Abstract
Background
Severity gradation of missense mutations is a big challenge for exome annotation. Predictors of deleteriousness that are most frequently used to filter variants found by next generation sequencing, produce qualitative predictions, but also numerical scores. It has never been tested if these scores correlate with disease severity.
Results
wANNOVAR, a popular tool that can generate several different types of deleteriousness-prediction scores, was tested on Fabry disease. This pathology, which is caused by a deficit of lysosomal alpha-galactosidase, has a very large genotypic and phenotypic spectrum and offers the possibility of associating a quantitative measure of the damage caused by mutations to the functioning of the enzyme in the cells. Some predictors, and in particular VEST3 and PolyPhen2 provide scores that correlate with the severity of lysosomal alpha-galactosidase mutations in a statistically significant way.
Conclusions
Sorting disease mutations by severity is possible and offers advantages over binary classification. Dataset for testing and training in silico predictors can be obtained by transient transfection and evaluation of residual activity of mutants in cell extracts. This approach consents to quantitative data for severe, mild and non pathological variants.
Electronic supplementary material
The online version of this article (10.1186/s12859-018-2416-7) contains supplementary material, which is available to authorized users.
Keywords: Rare disease, Clinical informatics, Variant analysis, Bioinformatics, Fabry disease
Background
Exome sequencing has become very popular for the diagnosis of genetic diseases.This is certainly due to high-throughput platforms that have greatly reduced the costs of sequences and to the tools for the analysis of data that are freely available to researchers. Pipelines for the processing [1] and the annotation of data have been proposed with the intent of “democratizing the ability to compile information on large amounts of genetic variations in individual laboratories” [2]. A critical step in the annotation process is represented by the evaluation of missense mutations. A popular annotation tool, wANNOWAR [3], can generate several different types of deleteriousness-prediction scores running SIFT [4], LRT [5], MutationAssessor [6], FATHMM [7], PROVEAN [8], VEST3 [9] metaSVM [10], metaLR [10], M-CAP [11], PolyPhen-2 [12], MutationTaster [13], CADD [14], DANN [15], fathmm-MKL coding [16], GenoCanyon [17], GERP++, [18, 19], phyloP7way vertebrate, phyloP20way mammalian [20], phastCons7way vertebrate, phastCons 20 way mammalian [21], SiPhy 29way logOdds [22]. However in the real world the situation is not simple. A continuum is observed, ranging from very sever to mild cases. The border between “disease mutation” and “non disease mutation” is artificial and dichotomizing continuous variables is problematic. We decided to address this point and challenge wANNOWAR [3] with a real example, Fabry disease (FD). Mutations that are responsible for this pathology, affect the functioning or the stability of lysosomial alpha-galactosidase (AGAL)(Uniprot: AGAL_HUMAN P06280; EC: 3.2.1.22), which is encoded by the gene GLA on the X chromosome.
AGAL is a dimer and its structure has been determined by X-ray crystallography [23–25]. More than 400 missense mutations have been described so far. This number is a surprisingly high value for a protein of 429 aminoacids and almost every amino acid has been found to be mutated. The large genotypic spectrum corresponds to the large phenotypic spectrum of FD, with respect to age at onset, rate of disease progression, severity of clinical manifestations. Patients with the late onset form of FD retain some AGAL activity and are asymptomatic until adult age when they develop cardiac and/or kidney problems [26–29]. When a severe mutation is diagnosed, enzyme replacement must be started even before the symptoms are manifested [30–32], for cases retaining some residual activity, a therapy with small molecules, can be possible [33–35]. Indeed for FD, as well as for other diseases which are due to deficits in lysosomal glycosidases, it is possible to employ iminosugars that stabilize the endogenous protein of the patient acting as pharmacological chaperones or reduce substrate accumulation [36–38]. Iminosugars represent a lucky case of drug repositioning because they were first derived to cure HIV and subsequently used to treat lysosomal storage disorders [24, 39–41].
The classification of FD genotypes is generally carried out on the base of clinical evaluation of patients [42]. Specialized databases such as fabry-database.org [43, 44] annotate mutations with qualitative phenotypes. However a more punctual classification of FD mutations is possible. In fact in order to test the effects of drugs on different mutations, a cell based assay has been developed [45, 46]. Expression vectors encoding mutant AGAL are transiently transfected into COS or HEK293 cells and the residual activity of the enzyme is measured in the extracts of cells that had been treated or not treated with the drug. Residual activity is normalized by the total amount of proteins in the cell (HEK293 or COS) and depends on the stability of the mutant as well as on its specific activity. The ratio between the normalized residual activity of a given mutant and that of wild type AGAL is measured under the same conditions. Part of these data, i.e. those obtained in the absence of the drug, can be “repositioned”, so to speak. They offer the unique possibility of associating a numerical value that correlates to the severity of the damage to hundreds of mutations and consent to evaluate the performance of the most popular predictors of deleterious variant in a realistic scenario of gradual disease severity.
