Abstract
HLA-E presented antigens are interesting targets for vaccination given HLA-Es’ essentially monomorphic nature. We have shown previously that Mycobacterium tuberculosis (Mtb) peptides are presented by HLA-E to CD8+ effector T cells, but the precise phenotype and functional capacity of these cells remains poorly characterized. We have developed and utilized in this study a new protocol combining HLA-E tetramer with intracellular staining for cytokines, transcription factors and cytotoxic molecules to characterize these cells in depth. We confirm in this study the significantly increased ex vivo frequency of Mtb-peptide/HLA-E-TM+ CD8+ T cells in the circulation of patients with active tuberculosis (TB). HLA-E restricted CD8+ T cells from TB patients produced more IL-13 than cells from controls or subjects with latent tuberculosis infection (LTBI). Compared to total CD8+ T cells, HLA-E restricted cells produced more IFNγ, IL-4, IL-10, and granulysin but less granzyme-A. Moreover, compared to “classical” Mtb specific HLA-A2 restricted CD8+ T cells, HLA-E restricted CD8+ T cells produced less TNFα and perforin, but more IL-4. In conclusion, HLA-E restricted- Mtb specific cells can produce Th2 cytokines directly.
Keywords: Active TB, CD8+ T cells, Cytokines, Tetramers, Th2
Introduction
An estimated one fourth of the global population is infected with Mycobacterium tuberculosis (Mtb) [1]. According to WHO Global Tuberculosis control report 2016, there were 10.4 million new tuberculosis (TB) cases and 1.8 million TB deaths in 2015, including deaths resulting from TB disease among HIV-positive people [2].
TB is a potentially lethal disease, but curable if properly treated[2]. M. bovis BCG-vaccination protects infants from severe and disseminating TB, particularly TB-meningitis and military TB [3] but has poor efficacy against pulmonary TB in adults. A novel, effective TB vaccine is urgently required and its design depends on a detailed understanding of what exactly controls host immune protection.
Over recent years it has become evident that many cell subsets are involved in immune protection. Antigen-specific CD8+ effector T cells are detectable in most Mtb infected subjects and animals [4]. Depletion of CD8+ T cells in mice reduced survival after infection with Mtb, demonstrating their essential role in protection against TB [5]. Similarly, also MHC class I knockout mice showed an enhanced sensitivity to Mtb infection [6]. In humans, both “classical” class Ia (HLA-A,-B and -C) and “non-classical” class Ib (HLA-E,-F and -G) HLA molecules are known to activate CD8+ T cells by presenting cognate peptide to the TCR. MHC class Ia molecules can present endogenous, cytoplasmic antigens as well as exogenous antigens which are “cross-presented” by dendritic cells [7], including antigens derived from intracellular bacteria or viruses [8–11]. Mtb-specific CD8+ T cells in the blood of Mtb infected individuals differ in frequency, phenotype and functional activities in patients with active TB compared to subjects with latent tuberculosis infection (LTBI) [12, 13].
HLA-E is a highly conserved MHC class Ib molecule with rather unique properties. Primarily HLA-E is involved in prevention of lysis by NK cells through ligation with NKG2/CD94 complex. Moreover, HLA-E can also present antigens to CD8+ T cells and thus plays a role in both innate and adaptive immunity [11]. Its low allelic variability positions it as an interesting candidate antigen presenting molecule for peptide based vaccination [11,14]. HLA-E comprises four alleles (E*01:01; E*01:03; E*01:04; E*01:05), but only the first two are expressed as functional proteins. HLA-E*01:01 and *01:03 differ in a single amino acid outside the peptide binding groove, therefore they are anticipated to have a very similar peptide binding profile [15–17]. HLA-E is enriched in Mtb phagosomes compared to class Ia molecules and accessible for loading with Mtb peptides [18]. In addition to HLA-E’s low allelic variation, another advantage is that HLA-E is not down-regulated by the HIV-nef protein, as opposed to HLA class Ia molecules [19, 20]. In fact, certain components of HIV may even stabilize HLA-E cell surface expression to prevent NK mediated lysis of HIV infected cells [21].
In a first study we showed that Mtb peptides were presented by HLA-E to CD8+ T cells from LTBI as well as BCG-vaccinated donors. These T cells had both cytolytic and immunoregulatory activities [14]. In subsequent work we demonstrated that HLA-E-restricted clonal CD8+ T cells were able to inhibit Mtb outgrowth in infected human macrophages [22]. Qa-1, the mouse homolog of HLA-E, can also present Mtb derived antigens during infection [23]. Mice that lack Qa-1, are more susceptible to Mtb infection and this increased susceptibility to progress to disease was due to dysregulated T-cell responses [23].
HLA-E-restricted CD8+ T cells circulated in higher frequencies in the blood of active TB patients [14, 22, 24] compared to LTBI [15, 22, 24] and were associated with a Th2 cytokine profile, notably IL-13. However, although we detected production of Th2 cytokines in response to specific Mtb peptide stimulation, and Mtb peptide driven type-2 cytokine production was most abundant in donors with strong tetramer staining [22, 24], we were unable to demonstrate that HLA-E restricted CD8+ T cells were the source of these cytokines. In the current study combine Mtb-peptide/HLA-E tetramer staining with intracellular cytokine staining in active TB and LTBI subjects, to demonstrate that HLA-E-restricted CD8+ T cells actively produce type-2 cytokines. These results provide important new evidence on the role of HLA-E restricted CD8+ T cells in human infectious disease.
Results
Ex vivo analysis
Direct ex vivo binding of HLA-E tetramers (TM) loaded with p62 and p68 Mtb- peptides to CD8+ T cells was analyzed in peripheral blood mononuclear cells (PBMCs) of 26 tuberculin (PPD)-negative healthy subjects (HDs), 14 LTBI subjects, 24 patients with active TB and 5 HIV-TB co-infected patients (for detailed characteristics see Table 1).
Table 1.
Characteristics of subjects enrolled in the study
| Variable | HD | LTBI | TB | TB-HIV | Total |
|---|---|---|---|---|---|
| Total subjects, n | 26 | 14 | 24 | 5 | 69 |
| Age (range) | a) | 34–60 | 22–38 | 28–40 | |
| Clinical characteristics | a) | No disease, | Pulmonary TB (22)Miliary TB (1) | Miliary TB and | |
| Mantoux positive | Extra-pulmonary TB (1) | HIV co-infection | |||
| Miliary TB (1) | |||||
| Nationality | b) | Italian | Italian (6) | Italian (2) | |
| North African (18) | North African (3) | ||||
| Subjects used for ex vivo analysis, n | 26/26 | 14/14 | 24/24 | 5/5 | 69 |
| Subjects included in ex vivo analysis of memory compartment (TM+ events count >100) | 16/26 | 11/14 | 20/24 | 5/5 | 52 |
| Subjects used for expansion with PHA and HLA-E peptides | 26/26 | 12/14 | 17/24 | 4/5 | 60 |
| Subjects included in functional analysis upon expansion (TM+ events count >1000) | 14/26 | 6/13 | 15/17 panel A 14/17 panel B |
- | 35 |
| Subjects used for expansion with PHA and HLA-A2 peptides | 3/26 | 3/13 | 7/17 panel A | - | 13 |
| 6/17 panel B | 12 | ||||
| Subjects used for TCR Vβ16 analysis | 6/26 | 7/13 | 8/17 | - | 21 |
No information of the anonymous samples was available.
