Acknowledgments
Supported by National Institutes of Health Grants EY013462 and P30EY 008126 (Bethesda, MD, USA).
Disclosure: Z. Wang, None; K.L. Schey, None
References
- 1.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Fisher DA, Storm DR. Analysis of the palmitoylation and membrane targeting domain of neuromodulin (GAP-43) by site-specific mutagenesis. Biochemistry. 1993;32:10714–10719. doi: 10.1021/bi00091a023. [DOI] [PubMed] [Google Scholar]
- 3.Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808:2981–2994. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 5.Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mumby SM. Reversible palmitoylation of signaling proteins. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 7.Hornemann T. Palmitoylation and depalmitoylation defects. J Inherit Metab Dis. 2015;38:179–186. doi: 10.1007/s10545-014-9753-0. [DOI] [PubMed] [Google Scholar]
- 8.Valdez-Taubas J, Pelham H. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanai A, Huang K, Kang R, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. J Biol Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 12.Washbourne P, Cansino V, Mathews JR, Graham M, Burgoyne RD, Wilson MC. Cysteine residues of SNAP-25 are required for SNARE disassembly and exocytosis, but not for membrane targeting. Biochem J. 2001;357:625–634. doi: 10.1042/0264-6021:3570625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.el-Husseini Ael-D, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nat Rev Neurosci. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- 14.Ji Y, Leymarie N, Haeussler DJ, Bachschmid MM, Costello CE, Lin C. Direct detection of s-palmitoylation by mass spectrometry. Anal Chem. 2013;85:11952–11959. doi: 10.1021/ac402850s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin BR. Non-radioactive analysis of dynamic protein palmitoylation. Curr Protoc Protein Sci. 2013;73(unit 14.15) doi: 10.1002/0471140864.ps1415s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson LJ, Michel T. Mutagenesis of palmitoylation sites in endothelial nitric oxide synthase identifies a novel motif for dual acylation and subcellular targeting. Proc Natl Acad Sci U S A. 1995;92:11776–11780. doi: 10.1073/pnas.92.25.11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzo MM, Sánchez-Puig JM, Blasco R. Mutagenesis of the palmitoylation site in vaccinia virus envelope glycoprotein B5. J Gen Virol. 2012;93:733–743. doi: 10.1099/vir.0.039016-0. [DOI] [PubMed] [Google Scholar]
- 18.Roth AF, Wan J, Bailey AO, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc M, David F, Abrami L, et al. SwissPalm: protein palmitoylation database. F1000Res. 2015;4:261. doi: 10.12688/f1000research.6464.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cenedella RJ. Palmitoylation of ocular lens membrane proteins. Invest Ophthalmol Vis Sci. 1990;31:368–373. [PubMed] [Google Scholar]
- 22.Manenti S, Dunia I, Benedetti EL. Fatty acid acylation of lens fiber plasma membrane proteins: MP26 and α-crystallin are palmitoylated. FEBS Lett. 1990;262:356–358. doi: 10.1016/0014-5793(90)80228-b. [DOI] [PubMed] [Google Scholar]
- 23.Schey KL, Gutierrez DB, Wang Z, Wei J, Grey AC. Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry. 2010;49:9858–9865. doi: 10.1021/bi101415w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim JC, Walker KL, Sherwin T, Schey KL, Donaldson PJ. Confocal microscopy reveals zones of membrane remodeling in the outer cortex of the human lens. Invest Ophthalmol Vis Sci. 2009;50:4304–4310. doi: 10.1167/iovs.09-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wride MA. Lens fiber cell differentiation and organelle loss: many paths lead to clarity. Philos Trans R Soc Lond B Biol Sci. 2011;366:1219–1233. doi: 10.1098/rstb.2010.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp Eye Res. 2009;88:133–139. doi: 10.1016/j.exer.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrova RS, Schey KL, Donaldson PJ, Grey AC. Spatial distributions of AQP5 and AQP0 in embryonic and postnatal mouse lens development. Exp Eye Res. 2015;132:124–135. doi: 10.1016/j.exer.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grey AC, Jacobs MD, Gonen T, Kistler J, Donaldson PJ. Insertion of MP20 into lens fibre cell plasma membranes correlates with the formation of an extracellular diffusion barrier. Exp Eye Res. 2003;77:567–574. doi: 10.1016/s0014-4835(03)00192-1. [DOI] [PubMed] [Google Scholar]
