Abstract
Background
Neonatal sepsis is an inflammatory systemic syndrome, which is a major cause of morbidity and mortality in premature infants. We analyzed the expression profile data of E-MTAB-4785 to reveal the pathogenesis of the disease.
Methods
The expression profile dataset E-MTAB-4785, which contained 17 sepsis samples and 19 normal samples, was obtained from the ArrayExpress database. The differentially expressed genes (DEGs) were analyzed by the Bayesian testing method in limma package. Based on the DAVID online tool, enrichment analysis was conducted for the DEGs. Using STRING database and Cytoscape software, protein-protein interaction (PPI) network and module analyses were performed. Besides, transcription factor (TF)-DEG regulatory network was also constructed by Cytoscape software. Additionally, miRNA-DEG pairs were searched using miR2Disease and miRWalk 2.0 databases, followed by miRNA-DEG regulatory network was visualized by Cytoscape software.
Results
A total of 275 DEGs were identified from the sepsis samples in comparison to normal samples. TSPO, MAPK14, and ZAP70 were the hub nodes in the PPI network. Pathway enrichment analysis indicated that CEBPB and MAPK14 were enriched in TNF signaling pathway. Moreover, CEBPB and has-miR-150 might function in neonatal sepsis separately through targeting MAPK14 and BCL11B in the regulatory networks. These genes and miRNA might be novel targets for the clinical treatment of neonatal sepsis.
Conclusion
TSPO, ZAP70, CEBPB targeting MAPK14, has-miR-150 targeting BCL11B might affect the pathogenesis of neonatal sepsis. However, their roles in neonatal sepsis still needed to be confirmed by further experimental researches.
Keywords: Neonatal sepsis, Differentially expressed genes, Enrichment analysis, Protein-protein interaction network, Regulatory network
Background
As a kind of inflammatory systemic syndrome that is determined by a serious infection (bacterial or viral sepsis), neonatal sepsis is composed of early-onset sepsis (EOS) and late-onset sepsis (LOS) [1]. The leading risk factors of the disease are prematurity, prolonged rupture of membranes, and low birth weight [2]. Besides, neonatal sepsis is a main reason for morbidity and mortality in premature infants [3]. Therefore, investigating the molecular mechanisms of neonatal sepsis is urgent and important for developing novel therapies.
The key genes and miRNAs involved in the mechanisms of neonatal sepsis have been reported by several studies. For instance, interleukin (IL)-17A is recognized as a effector of IL-18–mediated injury and the outcomes of neonatal sepsis can be improved by disrupting the IL-18/IL-1/IL-17A axis [4]. Previous studies explore pancreatic stone protein (PSP) in neonatal sepsis, finding that PSP is a potential biomarker in combination with procalcitonin in EOS [5, 6]. The overexpression of CD64 is demonstrated to be a highly specific indicator for patients with neonatal sepsis, and CD64 index can be utilized to determine LOS before infants show signs of infection [7, 8]. MiR-15a/16 is a miRNA that may have a critical role in the regulation of gene expression at the post-transcriptional level, which is declared to be useful for the diagnosis and prognosis of neonatal sepsis [9]. In spite of the above studies, the genes and miRNAs acting in neonatal sepsis have not been fully revealed.
In 2014, Cernada et al. [10] used genome-wide expression profiles to assess the expression differences between infants with and without neonatal sepsis, finding that the expression profiles have differences in the early days of neonatal period. However, they have not performed comprehensive bioinformatics analysis to reveal the mechanisms of neonatal sepsis. Using the data deposited by Cernada et al. [10], we further conducted differential expression analysis, enrichment analysis, protein-protein interaction (PPI) network analysis and regulatory network analysis to better or to further investigate the structure and expression of some of the genes that seemed to influence the genesis of neonatal sepsis.
Materials and methods
Microarray data
The expression profile data of E-MTAB-4785 was downloaded from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/), which was sequenced on the platform of the Affymetrix GeneChip Human Gene 1.0 ST Array [HuGene-1_0-st-v1]. E-MTAB-4785 included 17 blood samples from infants with neonatal sepsis (gestational age = 27 ± 2 weeks, birth weight = 1030 ± 231, 11 males and 6 females) and 19 blood samples from infants without neonatal sepsis (gestational age = 28 ± 2 weeks, birth weight = 1130 ± 329, 13 males and 6 females). All very low birth weight (VLBW) infants were selected from the University and Polytechnic Hospital La Fe between April 2011 to September 2012. Sepsis infants were those with clinical signs of sepsis [11] and/or risk factors [12, 13], while nonseptic infants were those without clinical signs of infection. Venous blood was obtained from sepsis infants and nonseptic infants before starting antibiotics, and stored at − 20 °C for following RNA extraction. The expression profile data of E-MTAB-4785 was deposited by Cernada et al. [10]. The study of Cernada et al. got the approval of the local institutional review board, and informed consent of all participants were provided by their parents or representatives.