13 samples were obtained from anonymous bloodbank donors (Sanquin Bloodbank, the Netherlands); 13 samples were collected from the Dipartimento di Medicina Clinica e delle Patologie Emergenti, University Hospital, Palermo.
HD, Healthy Donor; LTBI, Latently TB Infected subjects; TB, active TB patients TB-HIV, TB-HIV co-infected patients.
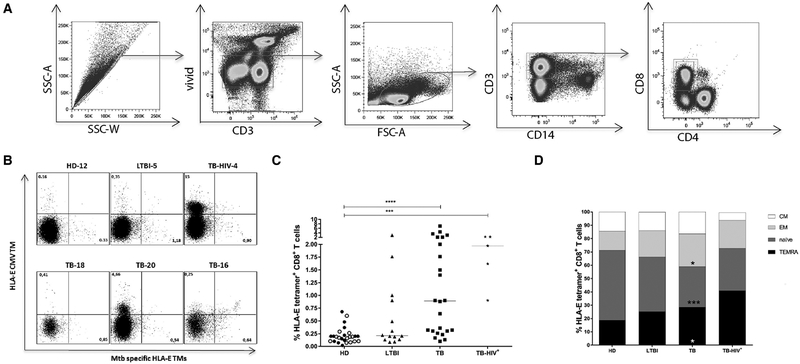
The gating strategy used to select TM+ CD8+ T cells is shown in Fig. 1A and comprised of sequential gating on single cells, live cells, lymphocytes, CD3+ cells and CD8+/CD4− cells. The CD94 blocking mAb was always included in the staining procedure to prevent HLA-E tetramer binding to the NKG2A/CD94 complex. Furthermore, an HLA-E CMV TM was included to exclude cells with aspecific binding. By definition T cells have single specificities and thus cells binding different TMs cannot do so through cognate TCR interactions and thus all double positive cells were excluded and only single Mtb-HLA-E TM+ cells were analyzed. Examples of TM staining for HD, LTBI, TB, and TB-HIV are shown and illustrate the variation in HLA-E TM binding (Fig. 1B).
Figure 1.
Ex vivo frequency of HLA-E restricted CD8+ T cells in HDs, LTBI, TB, and TB-HIV subjects. PBMCs were stained using TMs followed by viability staining and cell surface marker staining. Each experiment contained samples from different clinical groups, a total of 21 experiments were performed to analyze all samples. (A) Gating strategy: Initial gate was on single cells, followed by selection of live cells using VIVID cell viability dye and gating on lymphocytes on the basis of forward-side scatters. CD3+ cells were gated as T cells and further selected for CD8, but not CD4 expression. (B) Representative dot plot of ex vivo TM staining, with a control (CMV) TM on the y-axis and the combined Mtb TMs on the x-axis. Dot plots show results of one HD, one LTBI, one TB-HIV coinfected patient and 3 TB patients. (C) Frequency and (D) memory profile analysis of ex vivo HLA-E restricted CD8+ T cells in HDs, LTBI, TB, and TB-HIV patients. Memory populations were defined based on the expression of CCR7 and CD45RA. N = naïve T cells; EM = effector memory T cells; CM = central memory T cells; TEMRA = T effector memory recently activated cells. For ex vivo analysis, event counts below <100 were not considered a TM+ population and therefore not included in the downstream analysis of the memory compartment. 16 out 26 HDs (11 Dutch and 5 Italian HDs), 11 out 14 LTBI, 20 out 24 TB and 5 out 5 HIV-TB subjects were included in the analysis of memory subsets. Horizontal lines (C) represent median values. Black circles: Dutch HDs, open circles: Italian HDs, black triangles: LTBI; black squares: TB patients, black stars: HIV-TB. Bars represent mean values. Data shown in A-B is representative of 21 independent experiments with in total 71 samples and data shown in C and D are pooled from 21 independent experiments with —three to four donors per experiment. P-values were calculated using the Kruskal-Wallis test, including multiple test correction, ***p<0.001, ****p<0.0001. TB, LTBI and TB-HIV groups were each compared to the HD control group.
Confirming our own previous work, the ex vivo frequency of HLA-E-/Mtb-peptide TM+ CD8+ T cells was higher in patients with active TB than in HDs (p <0.0001) (Fig. 1C). As expected, the highest frequency of TM+ CD8+ T cells was found in the 5 HIV-TB patients (p = 0.0004 versus HD). Donors with LTBI showed a wide array of TM staining, but only a small proportion had a high frequency of HLA-E/Mtb TM+ cells. The median values of TM+ CD8+ T-cell frequencies were 0.22, 0.23, 0.89, and 1.9% in HDs, LTBI subjects, active TB and HIV-TB co-infected patients, respectively (Fig. 1C).
Thus, Mtb-peptide TM binding was highest to CD8+ T cells of patients with active TB and even increased with concomitant HIV infection.
Memory phenotypes of HLA-E-/Mtb-peptide TM+ CD8+ T cells
Next, the memory phenotype of HLA-E/Mtb-peptide TM+ CD8+ T cells was analyzed using CCR7 and CD45RA. To assess discrete populations rather than single cells, only samples with at least 100 TM+ events were included for memory subset analysis. Application of this cut-off permitted analysis of 16 out 26 HDs, 11 out 14 LTBI, 20 out 24 TB and 5 out 5 HIV-TB patients. The majority of circulating HLA-E/Mtb-peptide TM+ CD8+ T cells had an effector-memory profile, both TEMRA and TEM (p = 0.049) phenotypes were prominent in TB patients compared to HDs. In contrast, HLA-E/Mtb-peptide TM+ CD8+ T cells with a TEMRA phenotype were the most abundant in TB-HIV co-infected patients compared to other groups (Fig. 1D).
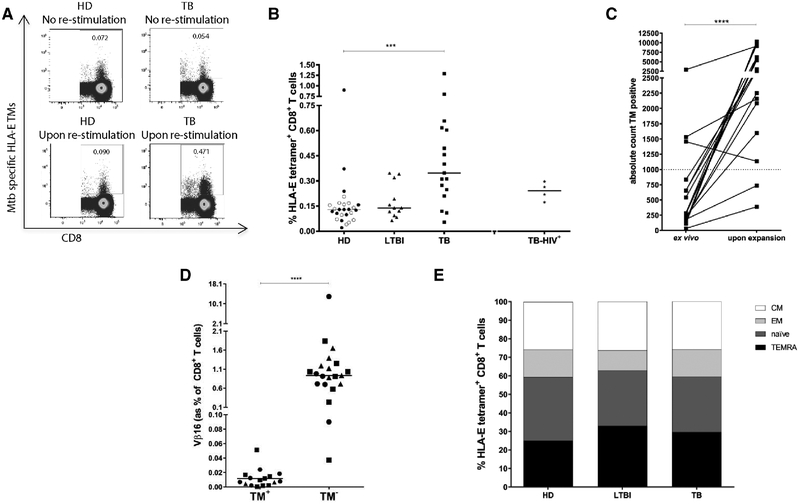
To characterize HLA-E restricted cells functionally in more depth, CD8+ T cells were expanded and stained with specific TMs combined with intracellular markers. Only few samples could not be included due to low cell yields or poor expansion; 26 out 26 HDs, 12 out 14 LTBI and 17 out 24 active TB were included. Of note but not unexpectedly, cells from the TB-HIV group were relatively impaired in their growth in culture, therefore only TM frequencies are shown but not the functional characteristics of these cells.