- 29.Petrova RS, Webb KF, Vaghefi E, Walker K, Schey KL, Donaldson PJ. Dynamic functional contribution of the water channel AQP5 to the water permeability of peripheral lens fiber cells. Am J Physiol Cell Physiol. 2018;314:C191–C201. doi: 10.1152/ajpcell.00214.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Schey KL. Identification of a direct Aquaporin-0 binding site in the lens-specific cytoskeletal protein filensin. Exp Eye Res. 2017;159:23–29. doi: 10.1016/j.exer.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Hill S, Luther JM, Hachey DL, Schey KL. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT) Proteomics. 2012;2:329–338. doi: 10.1002/pmic.201100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Schey KL. Proteomic analysis of lipid raft-like detergent-resistant membranes of lens fiber cells. Invest Ophthalmol Vis Sci. 2015;56:8349–8360. doi: 10.1167/iovs.15-18273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma ZQ, Dasari S, Chambers MC, et al. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowal L, Yang W, Freeman MR, Steen H, Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood. 2011;118:e62–e73. doi: 10.1182/blood-2011-05-353078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieuallen K, Christensen M, Brandriff B, Church R, Wang J, Lennon G. Assignment of the human lens fiber cell MP19 gene (LIM2) to chromosome 19q13.4, and adjacent to ETFB. Somat Cell Mol Genet. 1994;20:67–69. doi: 10.1007/BF02257488. [DOI] [PubMed] [Google Scholar]
- 38.Hoffman MD, Kast J. Mass spectrometric characterization of lipid-modified peptides for the analysis of acylated proteins. J Mass Spectrom. 2006;41:229–241. doi: 10.1002/jms.981. [DOI] [PubMed] [Google Scholar]
- 39.Godugu B, Neta P, Simón-Manso Y, Stein SE. Effect of N-terminal glutamic acid and glutamine on fragmentation of peptide ions. J Am Soc Mass Spectrom. 2010;21:1169–1176. doi: 10.1016/j.jasms.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer A, Rohrer J, Kornfeld S. Determination of the structural requirements for palmitoylation of p63. J Biol Chem. 1995;270:638–9644. doi: 10.1074/jbc.270.16.9638. [DOI] [PubMed] [Google Scholar]
- 41.Sefton BM, Buss JE. The covalent modification of eukaryotic proteins with lipid. J Cell Biol. 1987;104:1449–1453. doi: 10.1083/jcb.104.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Linder ME, Deschenes RJ. Model organisms lead the way to protein palmitoyltransferases. J Cell Sci. 2004;117:521–526. doi: 10.1242/jcs.00989. [DOI] [PubMed] [Google Scholar]
- 44.Bagchi M, Katar M, Lo WK, Maisel H. Paralemnin of the lens. J Cell Biochem. 2003;89:917–921. doi: 10.1002/jcb.10574. [DOI] [PubMed] [Google Scholar]
- 45.Gauthier-Campbell C, Bredt DS, Murphy TH, El-Husseini Ael-D. Regulation of dendritic branching and filopodia formation in hippocampal neurons by specific acylated protein motifs. Mol Biol Cell. 2004;15:2205–2217. doi: 10.1091/mbc.E03-07-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaskovic S, Blanc M, van der Goot FG. What does S-palmitoylation do to membrane proteins? FEBS J. 2013;280:2766–2774. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 47.Jensen HH, Login FH, Koffman JS, Kwon TH, Nejsum LN. The role of aquaporin-5 in cancer cell migration: a potential active participant. Int J Biochem Cell Biol. 2016;79:271–276. doi: 10.1016/j.biocel.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 48.Kumari SS, Varadaraj M, Yerramilli VS, Menon AG, Varadaraj K. Spatial expression of aquaporin 5 in mammalian cornea and lens, and regulation of its localization by phosphokinase A. Mol Vis. 2012;18:957–967. [PMC free article] [PubMed] [Google Scholar]
- 49.Grey AC, Walker KL, Petrova RS, et al. Verification and spatial localization of aquaporin-5 in the ocular lens. Exp Eye Res. 2013;108:94–102. doi: 10.1016/j.exer.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]