Abstract
Small intestine neuroendocrine tumors (SI-NETs) represent the most common histotype among small intestine neoplasms, and metastatic disease is usually present at diagnosis. A retrospective series of 52 sporadic primary surgically resected SI-NETs, which were metastatic at diagnosis, was analyzed by high-coverage target sequencing (HCTS) for the mutational status of 57 genes and copy number status of 40 genes selected from recently published genome sequencing data. Seven genes were found to be recurrently mutated: CDKN1B (9.6%), APC and CDKN2C (each 7.7%), BRAF, KRAS, PIK3CA, and TP53 (each 3.8%). Copy number analysis showed frequent allelic loss of 4 genes located on chromosome 18 (BCL2, CDH19, DCC, and SMAD4) in 23/52 (44.2%) and losses on chromosomes 11 (38%) and 16 (15%). Other recurrent copy number variations were gains for genes located on chromosomes 4 (31%), 5 (27%), 14 (36%), and 20 (20%). Univariate survival analysis showed that SRC gene copy number gains were associated with a poorer prognosis (p = 0.047). Recurrent copy number variations are important events in SI-NET and SRC may represent a novel prognostic biomarker for this tumor type.
Electronic supplementary material
The online version of this article (10.1007/s00428-018-2450-x) contains supplementary material, which is available to authorized users.
Keywords: Neuroendocrine tumors, Small intestine, SRC, Prognostic markers
Introduction
Small intestinal neuroendocrine tumor (SI-NET) is the most frequent cancer type of the small bowel [21]. Despite slow-growing [18, 19], SI-NET is a deadly disease usually diagnosed at metastatic stage [12]. Many patients are asymptomatic, and the disease is often diagnosed incidentally. Less frequently SI-NETs are responsible for a carcinoid syndrome, which usually occurs in patients with liver metastases, due to excessive production and release of hormones and substances such as serotonin and prostaglandins [5].
The genetics of SI-NET remains elusive. Genome sequencing of the two independent series so far published provided dissonant information regarding recurrent somatic gene mutations, suggesting multigene potential mechanisms of carcinogenesis [2, 10]. In particular, a combination of whole-genome and exome sequencing performed on 50 SI-NETs showed somatic mutations of the CDKN1B gene in 14% of cases [10], which were not found in the exome sequencing study of 48 SI-NET [2]. This latter study identified mutations in several other cancer genes, although none was recurrently altered [2]. A single study reported a relatively high frequency of APC gene alterations (7 of 30 cases; 23%) [4], but only one mutation was found in one [10] of the two genome sequencing studies [2, 10].
At variance with gene mutations, chromosomal and gene copy number alterations characterize a significant portion of SI-NETs [2, 10, 13]. Consistent chromosomal and gene copy number alterations reported by the same two genome sequencing studies included loss of chromosome 18 in about 50% and of chromosome 16 in up to 18% of cases, whereas gains were in chromosomes 4, 5, 7, 14, and 20 occurring in a range from 10 to 30% of cases [2, 10]. Allelic loss of chromosome 18 is the most frequent anomaly that has been consistently reported in several different studies [1, 7, 11, 14, 15, 24], whereas gains of chromosome 14 [1] and 20pter-p11.21 [11] have been associated with shorter patients’ survival.
In this paper, we studied a well-characterized cohort of 52 primary SI-NETs that were metastatic at diagnosis performing: (i) a targeted deep sequencing mutational analysis for 57 relevant genes; (ii) copy number variation (CNV) analysis of a selected number of 40 genes located in the most frequently altered chromosomes; (iii) survival analysis to investigate the potential clinical relevance of the results.
Materials and methods
Patients and samples
A retrospective series (1997–2012) of 52 sporadic surgically resected primary intestinal neuroendocrine tumors (47 ileal, 4 duodenal, and 1 jejunal) were retrieved from the formalin-fixed paraffin-embedded (FFPE) archives of the ARC-Net Biobank at Verona University Hospital and from the University of Genoa/IRCCS S. Martino-IST University Hospital. All cases were reclassified according to WHO 2010 [3] and staging was assessed according to AJCC/UICC 7th edition [9]. Matched normal small intestine samples were used to determine the somatic/germline nature of mutations.
In 25 cases, sufficient material was available for the construction of 1-mm core tissue microarrays (TMAs) using the Galileo CK3500 Arrayer (www.isenet.it), a semiautomatic and computer-assisted TMA platform. Three tissue cores per case were analyzed.
Clinico-pathological characteristics of the sample cohort along with the molecular analyses conducted in this study are reported in Table 1 and Supplementary Table 1.
Table 1.
Clinical-pathological features of the 52 SI-NETs considered in the study
| Clinico-pathological features | Total (52) | [%] | |
|---|---|---|---|
| Sex | 30M 22F | ||
| Age | 46.1 ± 24.0 | ||
| Dimension (cm) | 2.2 ± 1.3 | ||
| Differentiation | WD | 49 | [94.2] |
| PD | 3 | [5.8] | |
| Grade | G1 | 36 | [69.2] |
| G2 | 13 | [25.0] | |
| G3 | 3 | [5.8] | |
| Stage UICC/ENETS | IIIB | 22 | [42.3] |
| IV | 30 | [57.7] | |
| R | R1 | 4 | [7.7] |
| R0 | 48 | [92.3] | |
| Vascular invasion | Present | 35 | [67.3] |
| Absent | 17 | [32.7] | |
| Perineural invasion | Present | 32 | [61.5] |
| Absent | 20 | [38.5] | |
| Necrosis | Present | 3 | [5.8] |
| Absent | 49 | [94.2] | |
WD well differentiated, PD poorly differentiated, R1 positive resection margins, R0 negative resection margins
DNA extraction and qualification
DNA was obtained from matched tumor and normal FFPE tissues, after enrichment for neoplastic cellularity to at least 70% using manual microdissection of 10 consecutive 4-μm sections. DNA was purified using the QIAamp DNA FFPE Tissue Kit (Qiagen) and qualified as reported [25, 27]. Briefly, DNA was quantified using Qubit DNA HS Assay kit (ThermoFisher) and purity was evaluated using NanoDrop ND-2000 [25]; integrity was investigated using the BIOMED protocol, and only DNA samples producing fragments of at least 200 bp were deemed usable for NGS [27].