The median HLA-E TM+ CD8+ T-cell frequencies were 0.13,0.14, 0.34, and 0.23% in the blood of HDs, LTBI subjects, TB and HIV-TB co-infected patients, respectively. Thus, following Mtb peptide expansion the frequencies of HLA-E-TM+ Mtb-specific CD8+ T cells remained higher in active TB compared to LTBI and HDs. However it considerably decreased in TB-HIV co-infected patients, probably due to the poor expansion and survival of the cells in culture (Fig. 2A and B). In general, expansion resulted in a significant amplification of the number of TM+ cells to allow detailed characterization of their functional properties (Fig. 2C). The average TM+ cell counts increased from 750 directly ex vivo (for 24 active TB patients) to 4418 after expansion (for 17 TB patients), leading to an average expansion of 4.89-fold.
Figure 2.
HLA-E/Mtb TM+ CD8+ T-cell frequency after expansion. PBMCs were expanded using mitogenic stimulation, followed by magnetic bead separation of CD8+ T cells and specific peptide stimulation. Cells were stained with TMs first before staining with cell surface markers and intracellular cytokine staining. (A) Representative plots of TM staining of CD8+ T cells after control or combined p62 and p68 stimulation. Dot plots are representative of one HD and one TB patient that were analyzed in the same experiment. (B) Frequency of HLA-E/Mtb TM+ CD8+ T cells after PHA expansion and p62 and p68 peptide re-stimulation. 26 out 26 HDs (13 Dutch and 13 Italian HDs), 12 out 14 LTBI, 17 out 24 active TB and 4 out 5 HIV-TB had sufficient cells for in vitro expansion. Black circles: Dutch HDs, open circles: Italian HDs, black triangles: LTBI; black squares: TB patients, black stars: HIV-TB. Each symbol represents one patient. P-values were calculated using Kruskal–Wallis test including multiple-test correction. *** p<0.001. TB, LTBI and TB-HIV groups were compared to HD control group. —Two to three donors from different clinical groups were included per experiment. (C) Absolute count of TM+ events from TB patients before (ex vivo) and after PHA expansion and p62 and p68 re-stimulation. An average fold increase of 4.89 was achieved after expansion. Data were pooled from 13 culture experiments. The dashed horizontal line represents the cut off of 1000 cells required for in-depth analysis of the TM+ population, the p-value was calculated using a Wilcoxon signed rank test.****p<0.0001. (D) HLA-E restricted CD8+ T cells expanded with PHA and with p62 and p68 Mtb- specific peptides were characterized for expression of TCR-Vβ16. These results were obtained from six independent experiments in which 6 HDs (all Dutch), 7 LTBI and 8 active TB patients were included. Each symbol represents one sample and the p-value was calculated using a Wilcoxon-signed-rank-test. ****p<0.0001. (E) Memory profile analysis of HLA-E/Mtb TM+ CD8+ T cells upon PHA expansion and p62 and p68 re-stimulation. Samples with TM+ counts <1000 were not considered for analysis of the memory compartment: 14 out 26 HDs (10 Dutch and 4 Italian), 6 out 13 LTBI, 15 out 17 TB subjects were considered in the memory analysis. Bars represent mean values.
Phenotype after expansion of HLA-E-/Mtb-peptide TM+ CD8+ T cells
Previous characterization of T-cell receptors (TCR) recognizing the CMV UL40 epitope in HLA-E had revealed a strong preferential usage of the TCR variable β-region 16 (Vβ16) and it was suggested that expression of the Vβ16 TCR fragment was a hallmark of HLA-E restricted T cells [25]. Therefore we assessed the expression of Vβ16 on HLA-E- Mtb- specific CD8+ T cells. Although Vβ16 staining was observed in the total CD8+ T-cell population, we did not observe any preferential Vβ16 usage by TM+ CD8+ T cells in any of the study groups (Fig. 2D), showing that HLA-E/Mtb-specific CD8+ T cells have a clearly different TCR-Vβ repertoire compared to the previously published HLA-E/CMV-specific CD8+ T cells.
We next evaluated the memory phenotypes of the expanded HLA-E TM+ CD8+ T cells of 14 out 26 HDs (10 Dutch and 4 Italian), 6 out 13 LTBI, 15 out 17 TB, that had more than >1000 TM+ events. Memory profile analysis of TM+ CD8+ T cells upon stimulation was not significantly different between TEMRA, naïve (TN), effector memory (TEM) and central memory (TCM) compartments. HLA-E TM+ CD8+ T cells with an effector phenotype were the main population in all groups, consistent with the in vitro expansion (Fig. 2E). When compared to the ex vivo memory distribution we observed consistently more TEM and less TN, which fits with the expansion observed in the cultures. TEMRA and TCM frequencies were not different from ex vivo after the in vitro expansion.
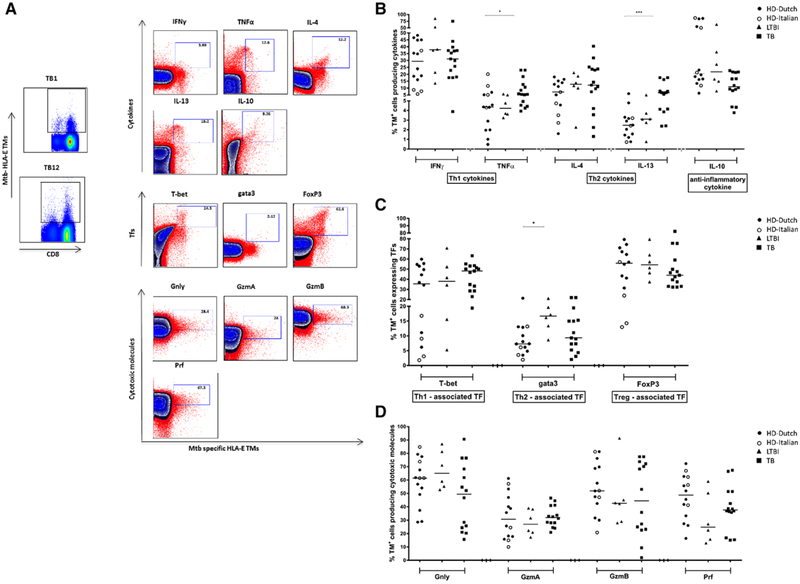
Cytokine profile of HLA-E/Mtb- specific CD8+ T cells
We evaluated expression of cytokine as well as cytotoxic molecules by intracellular staining of HLA-E/Mtb TM+ CD8+ T cells (Fig. 3A). Peptide stimulated cells are shown in comparison with unstimulated, but expanded cells. In general, TM+ cells produced both Th2 and Th1 cytokines as well as IL-10, and TM+ cells from the different clinical groups appeared relatively similar. TM+ CD8+ T cells from active TB patients produced more TNF-α and IL-13 compared to healthy controls (Fig. 3B). Approximately 30% of TM specific CD8+ T cells produced IFNγ in HDs, LTBI, active TB groups. Also transcription factors for the different lineages were expressed by HLA-E restricted CD8+ T cells. GATA3 was expressed more frequently in donors with LTBI (p = 0.042; Fig. 3C). Cytolytic molecules were expressed in most TM+ cells, however the range of expression between individuals was quite large and different combinations of cytolytic molecules were expressed between different individuals (Fig. 3D).
Figure 3.
