Abstract
Swimmer’s itch (cercarial dermatitis) is a nuisance encountered by bathers and recreational water users worldwide. The condition is caused by the penetration of larval digenean trematodes (cercariae) of the family Schistosomatidae, into the skin, following their release into freshwater from pulmonate snails that serve as the intermediate hosts for these parasites. This study utilizes qPCR-based cercariometry to monitor and quantify cercariae from water samples collected at 5 lakes in northern Michigan. The resolution provided by qPCR facilitated assessment of the environmental and biological drivers of swimmer’s itch-causing cercariae concentrations, allowing us to demonstrate that cercarial abundance is greatest at the top of the water column, in locations with prevailing on- and alongshore winds.
Electronic supplementary material
The online version of this article (10.1007/s10393-018-1362-1) contains supplementary material, which is available to authorized users.
Keywords: Swimmer’s itch, qPCR, Recreational water, Exposure, Cercariometry, Avian schistosomes, eDNA
Introduction
Parasites of the family Schistosomatidae utilize avian or mammalian definitive hosts and snail intermediate hosts to complete their life cycles. Adult worms reside in the avian or mammalian hosts and produce embryonated eggs that typically exit the definitive host via feces or urine into the aquatic environment, where they hatch into a free-living larval stage (miracidium). Miracidia penetrate the snail host and develop into sporocysts. Sporocysts produce cercariae asexually, which are then shed from infected snails (Cort 1928; Blair and Ottesen 1979; Blair and Islam 1983). Cercariae then seek out their definitive host to complete their life cycle. The number of cercariae released each day can vary significantly between hundreds to tens of thousands of larval parasites and can be influenced by factors such as temperature, duration of infection, and immunogenetic determinants of snail-schistosome compatibility (Sluiters et al. 1980; McCarthy 1999; Zbikowska 2004; Coady et al. 2006; Gordy et al. 2015; Soldánová et al. 2016; Pila et al. 2016). While the cercariae of some species of schistosome (those of the genus Schistosoma) are known to infect humans, species of the genera Trichobilharzia, Gigantobilharzia, Dendritobilharzia, Anserobilharzia, and Schistosomatium cannot, but are able to penetrate human skin and lead to a condition known as swimmer’s itch (Cort 1928; Batten 1956; Kolářová et al. 1999).
Swimmer’s itch, or cercarial dermatitis, is caused by epidermal penetration of humans by larval cercariae in surface waters (Brant and Loker 2009). The resultant immune response leaves itchy papules that can last for weeks (Baird and Wear 1987; Verbrugge et al. 2004; Soldánová et al. 2013). Swimmer’s itch is a prominent problem in recreational lakes in northern Michigan, where the predominant species responsible for swimmer’s itch is Trichobilharzia stagnicolae, hosted by the common merganser (Mergus merganser) and the snail Stagnicola emarginata (Blankespoor and Reimink 1991; Keas and Blankespoor 1997).
Monitoring efforts for swimmer’s itch typically rely on assessments of infection prevalence of relevant parasites in snails. This method requires collecting a large number of snails from multiple sites around a lake and allowing the snails to shed parasites. Shed cercariae are then morphologically identified using a stereomicroscope. While this method assesses the success of a specific parasite species in infecting a snail species such as S. emarginata, it has a number of downsides. Notably, avian schistosomes typically show a low prevalence in snail intermediate hosts (Crews and Esch 1986; Brown et al. 1988; Gordy et al. 2016), making detection difficult, and collecting and screening snails is time-consuming. Furthermore, as cercariae are essential both for transmission of the parasite, and cause swimmer’s itch, quantifying their abundance in the water is key to understanding both the ecological and public health aspects of this parasite. A quantitative polymerase chain reaction assay for the detection of avian schistosomes, or swimmer’s itch-causing cercariae, in recreational water was validated and published in 2015 (Narayanan et al. 2015). This assay can detect species of the family Schistosomatidae, including Trichobilharzia spp., Schistosomatium douthitti, Allobilharzia spp., Gigantobilharzia, spp., and Dendritobilharzia spp. qPCR cercariometry offers a notable advantage to traditional snail infection prevalence methodologies when the end goal is assessment of swimmer’s itch risk. Cercariae concentration and distribution in the water body may be influenced by environmental factors such as wind and water movement, or predation, inactivation due to UV light, or death (Haas 1994; McCarthy 1999). As qPCR cercariometry enables the quantification of cercariae in the water and at recreation sites, it measures the potential exposure of bathers to cercariae. Some questions still remain regarding the accuracy and reproducibility of using qPCR to measure trematodes (or for that matter, eukaryotic organisms) in water. Additionally, approaches to quantify to an organismal level remain in their infancy for eukaryotic targets.