RNA extraction and qualification
RNA was obtained from 10 consecutive 6-μm FFPE sections using RecoverAll total nucleic acid isolation kit protocol (ThermoFisher). RNA was quantified using Qubit RNA BR Assay Kit (ThermoFisher) and qualified using Agilent RNA 6000 Nano Kit (Agilent Technologies). A RNA Integrity Number (RIN) over 5 was considered suitable.
High-coverage target sequencing
Matched tumor/normal DNA from all FFPE samples was subjected to targeted next-generation sequencing (NGS). Three multigene panels were used: the 50-gene Ion AmpliSeq Cancer Hotspot panel v2 (ThermoFisher) and two AmpliSeq custom panels.
The first panel explores mutation status of selected hot-spot regions of 50 cancer-related genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR/VEGFR2, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, and VHL. Details on target regions of the commercial panel are at http://www.thermofisher.com.
The second panel has been designed for the mutational analysis of 7 genes selected upon the results of published whole genome, exome, and targeted sequencing of SI-NETs series [2, 4, 10, 13]: ATRX, CDKN1B, CDKN2C, DAXX, H3F3A, MEN1, and TERT.
The third panel investigates copy number variation (CNV) status of 40 genes reported as altered in SI-NETs [2, 7, 10, 11, 13, 15]: AKT1, APC, AURKA, BCL2, BCL2L2, BRAF, CDH1, CDH19, CDKN1B, CDKN2A, DCC, EGFR, ERBB2, FBXW7, FGFR3, FHIT, FOS, GNAS, HRAS, KDR, KIT, MAP2K2, MDM2, MEN1, MET, MYC, MYCL1, PDGFRA, PIK3CA, PIK3CD, PTEN, RICTOR, SDHA, SDHB, SDHD, SMAD4, SMAD5, SOX12, SRC, and TP53. Details on target regions of the CNV custom panel are in Supplementary Table 2A.
Twenty nanograms of DNA were used for each multiplex PCR amplification. The quality of the obtained libraries was evaluated by the Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent Technologies). Emulsion PCR to construct the libraries of clonal sequences was performed with the Ion OneTouch™ OT2 System (ThermoFisher). Sequencing was run on the Ion Proton (PI, ThermoFisher) loaded with Ion PI Chip v2.
Sequencing data analysis
Base calling, alignment to the hg19 human reference genome, and variant calling were done using the Torrent Suite Software v.5.0 (ThermoFisher). Called variants were annotated using a custom pipeline based on vcflib (https://github.com/ekg/vcflib), SnpSift [6], the Variant Effect Predictor (VEP) software [17], and NCBI RefSeq database. Filtering of variants was performed by: (i) removal of germline variants and (ii) visual verification of alignments on the IGV software v2.3 [26]. This latter step is key to remove false calls due to technique-dependent mispriming or sample age-related deamination, which cannot be ruled out by automated variant calling and filtering procedures (Supplementary Table 2B).
CNV analysis using next-generation sequencing
For all samples, a CNV baseline for AmpliSeq custom panels was performed using 10 genomic male DNA extracted from normal tissues included in FFPE samples. CNV was evaluated comparing BAM files of sequenced libraries to baseline through a custom workflow pipeline created on IonReporter 5.0 software. Copy number variation calling of genes included in the Ion AmpliSeq Cancer Hotspot panel v2 and custom panel was performed using followed criteria: (i) a median of the absolute values of all pairwise differences (MAPD) score under 1; (ii) a CNV confidence number major than 20; (iii) a tiles number major than 10. An orthogonal cross-validation using FISH or qPCR was performed. A chromosome integrity number score was evaluated for each sample dividing length of altered chromosomes to length of chromosome regions investigated.
CNV validation by quantitative-PCR
Q-PCR analysis of copy numbers was applied to all 52 SI-NETs for selected loci. All target and reference assays were purchased from ThermoFisher Scientific. RNaseP was used as endogenous control for normalization of analyzed loci. The following assays were used: AKT1 (Hs02893205), BCL2 (Hs01500302), CDH19 (Hs02826809), DCC (Hs02317964), FHIT (Hs03491211), MET (Hs04951661), PIK3CD (Hs04540050), SMAD4 (Hs06483146), SOX12 (Hs02822764), SRC (Hs07169853), SDHB (Hs00124581), and RNaseP (part number 4403326). The experimental procedure recommended by the manufacturer (Applied Biosystems) was followed. Twenty nanogram genomic DNA was used in the q-PCR reaction, and a negative control was analyzed in parallel. All q-PCR reactions were run in quadruplicate in a 7900HT qRT-PCR machine (Applied Biosystems) using standard cycling conditions of 10 min at 95 °C, followed by 40 cycles of [95 °C for 15 s and at 60 °C for 1 min]. Pooled normal FFPE DNA was used as calibrator and as reference unbiased genome. Microsatellite markers D18S484, D18S51, and D18S1110 were used to confirm LOH/homozygous deletion, respectively, in DCC, BCL2, and SMAD4 locus.