Data preprocessing and differentially expressed genes (DEGs) screening
After the raw data were read by the oligo package (http://www.bioconductor.org/packages/release/bioc/html/oligo.html) [14] in R, data preprocessing were performed using the Robust MultiArray Averaging (RMA) method [15], including background correction, quantile normalization and expression calculation. The average value of probes was obtained as the final gene expression value of their corresponding common gene symbol. The Bayesian testing method [16] in limma package (http://www.bioconductor.org/packages/release/bioc/html/limma.html) [17] was utilized to identify the DEGs between the two group samples. The p-values were conducted with multiple testing adjustment based on the Benjamini & Hochberg method [18]. The adjusted p-value < 0.05 and |log2fold-change (FC)| > 0.585 were set as the thresholds.
Functional and pathway enrichment analysis
The Gene Ontology (GO, http://www.geneontology.org) project describes gene products from biological process (BP), cell component (CC) and molecular function (MF) aspects [19]. The KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.ad.jp/kegg/kegg2.html) database involves not only genes but also their functional information [20]. Using the DAVID (Database for Annotation, Visualization and Integrated Discovery, version 6.8, http://david.abcc.ncifcrf.gov) online tool [21], GO functional and KEGG pathway enrichment analyses were carried out for the DEGs. The p-value < 0.05 was selected as the cut-off criterion.
PPI network and module analyses
Using the STRING database (version 10.0, http://string-db.org) [22], the PPI analysis was conducted for the DEGs, with the required confidence (combined score) > 0.4 as the threshold. Followed by the PPI network was visualized by the Cytoscape software (http://www.cytoscape.org/) [23]. Based on the CytoNCA plugin [24] in Cytoscape software, degree centrality (DC), betweenness centrality (BC) and closeness centrality (CC) of nodes were analyzed to identify hub nodes [25]. In addition, module analysis was conducted for the network using the MCODE plugin [26] in Cytoscape software.
Regulatory network construction
Using the iRegulon plugin [27] in Cytoscape software, the transcription factor (TF)-DEG pairs in the PPI network were predicted, with the Normalized Enrichment Score (NES) > 3 as the threshold. Then, TF-DEG regulatory network was visualized using Cytoscape software [23]. The sepsis-associated miRNAs were searched from the miR2Disease (http://www.mir2disease.org/) database [28], and then their targets were downloaded from the miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/genepub.html) database [29]. Through mapping the target genes into the DEGs, the sepsis-associated miRNA-DEG pairs were screened and then miRNA-DEG regulatory network was constructed by Cytoscape software [23].
Results
Analysis of DEGs
Through data preprocessing, the expression values of 18,718 genes were acquired. Compared with normal samples, there were a total of 275 DEGs in the sepsis samples, including 180 up-regulated genes and 95 down-regulated genes. Therefore, there were more up-regulated genes in relative to down-regulated genes.
Functional and pathway enrichment analysis
GO functional and KEGG pathway enrichment analyses separately were conducted for the up-regulated genes, and the top 5 terms in each category were listed in Table 1. The terms enriched for the up-regulated genes included cellular response to lipopolysaccharide (BP, p = 3.05E-06), extracellular exosome (CC, p = 3.26E-06), histone acetyltransferase binding (MF, p = 2.54E-03), and TNF signaling pathway (pathway, p = 3.65E-06). Meanwhile, the top 5 terms enriched for the down-regulated genes were listed in Table 2, including regulation of immune response (BP, p = 4.11E-15), T cell receptor complex (CC; p = 1.22E-10; which involved zeta-chain-associated protein of 70 kDa, ZAP70), transmembrane signaling receptor activity (MF, p = 4.20E-06), and T cell receptor signaling pathway (pathway; p = 4.90E-07; which involved ZAP70).
Table 1.