Methods
Missense GLA mutations with phenotypic annotation derived from clinical observation of patients were obtained from a disease specific database of clinical phenotypes and genotypes, fabry-database.org [43, 44] (dataset 1). The mutations (genomic Reference Sequence and protein Reference sequence) and the phenotypes are reported in the 1st, 10th and last column of Additional file 1, respectively.
Missense GLA mutations with residual activity annotation were obtained from Fabry_CEP [47, 48]. Relative residual activity is the ratio between the activity measured in cell extracts for a given mutant and the activity of wild type AGAL tranfected into suitable eukaryotic vectors × 100. When residual activity for a given mutation had been measured by more than one lab, the average value was considered (dataset 2). The mutations (genomic Reference Sequence and protein Reference sequence) and the residual activities are reported in the 1st, 10th and last column of Additional file 2, respectively.
The nucleotide numbering on coding DNA Reference Sequence was obtained for each mutation from the appropriate reference link in fabry-database.org or FABRY_CEP.
Nucleotide mutations were mapped onto the reference genome Ensembl GRCh37 release 91 [49].
We performed statistical analysis and data visualisation using the R environment for statistical computing [50].
We calculated descriptive statistics and drew box-and-whiskers plots of residual activity for severe and mild mutations subpopulations using the graphics::boxplot() function on the intersection of the two datasets.
We manually created a confusion matrix using data from the first dataset (175 mutations, from Fabry-database.org, Additional file 1), and measured the goodness of wANNOVAR qualitative predictors using the following indexes:
Raw accuracy:
Balanced Accuracy:
F1 score:
Matthew’s correlation coefficient:
We coded an R function for the simultaneous calculation of these indexes.
Using the second dataset (280 mutations, manually built), we expressed the correlation between the rank score of tools and residual activity as Pearson’s r, and then tested for no correlation using the stats::cor.test() function with ‘less’ alternative (i.e. negative correlation) and the ‘pearson’ method. We drew box-and-whiskers plots of residual activity for every wANNOVAR prediction and conservation tool using the graphics::boxplot() function on the second dataset (280 mutations, manually built).
On the same dataset, we used the graphics::barplot() function for drawing the rank scores of the mutations whose activity is equal or higher than that of wild-type for every wANNOVAR prediction and conservation tool.
Results
In some cases manifestations of FD occur at an early age with general, neurological, cardiovascular and renal signs, in other cases in adulthood and with a limited subset of symptoms. For this reason a qualitative phenotypic classification of mutations based on the symptoms observed in the patients, has been attempted and classic or severe ones have been distinguished from mild, late onset or variant forms [42].
Fabry-database.org [43, 44] provides a list of mutations and their qualitative phenotypic classification. Since FD is X linked and the association between genotype and phenotype is clearer in males [51], only the 175 hemizygous cases have been gathered from Fabry-database.org and form the first dataset analysed in this paper. The variants were annotated with wANNOVAR [3] and the output is provided in Additional file 1 with the original qualitative phenotypic description in the last column. In the first place it can be noticed that only 51 cases are also present in ClinVar, which is a public archive of reports of the relationships among human variations and phenotypes [52].
To test whether it is possible to broadly distinguish FD mutations collected from Fabry-database.org by the qualitative predictions provided by wANNOVAR annotation, the observed phenotypes were reduced to two classes, a severe group POS of 152 cases, which clusters mutations originally defined as “severe” or “classic”, and a mild group NEG of 23 cases, which clusters those mutations originally defined as “mild”, “late onset”, “variant” or “atypical variant” in fabry-database.org. For the predicted phenotypes, if the tool provides binary classification, like in the case of SIFT [4], the more deleterious one, D in the case of SIFT, is considered as predicted POS, the other one, T in the case of SIFT is considered as predicted NEG. If the tool provides multiple classes, as in the case of PolyPhen-2 [12], the most deleterious one, D in the case of PolyPhen-2, is considered as predicted POS, the other ones, P and B in the case of PolyPhen-2, is considered as predicted NEG. The results are summarized in Table 1. Since the two classes have different sizes, Matthews correlation coefficient should be preferred for the evaluation of predictors [53].
Table 1.