Fluorescent in situ hybridization analysis
FISH analysis was performed to evaluate CNV status of chromosome 18q, 14q, and SRC gene. The LSI IGH/BCL2 dual color, dual fusion translocation probe (Vysis Inc./Abbott) was used for chromosome 18q. The TelVysion 14q probe (Vysis Inc./Abbott) was used to evaluate chromosome 14q status as reported in a previous work [1]. A custom FISH probe was developed to evaluate the status of SRC gene (chr20q11.23). The custom FISH probe labeled SRC (chr20q11.23) in Spectrum Red and the control locus 20p11.21 in Spectrum Green (Empire Genomics). FISH analysis was performed according to the manufacturer’s protocols.
Expression analysis for SRC gene
An ampliseq RNA custom panel was built to analyze the expression levels of SRC and included a series of genes to normalize them (ACTB, CDH17, GC, HPRT1, KCNJ3, KIF12, MIA2, MUC13, RNF213, RPRM, SOX21). In brief, 1 μg of RNA was retro-transcribed and submitted for library construction. The quality of the obtained libraries was evaluated by the Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent Technologies). Emulsion PCR to construct the libraries of clonal sequences was performed with the Ion OneTouch™ OT2 System (Life Technologies). Sequencing was performed on Ion PGM loading 318v2 chip, reads were aligned to reference genome (hg19 Homo sapiens RNA Canonical Transcript). Normalization was performed dividing the number of reads obtained for SRC transcript by the sum of reads obtained for the other genes and expressed as reads per thousands of reads.
Immunohistochemistry
Immunohistostaining was performed with Leica Microsystems Bond-Max Autostainer System using the antibodies listed in Supplementary Table 3 on 3-μm FFPE sections from tissue blocks containing representative core samples. Immunolabeling for all antibodies was performed according to manufacturer protocols; normal intestinal tissue was used as positive control. Sections incubated without the primary antibodies served as negative control.
Statistical analysis
One-way ANOVA, Kruskal-Wallis test, Fisher’s test with Monte Carlo simulation, and Fisher’s exact test were used as appropriate; correction for multiple comparisons was performed according to Benjamini-Hochberg. For comparison of Kaplan-Meier survival curves, Mantel-Cox test was used; for multivariable survival analysis, stepwise Cox proportional hazards regression was used; selection of the best model was performed using the “enter” algorithm. For all the analyses, a p value below 0.05 was considered as significant. All analyses were performed using Medcalc for Windows version 15.6 (MedCalc Software, Ostend, Belgium) and R v. 3.2.1; multivariable Cox regression was done with R using survival library v.2.38-2.
Results
Clinico-pathological characteristics of the series
Clinico-pathological data are detailed in Table 1 and Supplementary Table 1. Tumor grade was G1 in 36 (69.2%), G2 in 13 (25.0%), and G3 in 3 cases (5.8%). Vascular and perineural invasions were present in 35 (67.3%) and 32 (61.5%) cases, respectively. The ENETS and UICC pathologic stages overlapped and were: stage IIIB in 22 and IV in 30.
Mutational profiles of small intestinal neuroendocrine tumors
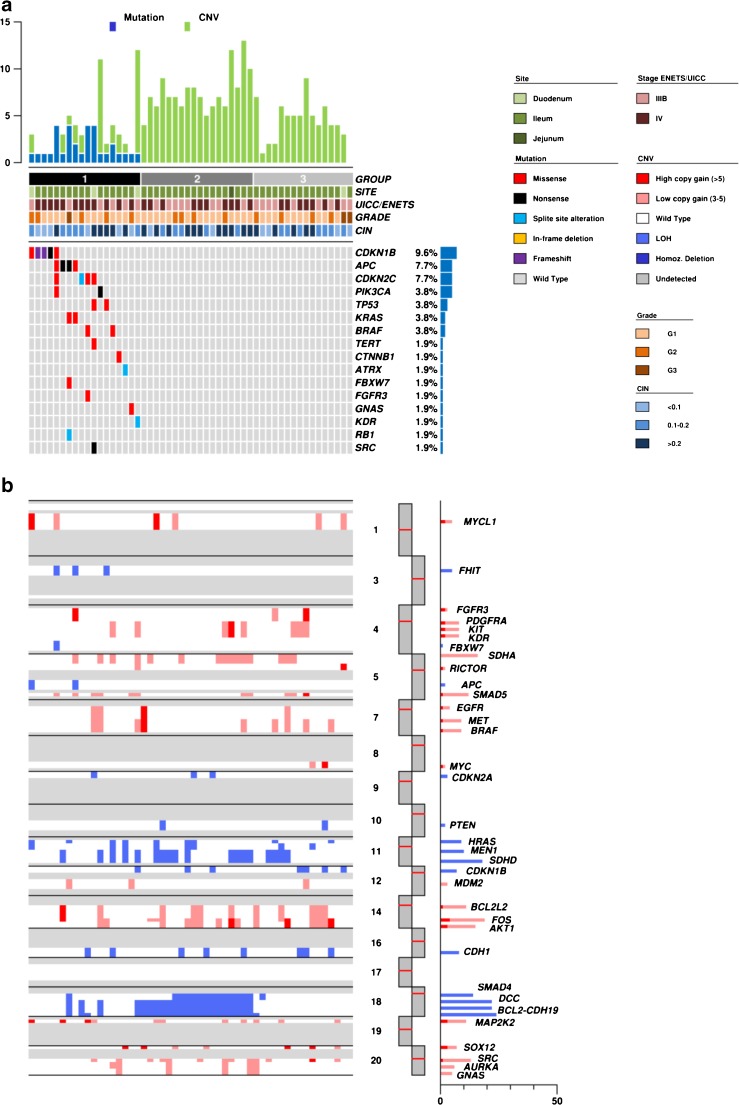
DNA from all samples was successfully amplified in multiplex PCR for the 57 selected genes and adequate libraries for deep sequencing were obtained. The mean read length was 103 base pairs and a mean coverage of 5893× was achieved, with 97.2% target bases covered more than 100×. A minimum coverage of 20× was obtained in all cases. A mean of 0.6 mutation was detected in all series for the 57 genes analyzed. In the whole cohort, 18 (34.6%) samples showed somatic mutations: 12 (23.1%) affected by one mutation; 6 (11.5%) affected by more than one. The remaining 34 (65.4%) tumors resulted wild type for the 57 genes assayed (Fig. 1a, Table 2).