The functions and pathways enriched for the up-regulated genes
| Category | Description | P-value | Gene number |
|---|---|---|---|
| BP | GO:0071222~cellular response to lipopolysaccharide | 3.05E-06 | 10 |
| GO:0051384~response to glucocorticoid | 1.75E-05 | 8 | |
| GO:0006955~immune response | 6.59E-05 | 14 | |
| GO:0045766~positive regulation of angiogenesis | 1.31E-04 | 8 | |
| GO:0045087~innate immune response | 3.22E-04 | 22 | |
| CC | GO:0070062~extracellular exosome | 3.26E-06 | 51 |
| GO:0005615~extracellular space | 3.15E-05 | 29 | |
| GO:0005886~plasma membrane | 1.58E-04 | 59 | |
| GO:0016020~membrane | 2.03E-03 | 32 | |
| GO:0031093~platelet alpha granule lumen | 1.08E-02 | 4 | |
| MF | GO:0035035~histone acetyltransferase binding | 2.54E-03 | 4 |
| GO:0001077~transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 9.14E-03 | 8 | |
| GO:0002020~protease binding | 1.40E-02 | 5 | |
| GO:0016787~hydrolase activity | 1.56E-02 | 5 | |
| GO:0019899~enzyme binding | 1.88E-02 | 9 | |
| Pathway | hsa04668:TNF signaling pathway | 3.65E-06 | 10 |
| hsa05150:Staphylococcus aureus infection | 3.23E-04 | 6 | |
| hsa05142:Chagas disease (American trypanosomiasis) | 7.74E-03 | 6 | |
| hsa04380:Osteoclast differentiation | 1.90E-02 | 6 | |
| hsa04910:Insulin signaling pathway | 2.39E-02 | 6 |
BP biological process, CC cell component, MF molecular function
Table 2.
The functions and pathways enriched for the down-regulated genes
| Category | Description | P-value | Gene number |
|---|---|---|---|
| BP | GO:0050776~regulation of immune response | 4.11E-15 | 15 |
| GO:0007166~cell surface receptor signaling pathway | 2.56E-06 | 11 | |
| GO:0050852~T cell receptor signaling pathway | 8.71E-05 | 7 | |
| GO:0072678~T cell migration | 3.56E-04 | 3 | |
| GO:0045059~positive thymic T cell selection | 8.47E-04 | 3 | |
| CC | GO:0042101~T cell receptor complex | 1.22E-10 | 7 |
| GO:0005886~plasma membrane | 1.40E-04 | 35 | |
| GO:0042105~alpha-beta T cell receptor complex | 2.26E-04 | 3 | |
| GO:0001772~immunological synapse | 5.25E-04 | 4 | |
| GO:0009897~external side of plasma membrane | 4.83E-03 | 6 | |
| MF | GO:0004888~transmembrane signaling receptor activity | 4.20E-06 | 9 |
| GO:1990405~protein antigen binding | 4.46E-04 | 3 | |
| GO:0030246~carbohydrate binding | 1.62E-03 | 6 | |
| GO:0023024~MHC class I protein complex binding | 1.40E-02 | 2 | |
| GO:0032393~MHC class I receptor activity | 2.31E-02 | 2 | |
| Pathway | hsa04660:T cell receptor signaling pathway | 4.90E-07 | 8 |
| hsa04672:Intestinal immune network for IgA production | 7.58E-05 | 5 | |
| hsa05340:Primary immunodeficiency | 5.41E-04 | 4 | |
| hsa04514:Cell adhesion molecules (CAMs) | 5.95E-04 | 6 | |
| hsa04650:Natural killer cell mediated cytotoxicity | 2.80E-03 | 5 |
BP biological process, CC cell component, MF molecular function
PPI network and module analyses
A PPI network was visualized for the DEGs, which involved 144 nodes and 311 interactions (Fig. 1). According to the DC, BC and CC scores of nodes, translocator protein (TSPO), mitogen-activated protein kinase 14 (MAPK14), and ZAP70 were the hub nodes in the PPI network (Table 3). Based on the MCODE plugin, a total of 5 modules were identified from the PPI network. Among which, the genes involved in module 1 (which had the highest score, score = 3.333) were mainly enriched in TNF signaling pathway (p = 4.59E-03), pathways in cancer (p = 5.42E-03), and osteoclast differentiation (p = 6.97E-03) (Table 4).
Fig. 1.