In this study, we have expanded upon qPCR cercariometry as a method to quantify cercariae abundance and use it to monitor for avian schistosome cercariae in lakes in northern Michigan. This paper defines appropriate sample collection methodology, and documents methods for estimating DNA losses due to DNA extraction, which enables a robust estimation of the number of cercariae present in samples. We have assessed the accuracy and reproducibility of the method in controlled laboratory experiments, and in the field. Furthermore, we have applied this method to monitor for swimmer’s itch-causing cercariae at a variety of lakes in northern Michigan, and used these data to assess the effects of wind direction, time of day, and seasonality on cercariae abundance.
Methods
Environmental Sampling
Water Sampling
Twenty-five-liter water samples were collected at each location and passed through a 20 cm × 80 cm × 20 micron zooplankton net (Aquatic Research Instruments). Samples were collected liter by liter across the entire swath of the beach, in approximately waist deep water, with the goal of maximizing the likelihood of capturing free-swimming cercariae. Debris from inside the net were washed down using well water followed by a 95% ethanol wash and collection in 50-mL conical tubes. This 50-mL sample was then passed through a 0.45-µM polycarbonate filter (Pall), the filter paper removed from the bed, and stored in 1 mL of 95% ethanol for transport to the University of Alberta (Canada) for DNA extraction and qPCR analysis.
Environmental Variable Measurements
Wind direction, and effect (i.e., onshore, offshore, alongshore), as well as with time of day were also recorded at each sampling site.
Snail, Miracidia, and Cercariae Collection
Cercariae, miracidia, and snails were collected from Higgins Lake, Lake Leelanau, and Crystal Lake in 2016. Cercariae shed from snails were pipetted into collection tubes and preserved with 95% ethanol. Tissue from each representative snail species was also preserved in 95% ethanol. Miracidia were collected from fresh bird feces deposited on the beach by diluting fecal samples in well water, hatching the miracidia under fluorescent lights, and pipetting into 95% ethanol. The species and approximate age of the bird from which the feces were deposited were recorded at the time of collection. Ethanol preserved samples were stored at ~ − 10 °C and then cold-shipped to the University of Alberta for further processing.
Method Robustness
Three consecutive samples (25 L each) at 10 sampling locations on Crystal Lake were collected and evaluated to assess the precision of the entire method in the field. Additionally, three samples containing an unknown number of cercariae were sent from researchers in Michigan to the University of Alberta where qPCR was performed in a single blind trial to assess the accuracy of the qPCR assay in predicting the true number of cercariae in a sample.
Time of Day Sampling
Eight locations were selected on Big and Little Glen Lakes for sampling on July 25, 2017. Two teams collected water samples at each of 4 locations at approximately 8:00, 8:15, 8:30, and 8:45 a.m. The 4-site collection sequence was repeated beginning at 12:00 p.m., 4:00 p.m., and 8:00 p.m.
Depth of Water Sampling
A stacked water column trap was designed and built to capture stacked subsamples of water. The water column trap can be used at depths up to 1.5 m deep. The trap consists of 3, 4-inch PVC ball valves connected in series by 2, 4 × 6 inch threaded nipples (Menards #MA0013H, #CC08350). Each section corresponded to a depth of 50 cm. The column trap was lowered into the water with valves open. Each valve was then manually closed, trapping 550 mL of lake water in each connector nipple. The subsamples were then emptied into separate buckets by opening the valves, one by one beginning at the bottom. A total of 30 full columns were collected. The buckets were then emptied into a 20-μm mesh plankton tow, the net washed with 95% ethanol, and the sample concentrated to ~ 50 mL. The depth of the water where sampling occurred was approximately 1.2 m. Therefore, the topmost section of the trap was not completely covered by the water, and the topmost section of the trap only collected the top 30 cm of the water column (Supplementary Figure 1).