Fig. 1.
Genetic landscape of 52 SI-NET. a The upper histogram shows the number of mutations (blue) and CNV (green) in recurrently altered genes for each sample. The central matrix shows 16 of 57 genes that were mutated in the whole cohort; alterations are annotated by different colors according to their impact on the gene product as illustrated in the panel on the right. b Summary of somatic copy number variations determined for 52 human SI-NETs. Significant gains (red) and losses (cyan) were determined for the chromosomal regions and are plotted according to their frequencies on the right
Table 2.
Mutated genes in the 52 SI-NETs
| GENE | Total (52) | [%] | M | N | S | D | F |
|---|---|---|---|---|---|---|---|
| APC | 4 | [7.7] | 1 | 3 | – | – | – |
| ATRX | 1 | [1.9] | – | – | 1 | – | – |
| BRAF | 2 | [3.8] | 2 | – | – | – | – |
| CDKN1B | 5 | [9.6] | 2 | 1 | – | – | 2 |
| CDKN2C/INK4C | 4 | [7.7] | 3 | – | 1 | – | – |
| CTNNB1 | 1 | [1.9] | 1 | – | – | – | – |
| FBXW7 | 1 | [1.9] | 1 | – | – | – | – |
| FGFR3 | 1 | [1.9] | 1 | – | – | – | – |
| GNAS | 1 | [1.9] | 1 | – | – | – | – |
| KRAS | 2 | [3.8] | 2 | – | – | – | – |
| PIK3CA | 2 | [3.8] | 1 | 1 | – | – | – |
| RB1 | 1 | [1.9] | – | – | 1 | – | – |
| SRC | 1 | [1.9] | – | 1 | – | – | – |
| TERT | 1 | [1.9] | 1 | – | – | – | – |
| TP53 | 2 | [3.8] | 2 | – | – | – | – |
M missense mutation, N nonsense mutation, S splice site alteration, D deletion, F frameshift mutation
The most frequently mutated genes were CDKN1B (9.6%), followed by APC and CDKN2C (each 7.7%), BRAF, KRAS, PIK3CA, and TP53 (each 3.8%). The details of the somatic mutations detected are reported in Supplementary Table 4. Five mutations in tumor suppressor genes (FGFR3, TP53, CDKN2C, APC, RB1) had a high allelic frequency (> 60%) compatible with homozygous alteration in a large fraction of tumor cells. Fourteen mutations, prevalently in oncogenes or haplo-insufficient tumor suppressor genes, had a frequency between 25 and 57%, compatible with heterozygous alteration in all tumor cells or with homozygous alteration in a consistent (30–50% of total tumor) subpopulation of neoplastic cells. Finally, 11 mutations had a frequency equal to or below 20%, compatible with heterozygous or homozygous alteration of smaller subclones. Notably, the same tumor (e.g., case G5) often bore different mutations with both high and low frequencies, suggesting molecular heterogeneity.
Copy number variations in selected genes
Copy number variations were evaluated using NGS and validated by q-PCR and FISH. The results are summarized in Table 3, detailed in Supplementary Table 5 and illustrated Fig. 1b and Supplementary Fig. 1.
Table 3.
Copy number variations for selected genes in 52 SI-NETs. Genes are listed per alphabetical order
| Gene | Chromosomal location | Gain or loss | Total (52) | [%] |
|---|---|---|---|---|
| AKT1 | 14 | Gain | 16 | [30.8] |
| APC | 5 | LOH | 2 | [3.8] |
| AURKA | 20 | Gain | 6 | [11.5] |
| BCL2 | 18 | LOH | 22 | [42.3] |
| BCL2L2 | 14 | Gain | 11 | [21.2] |
| BRAF | 7 | Gain | 9 | [17.3] |
| CDH1 | 16 | LOH | 8 | [15.4] |
| CDH19 | 18 | LOH | 24 | [46.2] |
| CDKN1B | 12 | LOH | 7 | [13.5] |
| CDKN2A | 9 | LOH | 3 | [5.8] |
| DCC | 18 | LOH | 22 | [42.3] |
| EGFR | 7 | Gain | 4 | [7.7] |
| ERBB2 | 17 | Gain | 0 | [0] |
| FBXW7 | 4 | LOH | 1 | [1.9] |
| FGFR3 | 4 | Gain | 3 | [5.8] |
| FHIT | 3 | LOH | 3 | [5.8] |
| FOS | 14 | Gain | 19 | [36.5] |
| GNAS | 20 | Gain | 5 | [9.6] |
| HRAS | 11 | LOH | 9 | [17.3] |
| KDR | 4 | Gain | 15 | [28.8] |
| KIT | 4 | Gain | 15 | [28.8] |
| MAP2K2 | 19 | Gain | 11 | [21.2] |
| MDM2 | 12 | Gain | 3 | [5.8] |
| MEN1 | 11 | LOH | 7 | [13.5] |
| MET | 7 | Gain | 9 | [17.3] |
| MYC | 8 | Gain | 2 | [3.8] |
| MYCL1 | 1 | Gain | 6 | [11.5] |
| PDGFRA | 4 | Gain | 15 | [28.8] |
| PIK3CA | 3 | Gain | 0 | [0] |
| PIK3CD | 1 | LOH | 0 | [0] |
| PTEN | 10 | LOH | 2 | [3.8] |
| RICTOR | 5 | Gain | 2 | [3.8] |
| SDHA | 5 | Gain | 15 | [28.8] |
| SDHB | 1 | LOH | 0 | [0] |
| SDHD | 16 | LOH | 18 | [34.6] |
| SMAD4 | 18 | LOH | 15 | [28.8] |
| SMAD5 | 5 | Gain | 11 | [21.2] |
| SOX12 | 20 | Gain | 7 | [13.5] |
| SRC | 20 | Gain | 13 | [25.0] |
| TP53 | 17 | LOH | 0 | [0] |
A mean of 6.5 CNV was found in the series for the 40 genes investigated. Forty-eight (92.3%) samples showed somatic CNV: 1 (1.9%) affected by one CNV and 47 (90.4%) affected by more than one CNV. Four (7.7%) samples showed no CNV event. The most frequent event was loss of a single copy (LOH) of the CDH19 gene located on 18q22 locus in 24/52 (46.2%). Other genes analyzed and located on chromosome 18q (BCL2, DCC, SMAD4) were mostly affected by LOH. In particular, 23 samples (44.2%) showed copy loss of one or more genes located on this chromosomal region. LOH of DCC, BCL2, and SMAD4 were validated using microsatellite marker quantification between tumor and matched normal samples for each specific locus. IHC analysis for CDH1, SMAD4, and CDKN1B was possible for the 25 cases included in the TMAs; in all samples affected by the loss of one gene copy, a significant decrease in the corresponding protein expression was observed (Supplementary Fig. 2). Frequent copy gains were detected in FOS gene (19/52, 36.5%) followed by AKT1 (16/52; 30.8%) and KIT, KDR, and PDGFRA locus (15/52, 28.8%).