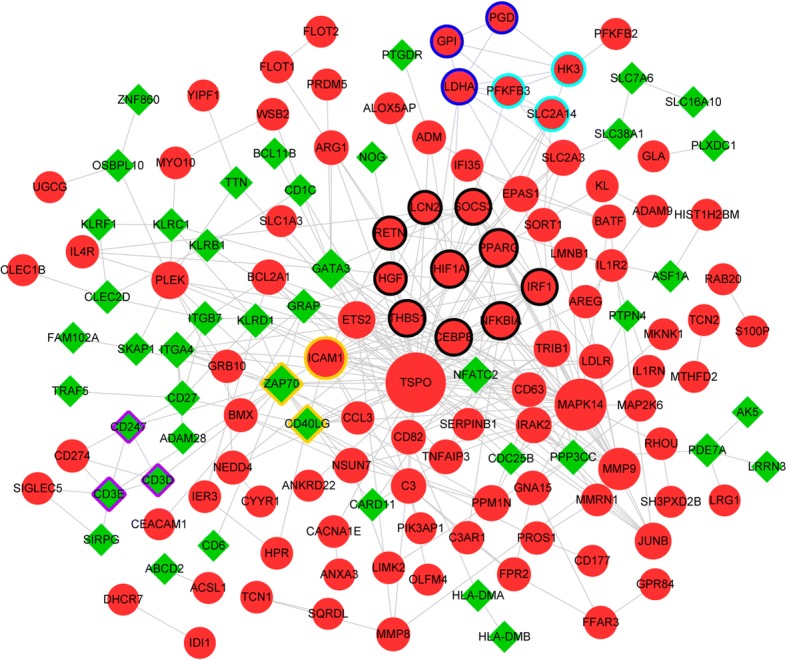
The protein-protein interaction (PPI) network constructed for the differentially expressed genes (DEGs). The red circles and green diamonds separately stand for up-regulated genes and down-regulated genes. The bigger nodes have higher connectivity degrees. Different edge colors represent different modules (module 1: black; module 2: yellow; module 3: dark blue; module 4: purple; module 5: light blue)
Table 3.
The top 15 nodes in the protein-protein interaction (PPI) network
| Node | DC | Node | BC | Node | CC |
|---|---|---|---|---|---|
| TSPO | 42 | TSPO | 8787.401 | TSPO | 0.084416 |
| MAPK14 | 30 | MAPK14 | 3945.718 | MAPK14 | 0.082755 |
| ZAP70 | 18 | ZAP70 | 2945.19 | ZAP70 | 0.081621 |
| ICAM1 | 18 | PLEK | 1416.809 | ICAM1 | 0.081481 |
| MMP9 | 16 | CD40LG | 1228.869 | CD40LG | 0.081066 |
| GATA3 | 13 | GATA3 | 1149.809 | NFATC2 | 0.080882 |
| PPARG | 13 | PPP3CC | 1065.391 | THBS1 | 0.080609 |
| NFKBIA | 12 | SLC2A3 | 878.1572 | MMP9 | 0.080609 |
| HIF1A | 12 | PDE7A | 786 | NFKBIA | 0.080473 |
| CD40LG | 11 | LDHA | 778.0661 | GATA3 | 0.080292 |
| THBS1 | 11 | NFKBIA | 706.2174 | HIF1A | 0.080292 |
| CEBPB | 11 | GNA15 | 660.3287 | CEBPB | 0.080247 |
| SOCS3 | 10 | HIF1A | 614.6019 | PPARG | 0.080157 |
| IRF1 | 10 | CD3E | 583.8388 | SOCS3 | 0.080157 |
| PLEK | 9 | CD82 | 550.5645 | PLEK | 0.079844 |
DC degree centrality, BC betweenness centrality, CC closeness centrality
Table 4.
The pathways enriched for the genes involved in module 1
| Description | Gene number | P-value |
|---|---|---|
| hsa04668:TNF signaling pathway | 3 | 4.59E-03 |
| hsa05200:Pathways in cancer | 4 | 5.42E-03 |
| hsa04380:Osteoclast differentiation | 3 | 6.97E-03 |
| hsa05160:Hepatitis C | 3 | 7.18E-03 |
| hsa05205:Proteoglycans in cancer | 3 | 1.57E-02 |
| hsa05144:Malaria | 2 | 4.87E-02 |
Regulatory network analysis
A TF-DEG regulatory network was identified from the PPI network, in which the TF CCAAT/enhancer binding protein (C/EBP), beta (CEBPB) targeted 43 DEGs (such as MAPK14) (Fig. 2). Besides, pathway enrichment analysis was also conducted for the genes involved in the TF-DEG regulatory network, the enriched pathways included TNF signaling pathway (p = 4.89E-07, which involved CEBPB and MAPK14), Epstein-Barr virus infection (p = 2.09E-04), and osteoclast differentiation (p = 5.64E-04) (Table 5).