Accuracy Indexes
| Category | Predictor | Raw Accuracy | Balanced accuracy | F1-score | Matthew’s correlation coefficient |
|---|---|---|---|---|---|
| B | SIFT | 0.749 | 0.549 | 0.241 | 0.092 |
| B | LRT | 0.794 | 0.576 | 0.280 | 0.162 |
| B | MutationAssessor | 0.191 | 0.460 | 0.247 | −0.106 |
| B | FATHMM | 0.846 | 0.500 | 0.000 | 0.000 |
| B | PROVEAN | 0.737 | 0.557 | 0.258 | 0.103 |
| Meta | MetaSVM | 0.846 | 0.500 | 0.000 | 0.000 |
| Meta | MetaLR | 0.846 | 0.500 | 0.000 | 0.000 |
| Meta | M-CAP | 0.846 | 0.500 | 0.000 | 0.000 |
| ML | Polyphen2_HDIV | 0.771 | 0.592 | 0.310 | 0.175 |
| ML | Polyphen2_HVAR | 0.691 | 0.621 | 0.341 | 0.188 |
| ML | MutationTaster | 0.194 | 0.433 | 0.230 | −0.156 |
| ML | FATHMM-MKL | 0.829 | 0.505 | 0.063 | 0.022 |
Accuracy indexes measuring the ability to differentiate severe from mild GLA mutations for all the predictors used by wANNOVAR. Categories are B for “biologically based prediction method”, ML for “Machine Learning based prediction method”, and Meta for “Meta prediction method”
For most tools the values are quite low and in some cases no discrimination is possible.
A different way of ordering by severity, relies on the residual activity of AGAL mutants measured in vitro in HEK293 or COS cell transiently transfected with expression plasmids. Values for 280 mutations have been collected gathering results of several laboratories [33, 45, 54–62]. They form the second dataset analyzed in this paper. wANNOVAR annotation for these mutants can be found in Additional file 2 with the relative residual activity in the last column.
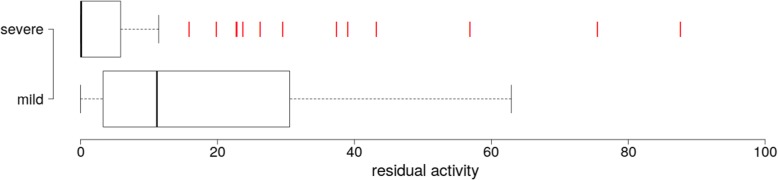
The intersection between the two datasets is formed by 67 mutations of the severe group POS and 12 of the mild group NEG, for which relative residual activity is available. The median residual activity of severe mutations POS is 0.1 (Fig. 1). This finding suggests that severe cases have null, or very close to null activity, when tested in transfected cells. The box plot in Fig. 1 shows 20% outliers with high residual activity in POS population that might represent an overestimation in the original literature.
Fig. 1.
Distribution of residual activities for phenotypically annotated GLA mutations. The boxplot shows the distribution of residual activity in the subpopulations of mutations with severe and mild effects. The red bars represent outliers
Contrary to what occurs in the first dataset of mutations whose phenotypic annotation is derived from clinical literature (Additional file 1), the second dataset, whose annotation is based on residual activity (Additional file 2), is balanced with half of the mutations with values above 0.
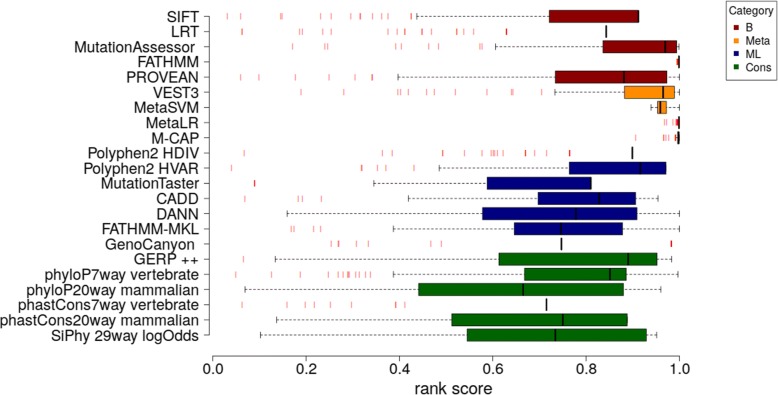
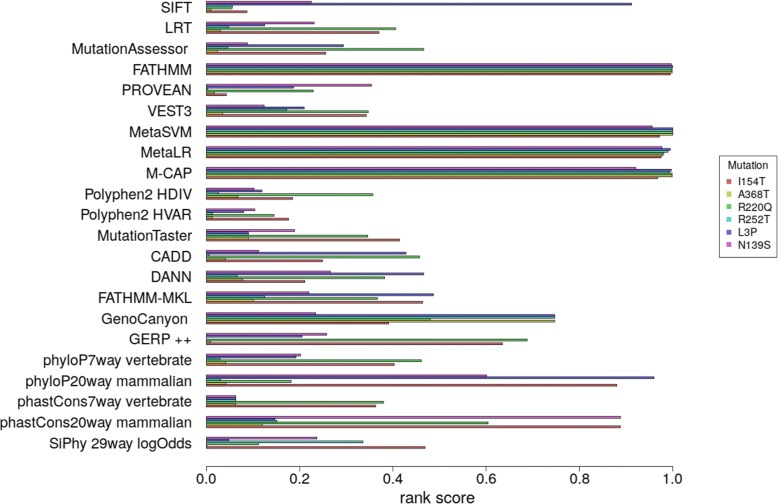
The box plot in Fig. 2 shows the distribution of rank scores for mutations showing 0 residual activity. Rank scores were created by wANNOVAR to make the functional prediction scores and conservation scores more comparable to each other and monotonic (a higher score indicating “more likely to be damaging”) [63]. As can be observed FATHMM [7], metaSVM [10], metaLR [10], M-CAP [11]correctly assign high scores to very severe cases. On the other side, the histograms in Fig. 3 show the rank scores assigned by the predictors to 6 non pathological mutation whose residual activity is comparable or higher than that of wild type. The same predictors, FATHMM, metaSVM [10], metaLR [10], M-CAP [11], give a constantly high score and tend to over-estimate the damage caused by a mutation. In Table 2 the correlation between the rank scores of the predicting tools and the residual activity of all the mutations in the second dataset (Additional file 2), is shown. Results obtained by some predictors used by wANNOVAR, for example VEST3 (Pearson correlation coefficient 0.71; p < 0.0001) and PolyPhen-2 (Pearson correlation coefficient − 0.62; p < 0.0001), demonstrate that the rank scores can correlate with severity in a statistically significant manner. Methods based on evolutionary and phylogenetic analysis perform very poorly.