Identification of molecular subgroups
To identify potential molecular subgroups in SI-NET, we separated samples according to their prevalent molecular features. This approach identified three distinct groups as illustrated in Fig. 1. Group 1 included 18 samples characterized by presence of somatic mutations. Groups 2 and 3 together included 34 samples, each with chromosome copy number alterations and no somatic mutations. In particular, group 2 comprised samples featured by LOH in at least 2 genes located on chromosome 18, while group 3 comprised samples with no mutations and low or no LOH affecting chromosome 18 genes.
Survival analysis
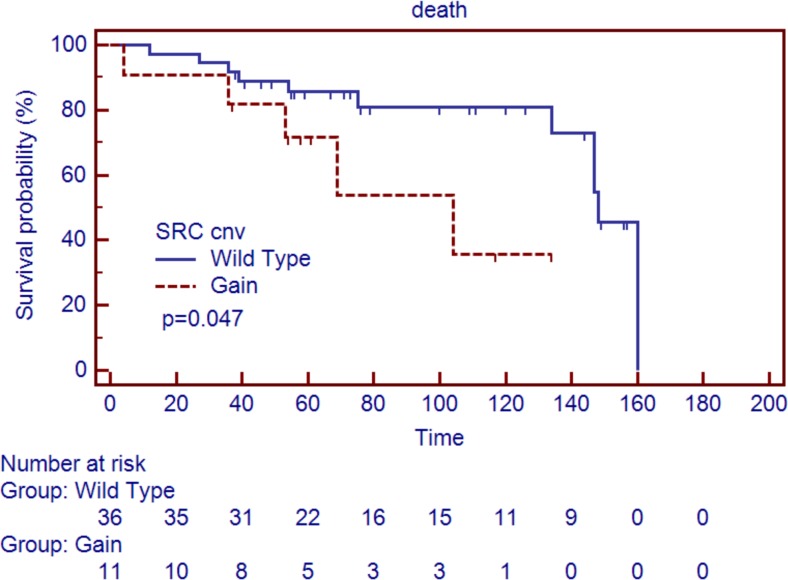
Follow-up data was available for 47 cases. Median survival was 71.0 (range 4–160) months and 16 (34.0%) subjects died of disease. To identify clinico-pathological markers of poor prognosis, we considered at univariate analysis: sex, age at diagnosis (under/over 50 years), and presence of nodal or distant metastases (stage IIIB vs. IV). The analysis showed no significant differences about prognosis performance among these markers. When matching clinical data and molecular features, copy gain of SRC was identified as the only molecular marker associated to poor prognosis (Fig. 2). Of interest, cases with SRC copy gain showed less perineural invasion (p = 0.019); no other correlation with clinicopathological features was found. Of note, a strong correlation between age at diagnosis over 50 years and copy gain at the FOS gene locus was observed (p = 0.001). Molecular subgroups as previously defined by Karpathakis et al. [13] did not associate with any difference in survival (p = 0.73).
Fig. 2.
Overall survival according to molecular and pathological features. Overall survival of SI-NETs (n = 47) is significantly affected by gain occurring in SRC locus gene (p = 0.047). Vertical axis indicates percent survival; horizontal axis shows time expressed in months. Kaplan–Meier and log-rank statistics were used to determine levels of significance
SRC expression analysis
To verify the impact of SRC copy gain on transcript levels, we performed targeted RNA sequencing on the 14 cases with available residual tissue for research purposes. These included 4 cases with SRC copy gains and 10 without locus alteration according to CNV analysis. The number of reads obtained for SRC was normalized to the sum of the reads obtained for a set of 11 control genes and expressed as normalized counts (reads per thousands of control reads). Cases were grouped according to their genomic SRC status (gain vs. wild-type) and Mann-Whitney test was performed to compare them (Supplementary Fig. 3). Normalized counts of SRC transcript were almost 3 times higher in the “Gain” group (median 38.4, 1st–3rd quartiles 31.0–82.2) than in the “Wild-type” group (median 12.6, 1st–3rd quartiles 3.4–19.0, p = 0.0020).
Discussion
The SI-NETs molecular landscape and its clinical implications are still to be fully elucidated.