Fig. 2.
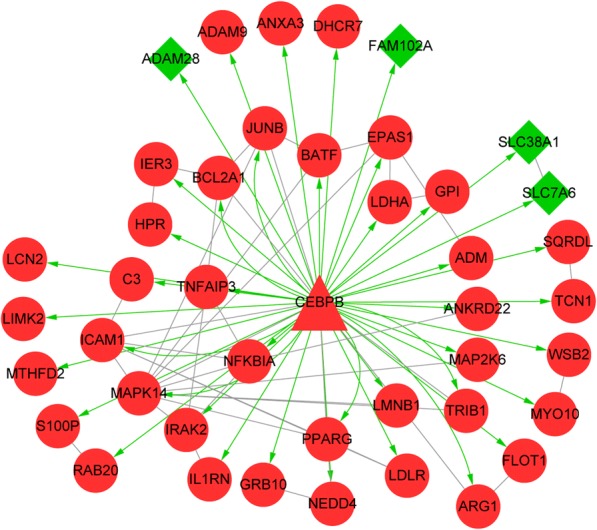
The transcription factor (TF)-differentially expressed gene (DEG) regulatory network identified from the protein-protein interaction (PPI) network. The red circles, green diamonds and red triangles represent up-regulated genes, down-regulated genes and TFs, respectively. Green arrows and grey lines separately indicate TF-DEG regulation relationships and PPIs
Table 5.
The pathways enriched for the genes involved in the transcriptional regulatory network
| Description | Gene number | P-value |
|---|---|---|
| hsa04668:TNF signaling pathway | 7 | 4.89E-07 |
| hsa05169:Epstein-Barr virus infection | 6 | 2.09E-04 |
| hsa04380:Osteoclast differentiation | 5 | 5.64E-04 |
| hsa04064:NF-kappa B signaling pathway | 4 | 2.19E-03 |
| hsa05145:Toxoplasmosis | 4 | 5.20E-03 |
| hsa04621:NOD-like receptor signaling pathway | 3 | 1.19E-02 |
| hsa05164:Influenza A | 4 | 1.51E-02 |
| hsa05152:Tuberculosis | 4 | 1.58E-02 |
| hsa05140:Leishmaniasis | 3 | 1.94E-02 |
| hsa05142:Chagas disease (American trypanosomiasis) | 3 | 3.93E-02 |
| hsa04620:Toll-like receptor signaling pathway | 3 | 4.07E-02 |
According to the miR2Disease database, has-miR-150 was obtained as a sepsis-associated miRNA. Followed by the targets of has-miR-150 were downloaded from the miRWalk 2.0 database. Among the targets, 8 genes (including 3 up-regulated genes and 5 down-regulated genes; such as B-cell CLL/lymphoma 11B, BCL11B) were differentially expressed in this study. In addition, the miRNA-DEG regulatory network is showed in Fig. 3.
Fig. 3.