Molecular Methodologies
DNA Sequencing
DNA extraction, PCR amplification of the Cytochrome c oxidase subunit 1 gene (cox1), Sanger sequencing, and alignments were completed as described in Gordy et al. (2016). The cox1 mitochondrial DNA sequences generated in this study are available on NCBI GenBank from accession numbers MG964019 to MG964043.
Extraction of Schistosome DNA from Water Samples
In 2016, ethanol was evaporated from filters using a vacuum centrifuge. Subsequently, 200 µL of lysis buffer AL (Qiagen) and 20 µL of proteinase K were then added to the tube containing the filter, along with 1-mm silica carbide beads, and bead beat for 10 min on high using a vortex before DNA extraction (as described by Webster (2009), using the DNeasy Extraction Kit [Qiagen]). In 2017, DNA extraction was accomplished in Michigan using the Biomeme Environmental DNA extraction kit (Biomeme, Philadelphia, PA, USA) according to the manufacturers instructions, with one deviation—samples were eluted into 100 µL of elution buffer. This change was made to reduce costs associated with shipping samples as the Biomeme extraction can be accomplished lakeside in Michigan.
qPCR Detection of Schistosome 18S rRNA
The 18S rRNA qPCR assay was run according to the method described in Narayanan et al. (2015). The sequences of the forward, probe, and reverse primer are, respectively, AGCCTTTCAGCCGTATCTGT, TCGGGAGCGGACGGCATCTTTA, AGGCCTGCCTTGAGCACT. Unlike the original method, samples were quantified to copies of plasmid DNA using a standard curve that consisted of 50,000, 5000, 500, 50, and 0.5 copies of cloned puc57 plasmid DNA (Genscript, New Jersey, USA) containing the avian schistosome 18S rRNA gene. The LOD95 of this technique is 3.4 copies per reaction (upper limit 7.9, lower limit 1.5) (Wilrich and Wilrich 2009). Samples and standards were performed in triplicate, with inhibition controls run for each sample in duplicate. Inhibition controls were performed as described in Rudko et al. (2017), where a plasmid inhibition control was spiked into water samples and subsequently detected by qPCR. Inhibition was defined as a 3-cycle threshold shift (USEPA 2012). Thermocycling (qPCR) was performed in a post-amplification room using the ABI 7500 Fast Real Time qPCR system.
Quantitation of Plasmid Copies Per Cercaria
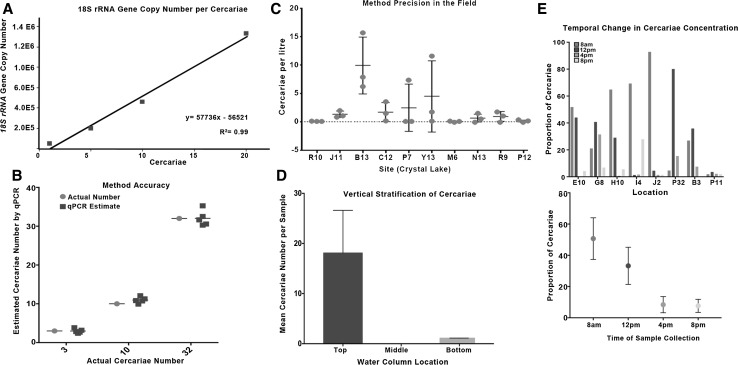
Hand counted stocks (stereo microscope [Zeiss]) of T. stagnicolae cercariae (1, 5, 10, and 20 cercaria(e)) were counted, and DNA extracted, according to Webster (2009). The average 18S rRNA copy number was determined using qPCR. Results were used to generate a standard curve to convert DNA copies per sample to cercariae per sample. Conversion equation: x = [(y + 56521)/57736] was derived and used to calculate the number of cercariae per sample. X is the number of cercariae, and y is the copy number per sample (Fig. 1A).