HLA-E/Mtb TM+ CD8+ T cells cytokine production, transcription factors, and cytolytic molecules. PBMCs were expanded using mitogenic stimulation, followed by magnetic bead separation of CD8+ T cells and specific peptide stimulation. Cells were stained with TMs first before staining with cell surface markers and intracellular cytokine staining. Events count below <1000 were not included for analysis of HLA-E restricted CD8+ T cells’ properties; 14 out 26 HDs (10 Dutch, 4 Italian), 6 out 13 LTBI, 15 out 17 active TB subjects were analyzed. P-values were calculated using a two-way ANOVA non-parametric Dunnet’s test multiple comparison of LTBI and TB with HD control group. Black circles: Dutch HD; Open circles: Italian HD; black triangles: LTBI; black squares: TB patients. (A) A representative plot from 21 independent experiments with —two to four samples per experiment show HLA-E restricted CD8+ T cells upon p62 and p68 peptide stimulation in a TB patient, dot plots show TM+ cells producing cytokines; transcription factors and cytotoxic molecules. Blue indicates expanded cells not re-stimulated with specific peptides (unstimulated control), red is the sample following overnight peptide stimulation. Data are shown for total CD8+ T cells, with the tetramer positive population on the x-axis. Donor TB1 is shown for IFN-γ, TNF-α, IL-4, IL-13 and the transcription factors, whereas donor TB12 is shown for IL-10 and the cytolytic molecules. (B) Frequency of TM+ CD8+ T cells producing IFNγ, TNFα, IL-4, IL-13, and IL-10 upon p62 and p68 peptide stimulation in HDs, LTBIs and TB samples. 15 out 17 TB patients were analyzed, only for IL-10 (included in Panel B) 14 out 17 TB patients were evaluated. (C) Frequency of TM+ CD8+ T cells expressing T-bet, gata3 and FoxP3 transcription factors upon expansion in HDs, LTBI and TB subjects. (D) Frequency of TM+ CD8+ T cells producing Granulysin (Gnly), Granzyme A (GzmA), Granzyme B (GzmB) and Perforin (Prf). upon expansion in HDs, LTBIs and TB samples. In Panel B (including cytotoxic molecules) 14 out 17 active TB patients were analyzed. Each symbol represents one sample. Data shown in B and D are pooled from 21 independent experiments with —two to four patients per experiment. the p-value was calculated using a two-way ANOVA with Dunnets’multiple comparison correction *p<0.05, **p<0.01, ***p<0.001.
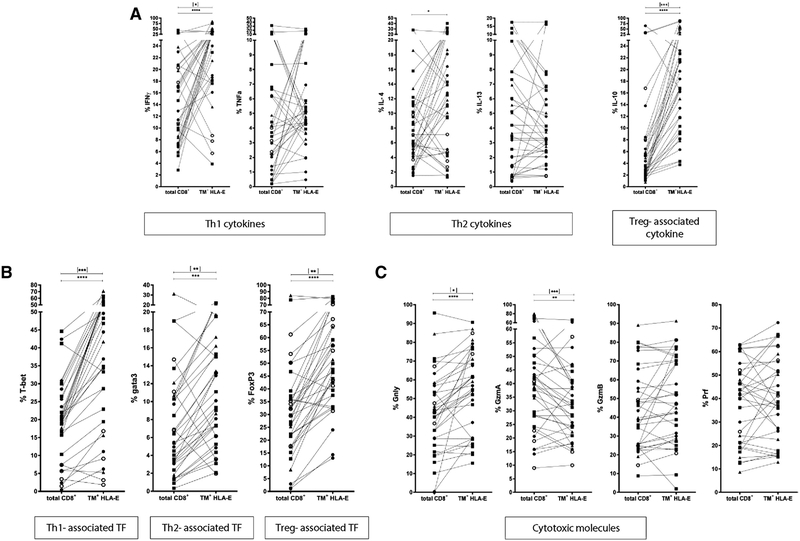
To investigate the relative specific properties of HLA-E-Mtb TM+ CD8+ T cells we compared them to the total pool of CD8+ T cells in terms of functional profiles and across all groups (Fig. 4); TB patients only are shown in Supporting Information Fig. 1 to show the relative abundance of specific cytokines in this group. HLA-E TM+ cells expressed significantly more IFN-γ compared to total CD8+ T cells in all samples (p < 0.0001; TB patients only p = 0.0127). Also IL-4 was detected at a higher frequency in the HLA-E TM+ populations compared to the total CD8+ T cells (p <0.0101), as was IL-10 (p <0.0001; TB patients only p = 0.0001) (Fig. 4A). TNF-α and IL-13 were not significantly differently expressed in the expanded TM+ cells compared to the total CD8+ T-cell population.
Figure 4.
Comparison of HLA-E TM+ with total CD8+ T cells. PBMCs were expanded usingmitogenic stimulation, followed by magnetic bead separation of CD8+ T cells and peptide stimulation. Cells were stained with TMs before staining cell surface markers and intracellular cytokine staining. Total CD8+ T-cell production of IFN-γ, TNF-α, IL-4, IL-13, and IL-10 (A), expression of T-bet, Gata3 and FoxP3 transcription factors (B), and Granulysin, Granzyme A, Granzyme B and Perforin (C) was compared with the proportion of cells within the HLA-E/Mtb TM+ CD8+ T cells upon p62 and p68 stimulation. Lines connect cytokine/transcription factor expression in the same patient sample. Data are pooled from 21 independent experiments with —two to four patients per experiment, p-values were calculated using the Wilcoxon-signed-rank-test. Black stars indicate p values calculated for all samples (n = 37) *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; stars in square brackets are p-values calculated for TB subjects only (n = 15). Circles: HD; squares: TB patients; triangles: LTBI.
In agreement with these data, the frequency of TM+ CD8+ T cells expressing T-bet, Gata3 and FoxP3 was higher compared to the total CD8+ T cells (T-bet p < 0.0001, TB samples only p = 0.0003; Gata3 p = 0.0009, TB samples only p = 0.0012; FoxP3 p <0.0001, TB samples only p = 0.0012) (Fig. 4B).
CD8+ T cells producing Granulysin (Gnly) were significantly increased in the TM+ population compared to total CD8+ T cells, while a decrease occurred in Granzyme A (GzmA) in TM+ CD8+ T cells (Gnly p < 0.0001, TB samples only p = 0.049; GzmA p = 0.0091, TB samples only p = 0.0002;) (Fig. 4C). Granzyme B (GzmB) and perforin (PRF) levels were not different between total and TM+ CD8+ T cells.