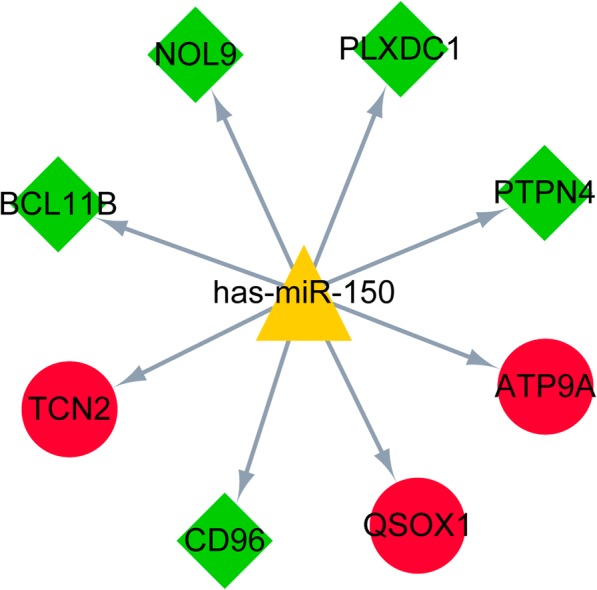
The microRNA (miRNA)-differentially expressed gene (DEG) regulatory network of has-miR-150 and its targets. The red circles, green diamonds and yellow triangles represent up-regulated genes, down-regulated genes and miRNAs, respectively
Discussion
In this study, there were 275 DEGs (including 180 up-regulated genes and 95 down-regulated genes) in the sepsis samples compared with normal samples. A PPI network for the DEGs was constructed, in which TSPO, MAPK14, and ZAP70 were hub nodes according to the DC, BC and CC scores. Besides, a total of 5 modules were screened from the PPI network. Furthermore, CEBPB-DEG regulatory network and has-miR-150-DEG regulatory network separately were constructed.
TSPO (also named the peripheral benzodiazepine receptor) functions in the regulation of inflammation and immune function, and its antagonist PK-11195 can relief cigarette smoke extract-induced inflammation [30]. Santoro et al. demonstrate that TSPO ligands can lower pro-inflammatory enzymes and oxidative stress in glial cells via the biosynthesis of neurosteroids, thus these compounds may be used for treating inflammatory-based neuropathologies [31]. The MAPK14/p38α signaling is considered as a central pathway for integrating useful signals in dendritic cells for inflammation and TH17 differentiation [32]. The p38 kinase plays roles in responses of T cells and inflammation, which mediates the production of crucial inflammatory regulators (including IL-1β; TNFα; and cyclooxygenase-2, COX-2) through cells of the innate immune system [33]. The p38 MAP kinase signal transduction pathway plays an essential role in regulating inflammation and proinflammatory cytokine production, and inhibiting p38α isoform is sufficient and necessary for anti-inflammatory effect [34]. Enrichment analysis showed that ZAP70 was enriched in the function of T cell receptor complex and the T cell receptor signaling pathway. The function deletion of the ZAP70 tyrosine kinase in humans can lead to a serious immunodeficiency, characterized by non-functional CD4+ T cells and lacking mature CD8+ T cells [35]. In ZAP70-deficient patients, circulating T cells are under inadequate supervision, no longer differentiate into TH2 T cells, are short of inhibitory growth controls, and show decreased apoptosis, finally developing into inflammation and autoimmunity [36]. These declared that TSPO, MAPK14, and ZAP70 might play critical roles in neonatal sepsis.
Through inducing the CEBP family, endoplasmic reticulum (ER) stress results in inflammatory responses in response to palmitic acid stimulation in pancreatic acinar cells, wherein CEBPB activation induces CEBPA activation [37]. As a key transcription factor affecting metabolic disturbances, CEBPB is critical for the maturation and differentiation of adipocytes and is overexpressed during proinflammatory conditions and ER stress [38]. Pathway enrichment analysis indicated that CEBPB and MAPK14 were enriched in TNF signaling pathway. In the TF-DEG regulatory network, MAPK14 was targeted by CEBPB, indicating that CEBPB targeting MAPK14 might function in neonatal sepsis through TNF signaling pathway.
Vasilescu et al. demonstrate that the levels of miR-150 in both plasma and leukocytes are associated with sepsis aggressiveness and thus can act as a biomarker of early sepsis [39]. Decreased miR-150 serum levels correlate with an adverse outcome in patients with sepsis and critical illness, and circulating miR-150 serum concentrations may be used as a potential prognostic marker in patients with critical illness [40]. Previous studies reported that BCL11B is not only a transcription factor specific in T-cell, but also contributes to the lineage fidelity and the genetic and functional programs of mature type 2 innate lymphoid cell (ILC2) [41, 42]. In the miRNA-DEG regulatory network, BCL11B was targeted by has-miR-150, suggesting that has-miR-150 might be involved in the pathogenesis of neonatal sepsis by targeting BCL11B.
Conclusions
In conclusion, a total of 275 DEGs were identified from the sepsis samples. Besides, TSPO, ZAP70, CEBPB targeting MAPK14, has-miR-150 targeting BCL11B might act in the pathogenesis of neonatal sepsis. However, further experimental researches are needed to confirm these results obtained from bioinformatics analysis.
Acknowledgements
None.