Fig. 2.
Distribution of rank scores for mutations with null residual activity. The boxplot show the distribution of the rank scores for all the predictors used by wANNOVAR. The red bars represent outliers. Predictor category label is B for “biologically based prediction method”, ML for “Machine Learning based prediction method”, Meta for “Meta prediction method” and Cons for “Conservation scoring tool”
Fig. 3.
Rank scores for mutations with residual activity equal or greater than wild type alpha-galactosidase. The histograms show the rank scores of the six mutations whose residual activity is greater or equal than the wild type alpha-galactosidase, for each of the wANNOVAR predictors. Mutations are color coded, and are detailed inset
Table 2.
Correlations
| Category | Name | Pearson’s r | p-value |
|---|---|---|---|
| B | SIFT | − 0.493 | 7.87E-19 |
| B | LRT | −0.486 | 2.76E-18 |
| B | MutationAssessor | −0.573 | 5.22E-26 |
| B | FATHMM | −0.054 | 1.85E-01 |
| B | PROVEAN | −0.546 | 1.86E-23 |
| Meta | VEST3 | −0.699 | 1.08E-42 |
| Meta | MetaSVM | 0.285 | 1.00E + 00 |
| Meta | MetaLR | −0.482 | 5.77E-18 |
| Meta | M-CAP | −0.255 | 8.09E-06 |
| ML | POLYPHEN2 HDIV | −0.672 | 1.67E-38 |
| ML | POLYPHEN2 HVAR | −0.648 | 4.53E-35 |
| ML | MutationTaster | −0.499 | 2.42E-19 |
| ML | CADD | −0.595 | 1.78E-28 |
| ML | DANN | −0.388 | 8.51E-12 |
| ML | FATHMM-MKL | −0.434 | 1.35E-14 |
| ML | GenoCanyon | −0.282 | 7.95E-07 |
| n | GERP++ | −0.405 | 9.34E-13 |
| Cons | phyloP7way vertebrate | −0.441 | 4.79E-15 |
| Cons | phyloP20way mammalian | −0.214 | 1.54E-04 |
| Cons | phastCons7way vertebrate | −0.486 | 2.55E-18 |
| Cons | phastCons 20 way mammalian | −0.256 | 7.35E-06 |
| Cons | SiPhy 29way logOdds | −0.389 | 7.65E-12 |
Pearson’s r correlation coefficient between rank scores and residual activities, together with the associated p-value for significance scoring, for all the predictors used by wANNOVAR. Bold text is used for the highest correlations. Categories are B for “biologically based prediction method”, ML for “Machine Learning based prediction method”, Meta for “Meta prediction method” and Cons for “Conservation scoring tool”
Discussion
The gene GLA offers an example of the critical points encountered when missense mutations are annotated. In the first place getting the phenotype associated to a mutation is difficult, most information is still missing in databases such as ClinVar [52] and is present only in specialized databases. Mild and severe mutations can be mis-classified in the literature. An example is provided by the mutation D313Y, that is reported as “classic” in fabry-database.org, but is regarded as “likely benign/uncertain significance” in ClinVar [52] and is relatively frequent in the population according to ExAC [64] and 1000Genomes [65] (Additional file 1). The residual activity of D313Y is as high as 75% than wild type (Additional file 2) thus suggesting that the interpretation of fabry-database.org, which is derived from the original source [66], is overestimated. Other examples are provided by the outliers in Fig. 1. Given these premises, it is not surprising that the tools provided by wANNOWAR cannot distinguish mild from severe mutations as they are defined in the literature.