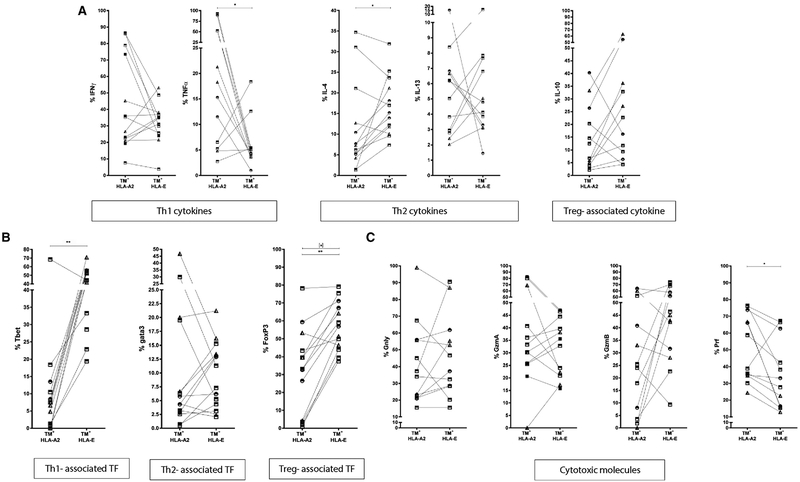
Comparison of HLA-E restricted CD8+ T cells to HLA-A2 restricted Mtb specific CD8+ T cells
Finally, we compared the functional phenotypes of “non-classical” HLA-E restricted Mtb-specific CD8+ T cells to that of “classical” HLA-A2 restricted, Mtb-specific CD8+ T cells in the same samples (Fig. 5). All donors were typed for HLA-A*0201 using specific antibody staining on PBMCs: 3 out 26 HDs, 3 out 13 LTBI and 7 out 17 TB patients were HLA-A*0201+. For Panel A (including cytokines) 7 out 17, and for Panel B (including cytotoxic molecules and IL-10) 6 out 17 active TB patients were analyzed after re-stimulation with HLA-E presented Mtb p62 and p68 peptides or with HLA-A2 presented Mtb-Ag85B peptides p74 and p79 (characteristics of Mtb-peptides: Table 2). The frequency of HLA-E-restricted CD8+ T cells producing TNF-α was lower than that of HLA-A2-restricted CD8+ T cells (all samples, p = 0.0342) (Fig. 5A). Th2 cytokines were more abundantly expressed in HLA-E restricted T cells (IL-4, all samples p = 0.0327) (Fig. 5A), especially in TB patients (Supporting Information Fig. 2), but this did not reach statistical significance due to the low number of HLA-A2+ TB patients that could be analyzed. In contrast, Tbet and FoxP3 expression was higher in HLA-E-restricted CD8+ T cells compared to HLA-A2-restricted CD8+ T cells (T-bet, all samples, p = 0.0017; FoxP3, all samples, p = 0.0012; FoxP3, TB patients only, p = 0.0156) (Fig. 5B). CD8+ T cells expressing Perforin (Prf) were higher and lower, respectively, in frequency among HLA-E restricted than HLA-A2- restricted Mtb specific CD8+ T cells (Prf p = 0.0161) (Fig. 5C). Together these data demonstrate that HLA-E restricted Mtb specific cells display unique functional properties compared to either total CD8+ T cells or HLA-A2 restricted Mtb specific T cells.
Figure 5.
Comparison of HLA-A*0201 and HLA-E TM+ CD8+ T cells. PBMCs were expanded using mitogenic stimulation, followed by magnetic bead separation of CD8+ T cells and specific peptide stimulation. Cells were stained with TMs first before staining with cell surface markers and intracellular cytokine staining. Frequency of HLA-A*0201/Mtb and HLA-E/Mtb TM+ CD8+ T cells producing IFNγ, TNFα, IL-4, IL-13, and IL-10 (A), expressing T-bet, Gata3 and FoxP3 transcription factors (B) and producing Granulysin, Granzyme A, Granzyme B and Perforin (C). 3 out 26 HDs (all Dutch), 3 out 13 LTBI, 7 out 17 active TB patients for panel A and 6 out 17 active TB patients for panel B were HLA-A*0201+. Data are pooled from 10 independent experiments with 1–3 HLA-A2+ samples per experiment. P-values were calculated using a Wilcoxon-signed-rank-test. p-values calculated for all samples (n = 14; except for cytotoxic molecules & IL-10 n = 13) *p<0.05, **p<0.01, ***p<0.001; stars in square brackets: p-values for TB subjects (n = 7). Circles: HD; squares: TB patients; triangles: LTBI.
Table 2.
Characteristics of peptides used in the study
| Epitope ID | 144950 | 144971 | 101382 | 32213 | 21078 | 59612 |
|---|---|---|---|---|---|---|
| Name code | p62 | p68 | TM-E control | p79 | p74 | TM-A2 control |
| Linear sequence | RMPPLGHEL | VLRPGGHFL | VMAPRTLLL | KLVANNTRL | GLPVEYLQV | SLYNTVATL |
| Source organism | MTB H37RV | MTB H37RV | CMV | MTB H37RV | MTB H37RV | HIV-1 |
| Source antigen | P49 protein, possible alanine rich dehydrogenase | Probable methyltransferase | hCMV UL40-derived peptide | Antigen 85-B precursor (Ag85B), 30 kDa extracellular protein, Mycolyl transferase 85B, Fibronectin-binding protein B | Antigen 85-A precursorMycolyl transferase 85A, Fibronectin-binding protein A | pl7 Gag (77–85) epitope |
| On-line source | http://www.iedb.org/epId/144950 | http://www.iedb.org/epId/144971 | http://www.iedb.org/epitope/101382 | http://www.iedb.org/epId/32213 | http://www.iedb.org/epId/21078 | http://www.iedb.org/epId/59612 |
| MHC restriction | HLA-E*01:03 | HLA-E*01:03 | HLA-E*01:03 | HLA-A2*02:01 | HLA-A2*02:01 | HLA-A2*02:01 |
| Tetramers | PE | PE | APC | APC and PE | APC and PE | PE |
CMV: CytomegaloVirus; HIV-1: human immunodeficiency virus type 1; MTB H37RV: Mycobacterium tuberculosis H37Rv strain.
Discussion
This is the first comprehensive analysis of human HLA-E restricted CD8+ T cells, permitting detailed characterization of functional capacities of these antigen specific cells. As we have previously shown [22, 24], PBMCs from TB patients produced significant levels of IL-4 and IL-13 following stimulation with HLA-E presented Mtb-peptides. In addition, HLA-E restricted Mtb-specific CD8+ T-cell clones displayed an unique Th2-like profile (secreting IL-4, IL-5, IL-13) upon peptide stimulation [22]. Although these studies strongly suggested an association between HLA-E restricted T cells and the production of Th2 cytokines, they did not formally prove that circulating HLA-E restricted Mtb specific T cells from TB patients produced Th2 cytokines. In this study, we developed a protocol that combined tetramer with intracellular (cytokine) staining and that allowed us to describe for the first time the characteristics of these unconventional, HLA-E restricted Mtb-specific CD8+ T cells in cohorts of active TB patients and LTBI subjects.
Firstly, the direct ex-vivo frequency of HLA-E restricted Mtb-TM+ CD8+ T cells was higher in patients with active TB compared to HDs. The majority of these cells had an effector-memory profile, suggesting that these cells were activated in the circulation of active TB patients, in contrast to healthy donors in which these cells mostly had a naïve phenotype. Expansion of cells allowed assessment of intracellular cytokines within the TM+ population. Most cytokines were expressed at similar levels by HLA-E restricted T cells among the different clinical groups. TNFα and IL-13 were abundantly expressed by TM+ CD8+ T cells from active TB patients compared to healthy controls. While the lineage-specifying transcription factor T-bet associated with Th1 function was expressed by the majority of HLA-E- TM+ CD8+ T cells from each group, a higher frequency of Gata3 expression was observed in TM+ CD8+ T cells from LTBI subjects. Gata3 expression was also detected previously in healthy PPD+ donors’ HLA-E restricted Mtb specific T-cell clones [22]. FoxP3 was detected in TM+ CD8+ T cells from all groups. CD4+ and CD8+ Tregs were present at the site of infection [26–28], and were increased in the circulation of patients with active TB compared to healthy controls [26–28]. Tregs were found to suppress T-cell IFN-γ production during active TB, which normalized after treatment [29]. In our previous work, in vitro stimulation with HLA-E Mtb-peptides p62 and p68 resulted in the induction of functionally active regulatory T cells as well as cytolytic effector cells [14]. This dualism of effector versus. immunoregulatory activities of HLA-E restricted T cells was also found for the T-cell clones [14, 22]. Not only the cytolytic effector functions but also the immunoregulatory properties of HLA-E restricted cells may be important in relation to protective immunity, by controlling pathogenic inflammation [23]. A negative consequence may be that immunoregulation contributes to pathogen persistence and chronic infection [11, 30].