Funding
None.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- (BP)
Biological process
- (CC)
Cell component
- (DC)
Degree centrality
- (EOS)
Early-onset sepsis
- (ER)
Endoplasmic reticulum
- (GO)
Gene Ontology
- (ILC2)
2 innate lymphoid cell
- (LOS)
Late-onset sepsis
- (MAPK14)
Mitogen-activated protein kinase 14
- (NES)
Normalized Enrichment Score
- (PPI)
Protein-protein interaction
- (PSP)
Pancreatic stone protein
- (RMA)
Robust MultiArray Averaging
- (TF)
Transcription factor
- (TNF-α)
Tumor necrosis factor-α
- (TSPO)
Translocator protein
- (VLBW)
Very low birth weight
Authors’ contributions
LH conceived and designed the research. LQ and HZ acquired the data. LH, LJ and LY analyzed and interpreted the data. LH drafted the manuscript. LH and LJ revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study of Cernada et al. got the approval of the local institutional review board, and informed consent of all participants were provided by their parents or representatives.
Consent for publication
The consent for publication of all participate were required.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Li Huang, Phone: +86-25-83262765, Email: hhuangli@hotmail.com.
Lixing Qiao, Email: qiaolixing@aliyun.com.
Huan Zhu, Email: zhuhuan1978@163.com.
Li Jiang, Email: jiangli77777@126.com.
Liping Yin, Email: 1035012153@qq.com.
References
- 1.Wynn JL. Defining neonatal sepsis. Curr Opin Pediatr. 2016;28(2):135–140. doi: 10.1097/MOP.0000000000000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma P, Berwal P, Nagaraj N, Swami S, Jivaji P, Narayan S. Neonatal sepsis: epidemiology, clinical spectrum, recent antimicrobial agents and their antibiotic susceptibility pattern. Int J Contemp Pediatr. 2015;2(3):176–180. doi: 10.18203/2349-3291.ijcp20150523. [DOI] [Google Scholar]
- 3.Polin RA. Management of neonates with suspected or proven early-onset bacterial sepsis. Pediatrics. 2012;129(5):1006–1015. doi: 10.1542/peds.2012-0541. [DOI] [PubMed] [Google Scholar]
- 4.Wynn JL, Wilson CS, Hawiger J, Scumpia PO, Marshall AF, Liu JH, et al. Targeting IL-17A attenuates neonatal sepsis mortality induced by IL-18. Proc Natl Acad Sci U S A. 2016;113(19):E2627–E2635. doi: 10.1073/pnas.1515793113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlapbach LJ. Pancreatic stone protein as a novel marker for neonatal sepsis. Intensive Care Med. 2013;39(4):754–763. doi: 10.1007/s00134-012-2798-3. [DOI] [PubMed] [Google Scholar]
- 6.Jiř Ž, Kýr M, Vavřina M, Fedora M. Pancreatic stone protein – a possible biomarker of multiorgan failure and mortality in children sepsis. Cytokine. 2014;66(2):106–111. doi: 10.1016/j.cyto.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Kipfmueller F, Schneider J, Prusseit J, Dimitriou I, Zur B, Franz AR, et al. Role of Neutrophil CD64 Index as a Screening Marker for Late-Onset Sepsis in Very Low Birth Weight Infants. Plos One. 2015;10(4):e0124634. doi: 10.1371/journal.pone.0124634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimoula A. Serial determinations of neutrophil CD64 expression for the diagnosis and monitoring of sepsis in critically ill patients. Clin Infect Dis. 2014;58(6):820–829. doi: 10.1093/cid/cit936. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Wang X, Liu X, Wang X, Xu J, Hou S, et al. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int J Clin Exp Med. 2015;8(4):5683–5690. [PMC free article] [PubMed] [Google Scholar]
- 10.Cernada M, Serna E, Bauerl C, Collado MC, Pérezmartínez G, Vento M. Genome-wide expression profiles in very low birth weight infants with neonatal sepsis. Pediatrics. 2014;133(5):1203–1211. doi: 10.1542/peds.2013-2552. [DOI] [PubMed] [Google Scholar]
- 11.Töllner U. Early diagnosis of septicemia in the newborn. Eur J Pediatr. 1982;138(4):331–337. doi: 10.1007/BF00442511. [DOI] [PubMed] [Google Scholar]
- 12.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD neonatal research network. Pediatrics. 2002;110(1):285–291. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 13.Sastre JL, Colomer BF, Cotallo GDC. Neonatal Sepsis of vertical Transmision. An epidemiological study from the “Grupo de Hospitales Castrillo”. J Perinat Med. 2000;28(4):309–315. doi: 10.1515/JPM.2000.039. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2013;10(5):568–571. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westfall PH. The Benjamini-Hochberg method with infinitely many contrasts in linear models. Biometrika. 2008;95(3):709–19.