Hence to train or test algorithms that can grade disease severity, datasets of quantitative measures of the damage caused by mutations to the proteins must be available. In this paper we used data produced by a cell based assay that measures relative residual activity in the cells. We showed that some of the popular tools used for exome analysis, are able to grade disease severity, even though they had not been trained or tested for this specific purpose. A summary of all the tools employed in this study is provided in Additional file 3. The best result was obtained with VEST3 [9] that uses a supervised machine learning algorithm, Random Forest based on 86 sequence features and trained with a positive class of missense variants from the Human Gene Mutation Database and a negative class of common missense variants detected in the Exome Sequencing Project population. In a recent paper Plon and co-workers [67] compared the performance of several algorithms using benign or pathogenic missense variants from the ClinVar database [52]. They found “poor concordance among algorithms, particularly for variants classified as benign by clinical laboratories”. Nevertheless they observed that VEST3 has the lowest rate of false positive calls, i.e. benign variants in ClinVar that are erroneously predicted as pathogenic. This finding suggests that the training protocol employed by VEST3 reduces over-prediction of deleterious variants. The second best result was obtained with PolyPhen-2 [12] that calculates bayesian probabilities and uses eight sequence-based and three structure-based predictive features. Since AGAL structure is known [23–25], it is possible that the incorporation of structure-based predictive features contributed to the good results obtained with PolyPhen-2. Two versions of the same program exist. PolyPhen-2 HumDiv is trained with a positive class of mutation causing Mendelian diseases from UniProt and a negative class of variants found in closely related mammalian homologs whereas PolyPhen-2 HumVar is trained with a positive class consisting of all human disease-causing mutations from UniProt and a negative class consisting of nsSNPs without annotated involvement in disease. HumDiv performed slightly better than HumVar. Among the tools that are not limited to exonic missense mutations, CADD is the best performing one. MutationAssessor is the best performing method based on biological principles with a combinatorial entropy formalism. In a previous paper we had shown that the flexibility of the residue where the mutation occurs is the best structural property to predict AGAL mutants residual activity [68]. Results obtained by VEST3, PolyPhen-2, CADD and MutationAssessor are better than those obtained with molecular dynamics (Pearson correlation coefficient R 0.50; p < 0.0001). Although the majority of disease mutations in GLA affect protein stability, methods based on a single structural property perform worse than those relying on several properties.
Admittedly our analysis has two major limitations. In the first place only the programs run by wANNOVAR [3] were considered leaving out those softwares that use three-dimensional structures, for example SDM [69], PoPMuSiC [70] and mCSM [71]. In the second place only one gene was considered. Yet GLA represents a unique case since, to the best of our knowledge, there are few data about residual activity of other mutant proteins. We hope that the effort that was put in place for GLA were extended.
Conclusions
Our paper aims at soliciting a combined effort to produce a large database where the residual activity measured in a cell-based test for diverse proteins is gathered. Indeed this is feasible if cDNA encoding mutants are expressed by transient transfection in suitable mammalian cells. In case of FD, it has been shown that this in vitro test recapitulates what can be observed ex vivo in the cells derived from patients. This approach is not limited to the variants already observed in the patients or in the healthy population and provides data for negative controls too, i.e. mutation that do not affect residual activity. One obvious limitation of the method is that the effect of exonic mutation affecting splicing cannot be evaluated. Once a large dataset from diverse genes is gathered, it could be used to train linear classifiers. We also suggests that programs relying on several features, including structure-based ones, are included in the tools used for the high throughput annotation of data deriving from exome sequencing.