In this paper, we focused on the major genetic SI-NET alterations in a cohort of 52 sporadic surgically resected advanced SI-NETs. Our study: (i) demonstrated that mutations in gene coding sequences are a relatively rare event; (ii) CNV is relatively frequent, affecting LOH of chromosome 18q in more than 40% of the samples; (iii) SRC gene gains are associated to a poorer patients’ survival.
Only 18 (34.6%) samples were characterized by somatic mutations in the 57 analyzed genes in the NGS mutational analysis. Our findings were concordant with those observed by Francis et al. [10] regarding mutation frequencies for CDKN1B (9.6%) and BRAF (3.8%) genes. Moreover, we found a significant mutational rate for APC (7.7%), confirming previous reports obtained by conventional sequencing methods [4, 23].
Unreported findings [4, 10, 22] were the mutations observed in PIK3CA (3.8%), TP53 (3.8%), KRAS (3.8%), and CDKN2C (7.7%) genes. Cases affected by these mutations did not show any peculiar clinico-pathological feature.
Unlike the poor mutational landscape observed in our series, copy number analysis in selected genes showed recurrent and frequent alterations in the whole cohort. As previously reported in several studies [1, 8, 11, 15, 20], in our series, we also observed copy loss of genes located on chromosome 18 (DCC, SMAD4, BCL2, and CDH19; 24/52, 46.2%). Again in line with previous studies [1, 2, 10, 11, 14–16], we identified copy number gains in genes located in chromosomes 4 (FGFR3, KDR, KIT, and PDGFRA), 5 (RICTOR, SDHA and SMAD5), 7 (EGFR and BRAF), 14 (AKT1, BCL2L2, and FOS), and 20 (AURKA, GNAS, SOX12, and SRC).
Among the principal genes affected by copy number variations, no mutually exclusive gene was observed between those located on chromosomes 14 and 18. This was at variance from what previously described by Hashemi et al. [11] but similar to what reported by Anderson et al. [1].
Abundance of genes affected by CNV suggests that aberrations in the genomic/chromosomal structure may play an important role in SI-NET biology. Also, survival analysis supported this hypothesis. Correlating clinical data and molecular features, we identified no correlation between prognosis and gene mutation, but did observe a significant relationship by univariate analysis between copy gain of SRC gene and poor patient prognosis (p = 0.047). Copy gains of the SRC gene in SI-NET were first identified by Banck et al. in a series of 48 cases, suggesting SRC as potential therapeutic target for this disease [2]. We also report that copy gain of the SRC gene is associated with elevated mRNA levels. This was also true for case #V22, presenting 5 copies of SRC and an apparently contradictive nonsense mutation. In this case, the relative abundance of the SRC transcript was one of the highest (45.8 normalized counts vs. a median of 12.6 normalized counts for SRC wild-type cases). Therefore, the nonsense mutation was likely a passenger event with no evident impact on SRC expression.
A recent integrative molecular analysis on 47 SI-NET by Karpathakis et al. [13] highlighted the presence of 3 molecular subgroups via matched copy number and methylation profile results. The most relevant included 26 samples (55% of whole cohort) characterized by loss of chromosome 18 with concurrent CDKN1B mutation and better survival compared to the other two molecular subgroups. In our study, no concurrence of chromosome 18 loss (44.2%) and CDKN1B mutation (9.6%) was observed. Moreover, classifying our cohort into the three groups of Karpathakis et al., we did not observe any statistically significant correlation with prognosis.
In our study, at variance with the current literature [1, 11], poor prognosis was not associated with gains of genes at 20p or 14q loci. To test CNV status of these loci, we analyzed SOX12 (locus 20p13), FOS (locus 14q24.3), and AKT1 (locus 14q32.33) genes as surrogate for such chromosome arms. Interestingly, FOS copy gain was strongly associated with another marker of poor prognosis in our study: advanced age at diagnosis (Fisher’s exact test p = 0.001). This might explain why Anderson et al. identified gain of 14q as a poor prognostic marker. In fact, in that study, all samples characterized by this alteration had an advanced age at diagnosis (i.e., over 50 years old) [1].
In conclusion, our study provides additional data to define the genetic landscape of sporadic SI-NETs, highlighting copy number variations as a central molecular event in this tumor type, not only for understanding SI-NET biology, but also with prognostic significance.
Electronic supplementary material
FISH images of representative CNV alterations in SI-NET samples. A) Diploid BCL2 status; B) Monosomy of BCL2; C) Diploid Tel14q status; D) Polysomy of Tel14q; E) Diploid SRC status; F) Polysomy of SRC. (PNG 2047 kb)
Immunohistochemical validation of the expression of E-cadherin (CDH1), p27 (CDKN1B) and smad4 (SMAD4) proteins in mutated and wild type tumors. Representative immunohistochemical staining of the products of the CDH1 (E-cadherin), CDKN1B (p27) and SMAD4 (smad4) genes in wild type tumors (left panels) and tumors characterized by LOH of the corresponding genes (right panels). The wild type tumors retained a normal protein expression, which was lost in LOH cases. (PNG 5738 kb)
Small intestinal neuroendocrine tumors with copy gain of SRC locus display enhanced levels of SRC mRNA. Normalized expression reads of 14 samples grouped according SRC copy number status in Gain (4 samples) and Wild-Type (10 samples). (PNG 104 kb)
(XLSX 117 kb)
Abbreviations
- NET
neuroendocrine tumor
- CNV
copy number variations
- dNET
duodenal NET
- FFPE
formalin-fixed paraffin embedded
- HCTS
high-coverage targeted sequencing
- iNET
ileal NET
- IHC
immunohistochemistry
- NET
jejunal NET
- LOH
loss of heterozygosity
- TMA
tissue micro-array
- WHO
World Health Organization
Author’s contributions
AS, MS, and RTL conceived the study. MS designed the study and validation experiments. VC supervised the validation experiments. RTL coordinated patients, sample data management, and supervised ethical protocols. LM, CP, LL, FG collected materials and clinical data. MF, BR, LM, MC, and AS analyzed histopathological data. MF, BR microdissected samples. MS and AM carried out sequencing and raw data analysis. AM and MS performed bioinformatic analysis. CV performed IHC analysis. CV and SP performed FISH analysis and data interpretation. MS and MF drafted the manuscript. AM, MF, VC, GP, CL, and RTL revised the manuscript. RTL finalized the manuscript. All authors approved the submitted version.