A specific TCR fragment, Vβ16, has been associated with the recognition of the CMV UL40 epitope in the context of HLA-E [25]. However, in contrast to previous suggestions, Vβ16 was not universally used in TCR mediated recognition of pathogen derived peptides presented by HLA-E, since none of our donors preferentially utilized the Vβ16 TCR fragment in their Mtb specific HLA-E restricted response. HLA-E-restricted, Mtb-specific CD8+ T cells were compared for expression of secreted molecules and transcription factors relative to the total CD8+ T-cell population within the same donor. HLA-E TM+ cells produced significantly more IFNγ, IL-4 and IL-10 compared to total CD8+ T cells. Increased expression of IFNγ and IL-10 in active TB patients may reflect recent activation. The frequency of TM+ CD8+ T cells expressing T-bet, Gata3 and FoxP3 was higher than in total CD8+ T cells. CD8+ T cells producing Granulysin were significantly higher in the HLA-E restricted population compared to total CD8+ T cells, contrasting with the decreased frequency of Granzyme A producing TM+ CD8+ T cells. Moreover, Mtb specific CD8+ T cells with classical (HLA-A2) and non-classical (HLA-E) restriction were compared. This revealed reduced TNF-α and perforin expression in HLA-E restricted T cells, but increased expression of T-bet, FoxP3 and IL-4.
During the first period of Mtb infection, key cytokines associated with protection are IFN-γ and TNF-α [31, 32]. After clearance or successful containment of Mtb, activated immune responses must be ceased to avoid inflammatory damage. This may be mediated by anti-inflammatory cytokines such as IL-10, IL-13, or by a Th2 environment. This Th1/Th2 ‘switch’ may be antigen-specific, depending on HLA restriction or the stage, early or late in infection, in which the antigens are presented [33]. The ex vivo frequency of HLA-E-restricted CD8+ T cells is higher in active TB but significantly decreased after 6 months of chemotherapy whereas the frequencies of HLA-A2-restricted CD8+ T cells increased after therapy [24]. This opposite trend of HLA-E and HLA-A2 restricted responses may be related to the different roles exerted by the two HLA molecules and by the different set of antigens presented during Mtb disease stages. This indicates that although both HLA-A2 restricted as well as HLA-E restricted CD8+ T cells are Mtb-specific, their restriction correlated with different functional properties. Future studies need to resolve the relative significance of effector versus. immunoregulatory activities within the HLA-E based antigen presentation system in infection and other human diseases.
Materials and methods
Recruited patients
Peripheral blood was collected from a total of 69 adults; 13 healthy donors, 14 with LTBI (age range 36–60), 24 with active TB disease (age range 22–38), and 5 with TB-HIV co-infection (age range 28–40) from the Dipartimento di Medicina Clinica e delle Patologie Emergenti, University Hospital, Palermo, Italy (for patients’ characteristics see Table 1). The study was approved by the Ethical Committee of the University Hospital, Palermo, where patients were recruited. All patients signed an informed consent and the study was performed according to the guidelines of the local ethics committee and in accordance with the principles of the Helsinki Declaration and those of “Good Clinical Practices”. All TB patients had clinical and radiological findings consistent with active pulmonary TB, the diagnosis was confirmed by bacteriological isolation of Mtb. None of the active TB patients had been vaccinated with BCG, or were treated with steroid or other immunosuppressive or anti-tubercular drugs at the time of sampling.
LTBI subjects were health-care workers with a positive tuber-culin (PPD) skin test and no symptoms and signs of active TB [34]. Skin tests were considered positive when the induration was larger than 10 mm at 72 h after injection of 5 U of PPD (Statens Serum Institut, Copenhagen, Denmark).
Besides the 13 healthy donors mentioned above, an additional 13 PPD negative anonymous bloodbank donors (Sanquin Bloodbank, the Netherlands) were recruited; These donors were tested for PPD responses in a 6 day lymphocyte-stimulation-assay [35, 36] with IFN-γ measurement by ELISA (U-CyTech, Utrecht, the Netherlands) as readout. Donors that produced less than 100 pg/mL IFN-γ in response to PPD were considered negative.
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood samples by Ficoll-Hypaque (Sigma) density centrifugation, frozen and stored in liquid nitrogen. Frozen samples were sent to the Department of Infectious Diseases at Leiden University Medical Center (LUMC) where the experiments were performed.
Preparation of MHC class I tetramers
Extracellular HLA class I, with a C-terminal BirA recognition site, and β2-microglobulin were expressed in Escherichia coli as insoluble aggregates in the form of inclusion bodies. Purified inclusion bodies were solubilized in urea and folded with peptide by dilution. Monomers were biotinylated using the BirA enzyme and purified by gelfiltration on a Hiload 16/60 Superdex 75 prep grade.
HLA class I tetramers were generated by adding SA-phycoerythrin (PE) or SA-allophycocyanin (APC) at an SA:MHC class I molar ratio of 1[2236]6 [37]. To ensure maximum saturation of the SA with peptide-MHC monomers (i.e. that tetramers are formed), SA was sequentially added in four steps at 20-min intervals. The resulting tetramers were stored in PBS, with a final concentration of 16% glycerol, 0,6% BSA, 0,02% azide and Complete protease inhibitors (Roche) at −80°C. Tetramers were generated for HLA-E*01:03 and HLA-A2*02:01.
The following peptides were used for in vitro stimulation of T cells and/or for TM generation: the HLA-E restricted Mtb peptide p62 (RMPPLGHEL), p68 (VLRPGGHFL), and CMV UL40 (VMAPRTLLL); the HLA-A2 restricted Ag85 Mtb peptides (GLPVEYLQV), (KLVANNTRL), and HIV Gag (SLYNTVATL) [14, 25, 38]. All peptides were synthesized by Peptide2.0 Inc (Chantilly, VA, USA).
Ex-vivo tetramer staining
PBMCs were thawed, counted (CASY cell counter, Roche, Woerden, The Netherlands) and rested for one hour in an incubator at 37°C with 5% CO2. Approximately 106 cells per sample were blocked for 10 min at RT with 10 μg/mL purified CD94 mAb (Clone HP-3D9, BD Bioscience) to block NKG2A/CD94 binding of HLA-E TMs, and then stained with HLA-E TMs for 15 minutes at 37°C. Both p62 and p68 HLA-E- TMs were PE-conjugated and merged in the same colour channel. CMV-specific-HLA-E- TM (APC-conjugated) was used as control. The CMV-TM was added in the same staining step as the Mtb-specific TMs. Table 2 summarizes the peptides and TMs used.
After incubation, samples were washed with PBS/0.1% BSA (Pharmacy LUMC, the Netherlands, Sigma-Aldrich, Zwijndrecht, the Netherlands), and stained with live/dead marker (Vivid fixable violet dye, Invitrogen, Thermo Fisher Scientific Inc., Bleiswijk, the Netherlands) according to the manufacturer’s protocol. Followed by Fluorochrome-conjugated antibodies: CD3 (Horizon-V500, clone UCHT1), CD8 (AlexaFluor700, clone RPA-T8), CD4 (PerCp-Cy5.5, clone RPAT4), CCR7 (PE-Cy7, clone 3D12) (all BDBio-sciences), and CD45RA (BV570, clone MI100), CD14 (Qdot655, clone TüK4) (both Invitrogen). Samples were stained for 30 min at 4°C, washed twice in PBS/0.1% BSA and then acquired on a LSRFortessa flow cytometer with Diva software (v6.2, BDBio-sciences). A minimum of 1 × 10e6 PBMC were acquired and analysis was performed on at least 100 000 CD8+ events for each sample.