Figure 1.
qPCR Method Validation. A Cercariae standard curve used to derive conversion equation for the conversion of qPCR copy number to number of cercariae. B Accuracy of the qPCR method. Actual number of cercariae, hand counted via microscopy, and the qPCR estimate of cercariae based on five replicate qPCR reactions. C Precision of the method in the field. Replicate 25-L water samples were analyzed at different sites on Crystal Lake in 2016. Error bars represent standard deviation around the mean. D Vertical stratification of cercariae concentrations. Top = 0–30 cm from surface, Middle = 50–100 cm, Bottom = 100–150 cm. e. Daily changes in cercariae concentration. Top: Changes in proportion of cercariae at each site from 8 a.m. to 8 p.m. Bottom: Average proportion of cercariae of all sites at each time point.
Extraction Efficiency
Extraction efficiency was determined by spiking 5 T. stagnicolae cercariae onto a clean 0.45-µM filter membrane and preserving the samples in 100% ethanol. Samples were analyzed in the same manner as described above for the water samples. Conversion equation was applied to determine the number of cercariae recovered through the method under ideal circumstances.
Statistics
The change in cercariae concentrations relative to the time of day, and wind direction relative to the shoreline, was assessed using a generalized linear model with a log link function, based on the negative binomial distribution. The outcome variable was cercariae concentration. Explanatory variables were lake, week (standardized to account for different days during the same week across years), time of day (morning or afternoon), and wind direction (onshore, offshore, and alongshore). In the initial model, the interaction effect of time of day and wind direction was also tested, but this interaction was found nonsignificant and was removed to improve model fit. Additionally, site on the lake, year, and lake were modeled as random effect variables in the original model with independent correlation matrices, but were found to be insignificant and were removed from the final model. All statistical analyses were performed using SPSS (version 24, IBM, Armonk, North Castle, New York, USA). T tests and graphs were executed using PRISM (ver. 7.0, GraphPad).
Results
Snail and Trematode Species Identifications
Mitochondrial cox1 DNA sequences confirmed T. stagnicolae (99.4–100% matches to GenBank vouchers) to be the common schistosome emerging from S. emarginata among Higgins Lake, Crystal Lake, and Lake Leelanau. Miracidia from M. merganser (common merganser duck), Anas platyrhynchos (mallard duck), and Branta canadensis (Canada goose) were collected. As suspected, Trichobilharzia sp. miracidia were found in the feces of mergansers and mallards. A miracidia, which most closely matched to a member of the Schistosomatidae family, was found in Canadian geese. The avian trematode Dendritobilharzia sp. miracidia were also found in the feces of mallard ducks (Table 1) (Brant et al. 2011).
Table 1.
Snail, Miracidia, and Cercariae Species Identified Based on Cox-1 Sequencing from Higgins Lake, Crystal Lake, and Lake Leelanau in 2016.
| Identification | Pairwise similarity (%) | Accession match |
|---|---|---|
| Crystal Lake | ||
| Cercariae | ||
| Diplostomum sp. 1 | 100.0 | KR271221.1 |
| Plagiorchis sp. | 99.3 | FJ477214.1 |
| Apatemon sp. 1 | 90.6 | HM064617.1 |
| Trichobilharzia Stagnicolae | 100.0 | FJ174489.1 |