Funding
This work was supported by Associazione Italiana Ricerca Cancro (AIRC grant n. 12182, to AS); Italian Cancer Genome Project (FIRB RBAP10AHJB to AS). The funding agencies had no role in the collection, analysis, and interpretation of data and in the writing of the manuscript.
Compliance with ethical standards
The materials used have been collected under Ethics Committee Approval (ECA) of both institutions. The study at the University of Verona received local ECA n. 55893/2017 in relation to Program 1885 protocol 52438 on 23 November 2010 which includes informed consent of the patient for the creation of a coordinated biobank for the collection of samples from all cancer patients that include neoplastic and associated local and distant normal tissue. An analogous approval was received by the local ethics committee of San Martino-Genova, ECA n.029/2012.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Michele Simbolo and Caterina Vicentinib are shared first authorship.
References
- 1.Andersson E, Sward C, Stenman G, Ahlman H, Nilsson O. High-resolution genomic profiling reveals gain of chromosome 14 as a predictor of poor outcome in ileal carcinoids. Endocr Relat Cancer. 2009;16:953–966. doi: 10.1677/ERC-09-0052. [DOI] [PubMed] [Google Scholar]
- 2.Banck MS, Kanwar R, Kulkarni AA, Boora GK, Metge F, Kipp BR, Zhang L, Thorland EC, Minn KT, Tentu R, Eckloff BW, Wieben ED, Wu Y, Cunningham JM, Nagorney DM, Gilbert JA, Ames MM, Beutler AS. The genomic landscape of small intestine neuroendocrine tumors. J Clin Invest. 2013;123:2502–2508. doi: 10.1172/JCI67963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosman F, Carneiro F, Hruban RH, Theise ND. Pathology and genetics of Tumours of the digestive system. Lyon: IARC Press; 2010. [Google Scholar]
- 4.Bottarelli L, Azzoni C, Pizzi S, D'Adda T, Silini EM, Bordi C, Rindi G. Adenomatous polyposis coli gene involvement in ileal enterochromaffin cell neuroendocrine neoplasms. Hum Pathol. 2013;44:2736–2742. doi: 10.1016/j.humpath.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Caplin ME, Buscombe JR, Hilson AJ, Jones AL, Watkinson AF, Burroughs AK. Carcinoid tumour. Lancet. 1998;352:799–805. doi: 10.1016/S0140-6736(98)02286-7. [DOI] [PubMed] [Google Scholar]
- 6.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham JL, Diaz de Stahl T, Sjoblom T, Westin G, Dumanski JP, Janson ET. Common pathogenetic mechanism involving human chromosome 18 in familial and sporadic ileal carcinoid tumors. Genes Chromosom Cancer. 2011;50:82–94. doi: 10.1002/gcc.20834. [DOI] [PubMed] [Google Scholar]
- 8.Delgado Verdugo A, Crona J, Maharjan R, Hellman P, Westin G, Bjorklund P. Exome sequencing and CNV analysis on chromosome 18 in small intestinal neuroendocrine tumors: ruling out a suspect? Horm Metab Res. 2015;47:452–455. doi: 10.1055/s-0034-1389992. [DOI] [PubMed] [Google Scholar]
- 9.Edge SB, Compton CC. The American joint committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 10.Francis JM, Kiezun A, Ramos AH, Serra S, Pedamallu CS, Qian ZR, Banck MS, Kanwar R, Kulkarni AA, Karpathakis A, Manzo V, Contractor T, Philips J, Nickerson E, Pho N, Hooshmand SM, Brais LK, Lawrence MS, Pugh T, McKenna A, Sivachenko A, Cibulskis K, Carter SL, Ojesina AI, Freeman S, Jones RT, Voet D, Saksena G, Auclair D, Onofrio R, Shefler E, Sougnez C, Grimsby J, Green L, Lennon N, Meyer T, Caplin M, Chung DC, Beutler AS, Ogino S, Thirlwell C, Shivdasani R, Asa SL, Harris CR, Getz G, Kulke M, Meyerson M. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet. 2013;45:1483–1486. doi: 10.1038/ng.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashemi J, Fotouhi O, Sulaiman L, Kjellman M, Hoog A, Zedenius J, Larsson C. Copy number alterations in small intestinal neuroendocrine tumors determined by array comparative genomic hybridization. BMC Cancer. 2013;13:505. doi: 10.1186/1471-2407-13-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jann H, Roll S, Couvelard A, Hentic O, Pavel M, Muller-Nordhorn J, Koch M, Rocken C, Rindi G, Ruszniewski P, Wiedenmann B, Pape UF. Neuroendocrine tumors of midgut and hindgut origin: tumor-node-metastasis classification determines clinical outcome. Cancer. 2011;117:3332–3341. doi: 10.1002/cncr.25855. [DOI] [PubMed] [Google Scholar]
- 13.Karpathakis A, Dibra H, Pipinikas C, Feber A, Morris T, Francis J, Oukrif D, Mandair D, Pericleous M, Mohmaduvesh M, Serra S, Ogunbiyi O, Novelli M, Luong T, Asa SL, Kulke M, Toumpanakis C, Meyer T, Caplin M, Meyerson M, Beck S, Thirlwell C. Prognostic impact of novel molecular subtypes of small intestinal neuroendocrine tumor. Clin Cancer Res. 2016;22:250–258. doi: 10.1158/1078-0432.CCR-15-0373. [DOI] [PubMed] [Google Scholar]