Data were analyzed with FlowJo software (version 9.9.6 Treestar Inc., Ashland, OR, USA). The analysis of each specific subset was performed using the gating strategy shown in Fig. 1A in compliance with the MIATA guidelines [39].
Generation of HLA-E restricted CD8+ T-cell lines
PBMCs were thawed, counted and 2 × 10e6 PBMCs were expanded in 24-well plates using 5 μg/mL PHA (Remel, Thermo Fisher Scientific Inc.), in Iscove’s modified Dulbecco’s medium (IMDM, Life Technologies-Invitrogen) supplemented with 10% human serum (Heat inactivated pooled serum from bloodbank donors) and incubated for 3 days at 37°C, 5% CO2. Medium was refreshed with 25 CU/mL IL-2 (Proleukin, Novartis Pharma, Amsterdam, the Netherlands). After 6 days, CD8+ T cells were sorted by positive bead selection (MACS, Miltenyi Biotec BV, Leiden, the Netherlands). Purity of all cells sorts was >98% as checked by flow cytometry (FACS Accuri Cytometer, BD Bioscience). Purified CD8+ T cells were subsequently stimulated with 10 μg/ml Mtb p62 (RMPPLGHEL) and p68 (VLRPGGHFL) peptides, recombinant human IL-15 and IL-7 (both 5 ng/mL final concentration Peprotech, Rocky Hill, NJ, USA), as well as irradiated (30 Gy) allogeneic feeder cells. At day 3, the culture was refreshed with complete IMDM medium with IL-2 (50 CU/ml). After 5 days, cultures were splitted and refreshed with complete IMDM with IL-2 (100 CU/mL). At day 7, cells were harvested and analyzed for tetramers, cytokines and surface markers by flowcytometry. Starting with a number of 3 – 4 × 106 PBMCs, the yield of CD8+ T cells after this procedure was about 80–100 × 106.
Intracellular staining (ICS) for flow cytometry
HLA-A2 TM analysis was only performed on donors that were HLA-A*0201, therefore 105 cells of all samples were typed with an anti-HLA-A2 antibody (FITC, clone BB7.2, BDBiosciences).
Expanded CD8+ T cells were stimulated for 6 h with 10 μg/mL Mtb- peptides and analyzed after 16 h incubation with BrefeldinA (3 μg/mL, Sigma-Aldrich) and monensin (1:1000, BioLegend) at 37°C in 5% CO2. Cells were harvested, washed with PBS/0.1% BSA, and blocked for 10 min at RT with a purified CD94 mAb and then stained with tetramers and CCR7 (PE-Cy7, clone 3D12, BDBiosciences) for 15 min at 37°C. After incubation, cells were washed once in PBS/0.1% BSA and subsequently stained for surface markers (30 min, 4°C) CD3 (BV570, clone UCHT1, BioLegend), CD8 (Alexa700, clone RPA-T8), CD45RA (Qdot655, clone MEM-56, Invitrogen) followed by a fix/perm procedure according to manufacturer’s instruction (BDBiosciences). Two different panels for intracellular markers were used. Panel A included: T-bet (BV605, clone 4BIO), IL-13 (PerCP-Cy5.5, clone JES10–5A2) (both BioLegend); IFN-γ (AlexaFluor700, clone B27), TNF-α (APC-H7, clone MAb11), IL4 (PE-C594, clone MP4–25D2), GATA3 (Alex-aFluor488, clone L50–823) (all BDBiosciences); FoxP3 (PE-Cy5, clone PCH101) (Ebioscience). Panel B: rabbit-anti-human Granulysin (kind gift of Dr. A. Krensky, Stanford, CA) followed by Goat-anti-Rabbit-FITC, Perforin (PEC594, clone δG9), Granzyme B (AlexaFluor700, clone GB11), Granzyme A (PerCP-Cy5.5, clone CB9) (BioLegend), IL10 (BV605, clone JES3–9D7) (BDBio-sciences). Cells were acquired on a LSRFortessa flow cytometer with Diva software (v6.2, BDBiosciences). A minimum of 3 × 106 events were acquired for all samples except for the cultures of the HIV-TB samples, there only 1 × 106 events could be acquired. Analysis of data was performed with FlowJo software (version9.9.6 Treestar Inc., Ashland, OR, USA) on a minimum of 2 × 106 CD8+ cells acquired for HD, LTBI and TB samples whereas only 5 × 105 CD8+ cells were analyzed in HIV-TB subjects.
Statistical analysis
Most data shown are individual patient data, lines indicate the median of the population. Data were analyzed using Kruskal–Wallis including multiple test correction, Wilcoxon-Signed Rank testing and 2-way ANOVA with Dunnet’s multiple comparisons. P-values < 0.05 were considered significant.
Supplementary Material
Acknowledgements:
We acknowledge assistance from Marco P. La Manna (University Hospital, Palermo, Italy) for help in sample organization and preparation. We thank Antonio Cascio and Paola Di Carlo (Dipartimento di Medicina Clinica e delle Patologie Emergenti, University Hospital, Palermo) for providing samples. This study was supported by funding from EC HORIZON2020 TBVAC2020 (Grant Agreement No. 643381) to S.A.J. and THMO; EC HORIZON2020 grant EMI-TB (Grant Agreement No. H2020-PHC-643558) to FD; EC-FP7 ADITEC (Grant Agreement No. 280873) to T.H.M.O.; The Netherlands Organization for Scientific Research (NWO-TOP Grant Agreement No. 91214038) to T.H.M.O. Research reported in this publication was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health under Award Number R21AI127133 to S.A.J. and T.H.M.O. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or any funder. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- LTBI
latent tuberculosis infection
- PPD
purified protein derivative
- Mtb
Mycobacterium tuberculosis
- TB
tuberculosis
- CMV
Cytomegalovirus
- HIV
Human Immunodeficiency Virus
- Th1
T helper 1 cytokines
- Th2
T helper 2 cytokines
- TM
tetramers
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site
Conflict of interest:
The authors declare no financial or commercial conflict of interest.