Additional files
wANNOVAR annotated GLA mutation with qualitative phenotypes. (XLSX 121 kb)
wANNOVAR annotated GLA mutation with relative residual activities. (XLSX 212 kb)
Summary of the predictors evaluated in this study and their main characteristics (XLSX 13 kb)
Acknowledgements
This paper is dedicated to our friend and colleague M.Malanga.
Funding
Publication costs for this manuscript were sponsored by a grant from MIUR PRIN 2015 2015JHLY35 (to MVC). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data are available.
About this supplement
This article has been published as part of BMC Bioinformatics Volume 19 Supplement 15, 2018: Proceedings of the 12th International BBCC conference. The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-19-supplement-15.
Abbreviations
- AGAL
lysosomial alpha-galactosidase
- FD
Fabry disease
Authors’ contributions
CC, VC and LL collected the data, GA conceived the experiments, MVC wrote the paper, BH-M carried out statistical analysis and conceived the paper. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chiara Cimmaruta, Email: chiara.cimmaruta@unina.it.
Valentina Citro, Email: vale.ctr@gmail.com.
Giuseppina Andreotti, Email: gandreotti@icb.cnr.it.
Ludovica Liguori, Email: lud.liguori@gmail.com.
Maria Vittoria Cubellis, Phone: +39 081-679118, Email: cubellis@unina.it.
Bruno Hay Mele, Email: bruno.haymele@unina.it.
References
- 1.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(11.10):11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49(7):433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19(9):1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34(1):57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, Bernstein JA, Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48(12):1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 14.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31(5):761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, Gaunt TR, Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31(10):1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Q, Hu Y, Sun J, Cheng Y, Cheung KH, Zhao H. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci Rep. 2015;5:10576. doi: 10.1038/srep10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper GM, Goode DL, Ng SB, Sidow A, Bamshad MJ, Shendure J, Nickerson DA. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7(4):250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25(12):i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garman SC, Garboczi DN. The molecular defect leading to Fabry disease: structure of human alpha-galactosidase. J Mol Biol. 2004;337(2):319–335. doi: 10.1016/j.jmb.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman RL, D’Aquino JA, Ringe D, Petsko GA. Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability. Biochemistry. 2009;48(22):4816–4827. doi: 10.1021/bi9002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guce AI, Clark NE, Salgado EN, Ivanen DR, Kulminskaya AA, Brumer H, 3rd, Garman SC. Catalytic mechanism of human alpha-galactosidase. J Biol Chem. 2010;285(6):3625–3632. doi: 10.1074/jbc.M109.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta A, Hughes DA: Fabry disease. 1993.
- 27.Germain DP. Fabry disease. Orphanet J Rare Dis. 2010;5:30. doi: 10.1186/1750-1172-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas AS, Mehta AB. Difficulties and barriers in diagnosing Fabry disease: what can be learnt from the literature? Expert Opin Med Diagn. 2013;7(6):589–599. doi: 10.1517/17530059.2013.846322. [DOI] [PubMed] [Google Scholar]
- 29.Citro Valentina, Cammisa Marco, Liguori Ludovica, Cimmaruta Chiara, Lukas Jan, Cubellis Maria, Andreotti Giuseppina. The Large Phenotypic Spectrum of Fabry Disease Requires Graduated Diagnosis and Personalized Therapy: A Meta-Analysis Can Help to Differentiate Missense Mutations. International Journal of Molecular Sciences. 2016;17(12):2010. doi: 10.3390/ijms17122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Germain DP, Waldek S, Banikazemi M, Bushinsky DA, Charrow J, Desnick RJ, Lee P, Loew T, Vedder AC, Abichandani R, et al. Sustained, long-term renal stabilization after 54 months of agalsidase beta therapy in patients with Fabry disease. J Am Soc Nephrol. 2007;18(5):1547–1557. doi: 10.1681/ASN.2006080816. [DOI] [PubMed] [Google Scholar]