- 14.Kim DH, Nagano Y, Choi IS, White JA, Yao JC, Rashid A. Allelic alterations in well-differentiated neuroendocrine tumors (carcinoid tumors) identified by genome-wide single nucleotide polymorphism analysis and comparison with pancreatic endocrine tumors. Genes Chromosom Cancer. 2008;47:84–92. doi: 10.1002/gcc.20510. [DOI] [PubMed] [Google Scholar]
- 15.Kulke MH, Freed E, Chiang DY, Philips J, Zahrieh D, Glickman JN, Shivdasani RA. High-resolution analysis of genetic alterations in small bowel carcinoid tumors reveals areas of recurrent amplification and loss. Genes Chromosom Cancer. 2008;47:591–603. doi: 10.1002/gcc.20561. [DOI] [PubMed] [Google Scholar]
- 16.Kytola S, Hoog A, Nord B, Cedermark B, Frisk T, Larsson C, Kjellman M. Comparative genomic hybridization identifies loss of 18q22-qter as an early and specific event in tumorigenesis of midgut carcinoids. Am J Pathol. 2001;158:1803–1808. doi: 10.1016/S0002-9440(10)64136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Modlin IM, Lye KD, Kidd M. A 5-decade analysis of 13,715 carcinoid tumors. Cancer. 2003;97:934–959. doi: 10.1002/cncr.11105. [DOI] [PubMed] [Google Scholar]
- 19.Modlin IM, Oberg K, Chung DC, Jensen RT, de Herder WW, Thakker RV, Caplin M, Delle Fave G, Kaltsas GA, Krenning EP, Moss SF, Nilsson O, Rindi G, Salazar R, Ruszniewski P, Sundin A. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol. 2008;9:61–72. doi: 10.1016/S1470-2045(07)70410-2. [DOI] [PubMed] [Google Scholar]
- 20.Nieser Maike, Henopp Tobias, Brix Joachim, Stoß Laura, Sitek Barbara, Naboulsi Wael, Anlauf Martin, Schlitter Anna M., Klöppel Günter, Gress Thomas, Moll Roland, Bartsch Detlef K., Heverhagen Anna E., Knoefel Wolfram T., Kaemmerer Daniel, Haybaeck Johannes, Fend Falko, Sperveslage Jan, Sipos Bence. Loss of Chromosome 18 in Neuroendocrine Tumors of the Small Intestine: The Enigma Remains. Neuroendocrinology. 2016;104(3):302–312. doi: 10.1159/000446917. [DOI] [PubMed] [Google Scholar]
- 21.Pan SY, Morrison H. Epidemiology of cancer of the small intestine. World J Gastrointest Oncol. 2011;3:33–42. doi: 10.4251/wjgo.v3.i3.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park C, Ha SY, Kim ST, Kim HC, Heo JS, Park YS, Lauwers G, Lee J, Kim KM. Identification of the BRAF V600E mutation in gastroenteropancreatic neuroendocrine tumors. Oncotarget. 2015;7:4024–4035. doi: 10.18632/oncotarget.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pizzi S, Azzoni C, Tamburini E, Bottarelli L, Campanini N, D'Adda T, Fellegara G, Luong TV, Pasquali C, Rossi G, Delle Fave G, Camisa R, Bordi C, Rindi G. Adenomatous polyposis coli alteration in digestive endocrine tumours: correlation with nuclear translocation of beta-catenin and chromosomal instability. Endocr Relat Cancer. 2008;15:1013–1024. doi: 10.1677/ERC-07-0230. [DOI] [PubMed] [Google Scholar]
- 24.Rindi G, Villanacci V, Ubiali A, Scarpa A. Endocrine tumors of the digestive tract and pancreas: histogenesis, diagnosis and molecular basis. Expert Rev Mol Diagn. 2001;1:323–333. doi: 10.1586/14737159.1.3.323. [DOI] [PubMed] [Google Scholar]
- 25.Simbolo M, Gottardi M, Corbo V, Fassan M, Mafficini A, Malpeli G, Lawlor RT, Scarpa A. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One. 2013;8:e62692. doi: 10.1371/journal.pone.0062692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamo A, Bertolaso A, van Raaij AW, Mancini F, Scardoni M, Montresor M, Menestrina F, van Krieken JH, Chilosi M, Groenen PJ, Scarpa A. Application of microfluidic technology to the BIOMED-2 protocol for detection of B-cell clonality. J Mol Diagn. 2012;14:30–37. doi: 10.1016/j.jmoldx.2011.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FISH images of representative CNV alterations in SI-NET samples. A) Diploid BCL2 status; B) Monosomy of BCL2; C) Diploid Tel14q status; D) Polysomy of Tel14q; E) Diploid SRC status; F) Polysomy of SRC. (PNG 2047 kb)
Immunohistochemical validation of the expression of E-cadherin (CDH1), p27 (CDKN1B) and smad4 (SMAD4) proteins in mutated and wild type tumors. Representative immunohistochemical staining of the products of the CDH1 (E-cadherin), CDKN1B (p27) and SMAD4 (smad4) genes in wild type tumors (left panels) and tumors characterized by LOH of the corresponding genes (right panels). The wild type tumors retained a normal protein expression, which was lost in LOH cases. (PNG 5738 kb)
Small intestinal neuroendocrine tumors with copy gain of SRC locus display enhanced levels of SRC mRNA. Normalized expression reads of 14 samples grouped according SRC copy number status in Gain (4 samples) and Wild-Type (10 samples). (PNG 104 kb)
(XLSX 117 kb)