References
- 1.Houben RM and Dodd PJ, The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 2016. 13: e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO, Global Tuberculosis report control 2016, World Health Organization, Geneva, 2016. [Google Scholar]
- 3.Brewer TF, Preventing tuberculosis with bacillus Calmette-Guerin vaccine: a meta-analysis of the literature. Clin. Infect. Dis 2000. 31(Suppl 3): S64–S67. [DOI] [PubMed] [Google Scholar]
- 4.Ernst JD, The immunological life cycle of tuberculosis. Nat. Rev. Immunol 2012. 12: 581–591. [DOI] [PubMed] [Google Scholar]
- 5.Mogues T, Goodrich ME, Ryan L, LaCourse R and North RJ, The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J. Exp. Med 2001. 193: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urdahl KB, Liggitt D and Bevan MJ, CD8+ T cells accumulate in the lungs of Mycobacterium tuberculosis-infected Kb−/−Db−/− mice, but provide minimal protection. J. Immunol 2003. 170: 1987–1994. [DOI] [PubMed] [Google Scholar]
- 7.Benvenuti F, The dendritic cell synapse: a life dedicated to T cell activation. Front Immunol. 2016. 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fresnay S, McArthur MA, Magder L, Darton TC, Jones C, Waddington CS, Blohmke CJ et al. , Salmonella Typhi-specific multi-functional CD8+ T cells play a dominant role in protection from typhoid fever in humans. J Transl Med 2016. 14: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mateus J, Lasso P, Pavia P, Rosas F, Roa N, Valencia-Hernandez CA, Gonzalez JM et al. , Low frequency of circulating CD8+ T stem cell memory cells in chronic chagasic patients with severe forms of the disease. PLoS Negl Trop Dis 2015. 9: e3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sissons JG and Oldstone MB, Killing of virus-infected cells: the role of antiviral antibody and complement in limiting virus infection. J. Infect. Dis 1980. 142: 442–448. [DOI] [PubMed] [Google Scholar]
- 11.Joosten SA, Sullivan LC and Ottenhoff TH, Characteristics of HLA-E restricted T-cell responses and their role in infectious diseases. J Immunol Res 2016. 2016: 2695396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ and Berry MP, The immune response in tuberculosis. Annu. Rev. Immunol 2013. 31: 475–527. [DOI] [PubMed] [Google Scholar]
- 13.Rozot V, Vigano S, Mazza-Stalder J, Idrizi E, Day CL, Perreau M, Lazor-Blanchet C et al. , Mycobacterium tuberculosis-specific CD8+ T cells are functionally and phenotypically different between latent infection and active disease. Eur. J. Immunol 2013. 43: 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joosten SA, van Meijgaarden KE, van Weeren PC, Kazi F, Geluk A, Savage ND, Drijfhout JW et al. , Mycobacterium tuberculosis peptides presented by HLA-E molecules are targets for human CD8 T cells with cytotoxic as well as regulatory activity. PLoS Pathog. 2010. 6: e1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel AS, Grotzke JE, Lines RA, Lewinsohn DA, McNabb AL, Streblow DN, Braud VM et al. , HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med 2002. 196: 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P and Marsh SG, The IPD and IMGT/HLA database: allele variant databases. Nucleic. Acids. Res 2015. 43: D423–D431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulbrecht M, Honka T, Person S, Johnson JP and Weiss EH, The HLA-E gene encodes two differentially regulated transcripts and a cell surface protein. J. Immunol 1992. 149: 2945–2953. [PubMed] [Google Scholar]
- 18.Grotzke JE, Harriff MJ, Siler AC, Nolt D, Delepine J, Lewinsohn DA and Lewinsohn DM, The Mycobacterium tuberculosis phagosome is a HLA-I processing competent organelle. PLoS Pathog. 2009. 5: e1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL and Baltimore D, The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999. 10: 661–671. [DOI] [PubMed] [Google Scholar]
- 20.Collins KL, Chen BK, Kalams SA, Walker BD and Baltimore D, HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 1998. 391: 397–401. [DOI] [PubMed] [Google Scholar]
- 21.Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, Rockstroh J et al. , HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir. Ther 2005. 10: 95–107. [DOI] [PubMed] [Google Scholar]
- 22.van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff TH and Joosten SA, Human CD8+ T cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLoS Pathog. 2015. 11: e1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bian Y, Shang S, Siddiqui S, Zhao J, Joosten SA, Ottenhoff THM, Cantor H et al. , MHC Ib molecule Qa-1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLoS Pathog. 2017. 13: e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caccamo N, Pietra G, Sullivan LC, Brooks AG, Prezzemolo T, La Manna MP, Di Liberto D et al. , Human CD8 T lymphocytes recognize Mycobacterium tuberculosis antigens presented by HLA-E during active tuberculosis and express type 2 cytokines. Eur. J. Immunol 2015. 45: 1069–1081. [DOI] [PubMed] [Google Scholar]
- 25.Hoare HL, Sullivan LC, Pietra G, Clements CS, Lee EJ, Ely LK, Beddoe T et al. , Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol 2006. 7: 256–264. [DOI] [PubMed] [Google Scholar]
- 26.Joosten SA, van Meijgaarden KE, Savage ND, de Boer T, Triebel F, van der Wal A, de Heer E et al. , Identification of a human CD8+ regulatory T cell subset that mediates suppression through the chemokine CC chemokine ligand 4. Proc Natl Acad Sci USA 2007. 104: 8029–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Zhou B, Li M, Deng Q, Wu X, Le X, Wu C et al. , CD4(+)CD25(+)FoxP3(+) regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin. Immunol 2007. 123: 50–59. [DOI] [PubMed] [Google Scholar]
- 28.Guyot-Revol V, Innes JA, Hackforth S, Hinks T and Lalvani A, Regulatory T cells are expanded in blood and disease sites in patients with tuberculosis. Am. J. Respir. Crit. Care Med 2006. 173: 803–810. [DOI] [PubMed] [Google Scholar]
- 29.Ribeiro-Rodrigues R, Resende Co T, Rojas R, Toossi Z, Dietze R, Boom WH, Maciel E et al. , A role for CD4+CD25+ T cells in regulation of the immune response during human tuberculosis. Clin. Exp. Immunol 2006. 144: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joosten SA and Ottenhoff TH, Human CD4 and CD8 regulatory T cells in infectious diseases and vaccination. Hum. Immunol 2008. 69: 760–770. [DOI] [PubMed] [Google Scholar]
- 31.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA and Bloom BR, An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med 1993. 178: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez-Pando R and Rook GA, The role of TNF-alpha in T-cell-mediated inflammation depends on the Th1/Th2 cytokine balance. Immunology 1994. 82: 591–595. [PMC free article] [PubMed] [Google Scholar]
- 33.Skeiky YA, Ovendale PJ, Jen S, Alderson MR, Dillon DC, Smith S, Wilson CB et al. , T cell expression cloning of a Mycobacterium tuberculosis gene encoding a protective antigen associated with the early control of infection. J. Immunol 2000. 165: 7140–7149. [DOI] [PubMed] [Google Scholar]
- 34.Menzies D, Interpretation of repeated tuberculin tests. Boosting, conversion, and reversion. Am. J. Respir. Crit. Care Med 1999. 159: 15–21. [DOI] [PubMed] [Google Scholar]
- 35.Commandeur S, Lin MY, van Meijgaarden KE, Friggen AH, Franken KL, Drijfhout JW, Korsvold GE et al. , Double- and mono-functional CD4(+) and CD8(+) T-cell responses to Mycobacterium tuber culosis DosR antigens and peptides in long-term latently infected individuals. Eur. J. Immunol 2011. 41: 2925–2936. [DOI] [PubMed] [Google Scholar]
- 36.Lin MY, Reddy TB, Arend SM, Friggen AH, Franken KL, van Meijgaarden KE, Verduyn MJ et al. , Cross-reactive immunity to Mycobacterium tuberculosis DosR regulon-encoded antigens in individuals infected with environmental, nontuberculous mycobacteria. Infect. Immun 2009. 77: 5071–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ et al. , Phenotypic analysis of antigen-specific T lymphocytes. Science 1996. 274: 94–96. [PubMed] [Google Scholar]
- 38.Geluk A, van Meijgaarden KE, Franken KL, Drijfhout JW, D’Souza S, Necker A, Huygen K et al. , Identification of major epitopes of Mycobacterium tuberculosis AG85B that are recognized by HLAA*0201-restricted CD8+ T cells in HLA-transgenic mice and humans. J. Immunol 2000. 165: 6463–6471. [DOI] [PubMed] [Google Scholar]
- 39.Britten CM, Janetzki S, Butterfield LH, Ferrari G, Gouttefangeas C, Huber C, Kalos M et al. , T cell assays and MIATA: the essential minimum for maximum impact. Immunity 2012. 37: 1–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.