- 31.Tondel C, Bostad L, Larsen KK, Hirth A, Vikse BE, Houge G, Svarstad E. Agalsidase benefits renal histology in young patients with Fabry disease. J Am Soc Nephrol. 2013;24(1):137–148. doi: 10.1681/ASN.2012030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rombach SM, Smid BE, Bouwman MG, Linthorst GE, Dijkgraaf MG, Hollak CE. Long term enzyme replacement therapy for Fabry disease: effectiveness on kidney, heart and brain. Orphanet J Rare Dis. 2013;8:47. doi: 10.1186/1750-1172-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giugliani R, Waldek S, Germain DP, Nicholls K, Bichet DG, Simosky JK, Bragat AC, Castelli JP, Benjamin ER, Boudes PF. A phase 2 study of migalastat hydrochloride in females with Fabry disease: selection of population, safety and pharmacodynamic effects. Mol Genet Metab. 2013;109(1):86–92. doi: 10.1016/j.ymgme.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Germain DP, Hughes DA, Nicholls K, Bichet DG, Giugliani R, Wilcox WR, Feliciani C, Shankar SP, Ezgu F, Amartino H, et al. Treatment of Fabry's disease with the pharmacologic chaperone Migalastat. N Engl J Med. 2016;375(6):545–555. doi: 10.1056/NEJMoa1510198. [DOI] [PubMed] [Google Scholar]
- 35.Benjamin ER, Della Valle MC, Wu X, Katz E, Pruthi F, Bond S, Bronfin B, Williams H, Yu J, Bichet DG, et al. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genet Med. 2017;19(4):430–438. doi: 10.1038/gim.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guce AI, Clark NE, Rogich JJ, Garman SC. The molecular basis of pharmacological chaperoning in human alpha-galactosidase. Chem Biol. 2011;18(12):1521–1526. doi: 10.1016/j.chembiol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haneef SA, Doss CG. Personalized Pharmacoperones for Lysosomal storage disorder: approach for next-generation treatment. Adv Protein Chem Struct Biol. 2016;102:225–265. doi: 10.1016/bs.apcsb.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Cox TM. Substrate reduction therapy for lysosomal storage diseases. Acta Paediatr Suppl. 2005;94(447):69–75. doi: 10.1080/08035320510028157. [DOI] [PubMed] [Google Scholar]
- 39.Platt FM, Neises GR, Dwek RA, Butters TD. N-butyldeoxynojirimycin is a novel inhibitor of glycolipid biosynthesis. J Biol Chem. 1994;269(11):8362–8365. [PubMed] [Google Scholar]
- 40.Tierney M, Pottage J, Kessler H, Fischl M, Richman D, Merigan T, Powderly W, Smith S, Karim A, Sherman J, et al. The tolerability and pharmacokinetics of N-butyl-deoxynojirimycin in patients with advanced HIV disease (ACTG 100). The AIDS Clinical Trials Group (ACTG) of the National Institute of Allergy and Infectious Diseases. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10(5):549–553. doi: 10.1097/00042560-199510050-00008. [DOI] [PubMed] [Google Scholar]
- 41.Hay Mele B, Citro V, Andreotti G, Cubellis MV. Drug repositioning can accelerate discovery of pharmacological chaperones. Orphanet J Rare Dis. 2015;10:55. doi: 10.1186/s13023-015-0273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whybra C, Bahner F, Baron K: Measurement of disease severity and progression in Fabry disease. 2006. [PubMed]
- 43.Saito S, Ohno K, Sese J, Sugawara K, Sakuraba H. Prediction of the clinical phenotype of Fabry disease based on protein sequential and structural information. J Hum Genet. 2010;55(3):175–178. doi: 10.1038/jhg.2010.5. [DOI] [PubMed] [Google Scholar]
- 44.Saito S, Ohno K, Sakuraba H. Fabry-database.org: database of the clinical phenotypes, genotypes and mutant alpha-galactosidase a structures in Fabry disease. J Hum Genet. 2011;56(6):467–468. doi: 10.1038/jhg.2011.31. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Katz E, Della Valle MC, Mascioli K, Flanagan JJ, Castelli JP, Schiffmann R, Boudes P, Lockhart DJ, Valenzano KJ, et al. A pharmacogenetic approach to identify mutant forms of alpha-galactosidase a that respond to a pharmacological chaperone for Fabry disease. Hum Mutat. 2011;32(8):965–977. doi: 10.1002/humu.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukas J, Knospe AM, Seemann S, Citro V, Cubellis MV, Rolfs A. In vitro enzyme measurement to test pharmacological chaperone responsiveness in Fabry and Pompe disease. J Vis Exp. 2017;130. [DOI] [PMC free article] [PubMed]
- 47.Andreotti G, Guarracino MR, Cammisa M, Correra A, Cubellis MV. Prediction of the responsiveness to pharmacological chaperones: lysosomal human alpha-galactosidase, a case of study. Orphanet J Rare Dis. 2010;5:36. doi: 10.1186/1750-1172-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cammisa M, Correra A, Andreotti G, Cubellis MV. Fabry_CEP: a tool to identify Fabry mutations responsive to pharmacological chaperones. Orphanet J Rare Dis. 2013;8:111. doi: 10.1186/1750-1172-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Team RC: R: a language and environment for statistical computing. 2013.