- 19.Consortium GO. Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43(D1):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. David bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server):W169–W175. [DOI] [PMC free article] [PubMed]
- 22.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huertacepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696(696):291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Bio Systems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 25.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2(6):826–834. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, et al. ENCODE whole-genome data in the UCSC genome browser. Nucleic Acids Res. 2010;38(Database issue):D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37(Database issue):D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 30.Jean R, Bribes E, Knabe L, Fort-Petit A, Vachier I, Bourdin A. TSPO is a new anti-inflammatory target in the airway of COPD. Rev Mal Respir. 2015;32(3):320. doi: 10.1016/j.rmr.2015.02.041. [DOI] [Google Scholar]
- 31.Santoro A, Mattace RG, Taliani S, Da PE, Simorini F, Costa B, et al. TSPO-ligands prevent oxidative damage and inflammatory response in C6 glioma cells by neurosteroid synthesis. Eur J Pharm Sci. 2016;88:124–131. doi: 10.1016/j.ejps.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Huang G, Wang Y, Vogel P, Kanneganti TD, Otsu K, Chi H. Signaling via the kinase p38α programs dendritic cells to drive TH17 differentiation and autoimmune inflammation. Nat Immunol. 2012;13(2):152–161. doi: 10.1038/ni.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schieven GL. The p38alpha kinase plays a central role in inflammation. Curr Top Med Chem. 2009;9(11):1038–1048. doi: 10.2174/156802609789630974. [DOI] [PubMed] [Google Scholar]
- 34.O'Keefe SJ, Mudgett JS, Cupo S, Parsons JN, Chartrain NA, Fitzgerald C, et al. Chemical genetics define the roles of p38 and p38β in acute and chronic inflammation. J Biol Chem. 2007;282(48):34663–34671. doi: 10.1074/jbc.M704236200. [DOI] [PubMed] [Google Scholar]
- 35.Picard C, Dogniaux S, Chemin K, Maciorowski Z, Lim A, Mazerolles F, et al. Hypomorphic mutation of ZAP70 in human results in a late onset immunodeficiency and no autoimmunity. Eur J Immunol. 2009;39(7):1966–1976. doi: 10.1002/eji.200939385. [DOI] [PubMed] [Google Scholar]
- 36.Roifman CM, Dadi H, Somech R, Nahum A, Sharfe N. Characterization of ζ-associated protein, 70 kd (ZAP70)–deficient human lymphocytes. J Allergy Clin Immunol. 2010;126(6):1226–1233. doi: 10.1016/j.jaci.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Hu G, Lu Y, Zheng J, Chen J, Wang X, et al. Palmitic acid aggravates inflammation of pancreatic acinar cells by enhancing unfolded protein response induced CCAAT-enhancer-binding protein β–CCAAT-enhancer-binding protein α activation. Int J Biochem Cell Biol. 2016;79:181–193. doi: 10.1016/j.biocel.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 38.Krieken SEVD, Popeijus HE, Mensink RP, Plat J. CCAAT/enhancer binding protein β in relation to ER stress, inflammation, and metabolic disturbances. Biomed Res Int. 2015;2015:324815. doi: 10.1155/2015/324815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with Sepsis. PLoS One. 2009;4(10):286–296. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roderburg C, Luedde M, Cardenas DV, Vucur M, Scholten D, Frey N, et al. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One. 2013;8(1):4926–4931. doi: 10.1371/journal.pone.0054612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Califano D, Cho J, Uddin M, Lorentsen K, Qi Y, Bhandoola A, et al. Transcription factor Bcl11b controls identity and function of mature type 2 innate lymphoid cells. Immunity. 2015;43(2):354–368. doi: 10.1016/j.immuni.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker JA, Oliphant CJ, Englezakis A, Yu Y, Clare S, Rodewald HR, et al. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med. 2015;212(6):875–882. doi: 10.1084/jem.20142224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


