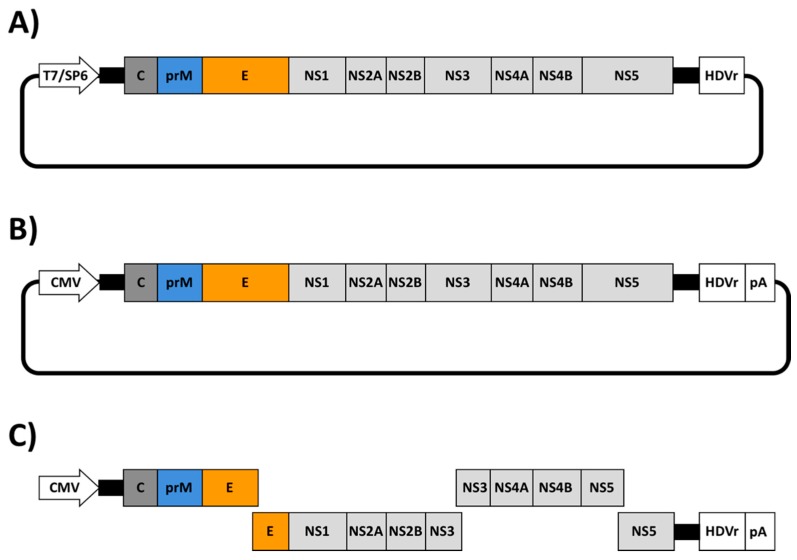
Figure 2.
Schematic representation of ZIKV reverse genetic approaches. (A) Infectious RNA transcripts from full-length cDNA clones: A full-length genomic cDNA clone containing the ZIKV genome flanked by a prokaryotic promoter (e.g., T7 or SP6) and the hepatitis delta virus ribozyme (HDVr) is usually assembled in a low-copy plasmid. In this approach, in vitro transcription is required to produce viral RNA that is transfected into susceptible cells to initiate viral replication and transcription in the cytoplasm of transfected cells. (B) Full-length infectious genomic cDNA clones: A full-length infectious genomic cDNA clone containing the viral genome flanked by a polymerase II-driven promoter from cytomegalovirus (CMV) and the HDVr followed by a polymerase II terminator and polyadenylation signal (pA) is assembled in a low-copy plasmid. In this case, the full-length cDNA clone is transcribed in the nucleus of transfected cells by the cellular RNA polymerase II. Primary transcripts are translocated to the cytoplasm where further amplifications steps are conducted by the viral polymerase. (C) Infectious Subgenomic Amplicons (ISA): The ISA approach can be used for the production of infectious viruses from genomic DNA material, including pre-existing infectious cDNA clones, viral RNA or de novo synthesized DNA genomic sequences. The entire viral genome is amplified by overlapping PCR reactions with each PCR product containing 30–40 base pairs overlapping regions [45]. The first and last PCR products are flanked by the CMV promoter and the HDVr followed by a polymerase II terminator and pA signal, respectively. Co-transfected cDNAs result in self-assembly in the cytoplasm of susceptible cells and virus production.