| Snail | ||
| Physella ancillaria | 99.56 | KM612195.1 |
| Unclassified Planorbidae | 100.00 | KM612069.1 |
| Miracidia | ||
| Trichobilharzia stagnicolae From hatch-year common merganser brood | 99.78 | KT831352.1 |
| Trichobilharzia stagnicolae From hatch-year common merganser brood | 100 | KT831352.1 |
| Trichobilharzia stagnicolae From hatch-year common merganser | 99.38 | FJ174490.1 |
| Higgins Lake | ||
| Cercariae | ||
| Diplostomum sp. 4 | 99.2 | KR271383.1 |
| Apatemon sp. 1 | 90.6 | HM064617.1 |
| Plagiorchis sp. | 99.5 | FJ477214.1 |
| Stagnicola elodes | 97.8 | KM612224.1 |
| Snail | ||
| Physella ancillaria | 99.84 | KM612168.1 |
| Elimia livescens | 100 | EF586916.1 |
| Campeloma decisum | 99.6 | KU905792.1 |
| Stagnicola elodes | 97.8 | KM612224.1 |
| Marstonia lustrica | 99.7 | AF520945.1 |
| Miracidia | ||
| Trichobilharzia stagnicolae From hatch-year common merganser | 99.8 | KT831352.1 |
| Lake Leelanau | ||
| Cercariae | ||
| Trichobilharzia stagnicolae | 99.8 | FJ174490.1 |
| Snails | ||
| Stagnicola elodes | 97.7 | KM612224.1 |
| Unclassified Planorbidae | 99.8 | KM612069.1 |
| Stagnicola elodes | 99.7 | HQ969867.1 |
| Physella gyrina | 98.5 | KT708102.1 |
| Pleurocera catenaria | 83 | EU414649.1 |
| Unidentified Planorbidae—Biomphalaria choanomphala/Biomphalaria tenagophila | 85 | HM768933.1/EF433576.1 |
| Miracidia | ||
| Trichobilharzia stagnicolae From hatch-year common merganser | 99.80 | FJ174492.1 |
| Dendritobilharzia sp. From hatch-year mallard | 99.0 | KX302892.1 |
| Schistosomatidae gen. spp From Canadian goose. | 84.0 | FJ174486.1 |
qPCR Method Validation
qPCR Validation Against Trematode Library
The qPCR assay was tested against a library of 25 North American trematodes (Gordy et al. 2016). Amplification was only observed in samples containing Trichobilharzia sp. (Table 2).
Table 2.
The qPCR Assay is Specific to Swimmer’s Itch-Causing Cercariae.
| Species | qPCR Result |
|---|---|
| Trichobilharzia stagnicolae* | + |
| Trichobilharzia szidati* | + |
| Cotylurus sp. | − |
| Diplostomum baeri | − |
| Diplostomum huronense | − |
| Diplostomum indistinctum | − |
| Diplostomum sp. 1 | − |
| Diplostomum sp. 2 | − |
| Diplostomum sp. 3 | − |
| Diplostomum sp. 8 | − |
| Drepanocephalus auritus | − |
| Echinostoma caproni | − |
| Echinostoma trivolvis | − |
| Neodiplostomum americanum | − |
| Notocotylidae sp. | − |
| Haematoloechus sp. | − |
| Icthyocotylurus sp. 3 | − |
| Ornithodiplostomum sp. 8 | − |
| Plagiorchis sp. | − |
| Pseudopsilostoma varium | − |
| Strigeidae gen. | − |
| Telorchis sp. | − |
| Apharyngostrigea pipientis | − |
| Bolbophorus sp. | − |
| Tylodelphys scheuringi | − |
The qPCR assay was tested against a library of purified trematode DNA from across North America. Asterisks indicate swimmer’s itch-causing species. (+) indicates the target product amplified, while (−) indicates that no target was amplified.
*Statistically significant (P < 0.05)
Extraction Efficiency
DNA losses occur during any DNA extraction due to inefficient lysis or DNA absorbance. DNA extraction efficiency using the DNeasy kit ranged from 0.1 to 7%, and averaged 4.2%. DNA extraction efficiency using the Biomeme eDNA kit employed during the 2017 collection year averaged 4.1%, and ranged from 0.4 to 10%. These values are not significantly different (p [two tailed] = 0.6, df = 19). Copy numbers were adjusted to reflect a recovery rate of 4.2% (Table 3).
Table 3.