- 51.Echevarria L, Benistan K, Toussaint A, Dubourg O, Hagege AA, Eladari D, Jabbour F, Beldjord C, De Mazancourt P, Germain DP. X-chromosome inactivation in female patients with Fabry disease. Clin Genet. 2016;89(1):44–54. doi: 10.1111/cge.12613. [DOI] [PubMed] [Google Scholar]
- 52.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chicco D. Ten quick tips for machine learning in computational biology. BioData Min. 2017;10:35. doi: 10.1186/s13040-017-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishii S, Suzuki Y, Fan JQ. Role of Ser-65 in the activity of alpha-galactosidase a: characterization of a point mutation (S65T) detected in a patient with Fabry disease. Arch Biochem Biophys. 2000;377(2):228–233. doi: 10.1006/abbi.2000.1743. [DOI] [PubMed] [Google Scholar]
- 55.Spada M, Pagliardini S, Yasuda M, Tukel T, Thiagarajan G, Sakuraba H, Ponzone A, Desnick RJ. High incidence of later-onset fabry disease revealed by newborn screening. Am J Hum Genet. 2006;79(1):31–40. doi: 10.1086/504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park JY, Kim GH, Kim SS, Ko JM, Lee JJ, Yoo HW. Effects of a chemical chaperone on genetic mutations in alpha-galactosidase a in Korean patients with Fabry disease. Exp Mol Med. 2009;41(1):1–7. doi: 10.3858/emm.2009.41.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filoni C, Caciotti A, Carraresi L, Cavicchi C, Parini R, Antuzzi D, Zampetti A, Feriozzi S, Poisetti P, Garman SC, et al. Functional studies of new GLA gene mutations leading to conformational Fabry disease. Biochim Biophys Acta. 2010;1802(2):247–252. doi: 10.1016/j.bbadis.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreotti G, Citro V, De Crescenzo A, Orlando P, Cammisa M, Correra A, Cubellis MV. Therapy of Fabry disease with pharmacological chaperones: from in silico predictions to in vitro tests. Orphanet J Rare Dis. 2011;6:66. doi: 10.1186/1750-1172-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukas J, Giese AK, Markoff A, Grittner U, Kolodny E, Mascher H, Lackner KJ, Meyer W, Wree P, Saviouk V, et al. Functional characterisation of alpha-galactosidase a mutations as a basis for a new classification system in fabry disease. PLoS Genet. 2013;9(8):e1003632. doi: 10.1371/journal.pgen.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreotti G, Citro V, Correra A, Cubellis MV. A thermodynamic assay to test pharmacological chaperones for Fabry disease. Biochim Biophys Acta. 2014;1840(3):1214–1224. doi: 10.1016/j.bbagen.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Citro V, Pena-Garcia J, den-Haan H, Perez-Sanchez H, Del Prete R, Liguori L, Cimmaruta C, Lukas J, Cubellis MV, Andreotti G. Identification of an allosteric binding site on human Lysosomal alpha-Galactosidase opens the way to new pharmacological chaperones for Fabry disease. PLoS One. 2016;11(10):e0165463. doi: 10.1371/journal.pone.0165463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lukas J, Scalia S, Eichler S, Pockrandt AM, Dehn N, Cozma C, Giese AK, Rolfs A. Functional and clinical consequences of novel alpha-Galactosidase a mutations in Fabry disease. Hum Mutat. 2016;37(1):43–51. doi: 10.1002/humu.22910. [DOI] [PubMed] [Google Scholar]
- 63.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37(3):235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eng CM, Resnick-Silverman LA, Niehaus DJ, Astrin KH, Desnick RJ. Nature and frequency of mutations in the alpha-galactosidase a gene that cause Fabry disease. Am J Hum Genet. 1993;53(6):1186–1197. [PMC free article] [PubMed] [Google Scholar]
- 67.Ghosh R, Oak N, Plon SE. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 2017;18(1):225. doi: 10.1186/s13059-017-1353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cubellis MV, Baaden M, Andreotti G. Taming molecular flexibility to tackle rare diseases. Biochimie. 2015;113:54–58. doi: 10.1016/j.biochi.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandurangan AP, Ochoa-Montano B, Ascher DB, Blundell TL. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017;45(W1):W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinform. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pires DE, Ascher DB, Blundell TL. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30(3):335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
wANNOVAR annotated GLA mutation with qualitative phenotypes. (XLSX 121 kb)
wANNOVAR annotated GLA mutation with relative residual activities. (XLSX 212 kb)
Summary of the predictors evaluated in this study and their main characteristics (XLSX 13 kb)
Data Availability Statement
All data are available.