Extraction Efficiencies of the Qiagen DNeasy DNA Extraction Kit, and the Biomeme eDNA Field Extraction Kit.
| Trial | Loss (%) | Recovery (%) |
|---|---|---|
| Qiagen DNeasy | ||
| 1 | 99.99 | 0.01 |
| 2 | 99.2 | 0.8 |
| 3 | 99.9 | 0.1 |
| 4 | 98.9 | 1.1 |
| 5 | 98.7 | 1.3 |
| 6 | 92.2 | 7.8 |
| 7 | 92.0 | 8.0 |
| 8 | 99.7 | 0.3 |
| 9 | 99.7 | 0.3 |
| 10 | 98.0 | 2.0 |
| 11 | 98.0 | 2.0 |
| 12 | 92.8 | 7.2 |
| 13 | 94.3 | 5.7 |
| 14 | 92.9 | 7.1 |
| 15 | 92.9 | 7.1 |
| Biomeme eDNA | ||
| 1 | 98.1 | 1.9 |
| 2 | 96.1 | 3.9 |
| 3 | 98.6 | 1.4 |
| 4 | 78.8 | 21 |
| 5 | 96.6 | 3.4 |
| 6 | 99.0 | 1.0 |
| 7 | 98.7 | 1.3 |
| 8 | 99.6 | 0.40 |
Accuracy and Precision of the qPCR Method
Sampling variability was assessed by three samples taken consecutively of 25-L water samples at ten beaches on the same day on August 1, 2016, from Crystal Lake (Fig. 2). The sites with the highest dispersion between consecutive samples (Sites B13, P7, Y13) differed by 10, 8, and 12 cercariae each, respectively (Fig. 1C). Additionally, a single blinded experiment was performed wherein known cercariae stocks were shipped from Michigan, USA, to the laboratory in Alberta, Canada to be analyzed. The DNeasy extraction, qPCR, coupled with analysis using conversion equation correctly predicted the number of cercariae in each stock all three times the experiment was performed (Fig. 1B). The copy number in a single T. stagnicolae cercariae averages 55,851 copies (σx = 65.85).
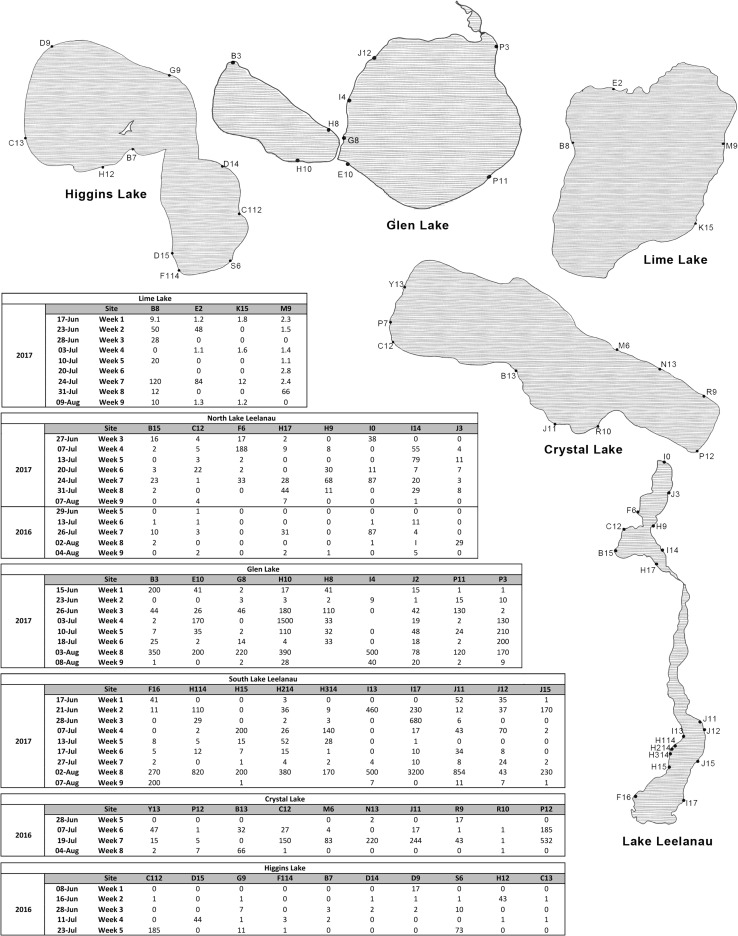
Figure 2.
Monitoring results. Lakes in northern Michigan were monitored weekly throughout 2016 and 2017. Results are in cercariae/25 L.
Environmental and Seasonal Drivers of Cercariae Abundance
Avian Schistosome Concentrations Vary in the Water Column
The vertical distribution of cercariae in the water column was assessed and demonstrates that the majority of swimmer’s itch-causing cercariae position themselves in the top 30 cm of the water column (Fig. 1D).
Time of Day Sampling
Eight (25 L) samples were collected at 4 time points throughout the day on July 25, 2017, to determine if the time of sample collection influences cercariae counts. On average, the highest numbers of cercariae were detected in the water at 8 a.m., and average numbers of cercariae at the same sampling point declined throughout the day (Fig. 1E).
Monitoring Results
Monitoring projects were conducted in 2016 and 2017. In 2016, Higgins Lake (44.495790, − 84.741729), Crystal Lake (44.661876, − 86.170675), and Lake Leelanau (44.981424, − 85.711727) were monitored, and in 2017, Lake Leelanau, Lime Lake (44.896239, − 85.841167), and Glen Lake (44.864639, − 85.962053) were monitored. Sampling sites were selected based on local knowledge—places where property owners commonly reported swimmer’s itch were sampled. South Lake Leelanau, Glen Lake, and Crystal Lake saw the highest numbers of cercariae detected in the water throughout their respective sampling periods, while Higgins, Lime Lake, and North Lake Leelanau have lower numbers of cercariae. However, it is important to note the high degree of variability in cercariae concentrations among all lakes; even lakes with lower concentrations of cercariae occasionally see extreme peaks in cercariae concentration, likely due to environmental factors such as wind or beach hydrology, which may concentrate cercariae in certain locations (Fig. 2).
Wind Direction Relative to the Shoreline Predicts Cercariae Concentrations
The effect of the factors, such as time of day, wind direction relative to the shore, and week (standardized between years, for exact dates see Fig. 2), was tested on cercariae concentrations. These effects were tested using a generalized linear model (Table 4). Results indicate that onshore and alongshore winds are predictive of higher concentrations of cercariae. The data also show that cercariae concentration changes temporally over the summer months, with weeks 4 and 8 of the sampling period predicting higher numbers of cercariae.
Table 4.
Generalized Linear Model Analysis of Environmental Factors Influencing Cercariae Concentrations.
| Main effects | P value |
|---|---|
| Wind direction | < 0.0001* |
| Lake | < 0.0001* |
| Week | < 0.0001* |
| Time of day (am/pm) | 0.59 |
| Parameter | β | Confidence interval | P value |
|---|---|---|---|
| Parameter estimates | |||
| Wind | |||
| Onshore wind | 0.89 | 0.23–1.5 | 0.008* |
| Alongshore wind | 1.2 | 0.57–1.8 | < 0.0001* |
| Offshore wind | 0.48 | − 0.24 to 1.2 | 0.19 |
| Lake | |||
| North Leelanau | − 1.213 | − 1.7 to − 0.7 | < 0.0001* |
| Lime Lake | − 1.825 | − 2.3 to − 1.3 | < 0.0001* |
| Higgins | − 1.406 | − 0.77 to − 0.72 | < 0.0001* |
| Glen Lake | − 0.077 | − 0.47 to 0.14 | 0.70 |
| South Leelanau | − 0.045 | − 0.42 to 0.50 | 0.85 |
| Crystal | 0.045 | − 0.42 to 0.50 | 0.85 |
| Week | |||
| Week 1 | − 0.74 | − 1.5 to 0.05 | 0.07 |
| Week 2 | − 0.58 | − 1.2 to 0.70 | 0.08 |
| Week 3 | − 0.13 | − 0.75 to 0.50 | 0.70 |
| Week 4 | 0.63 | 0.03 to 1.23 | 0.04* |
| Week 5 | − 0.12 | − 0.8 to 0.50 | 0.72 |
| Week 6 | − 0.42 | − 1.2 to 0.30 | 0.25 |
| Week 7 | 0.38 | − 0.43 to 1.2 | 0.36 |
| Week 8 | 2.53 | 1.9 to 3.2 | < 0.0001* |
| Week 9 | 0.738 | − 0.58 to 0.9 | 0.07 |
| Time of day | |||
| Morning (8 a.m.–12 p.m.) | 0.08 | − 0.20 to 0.36 | 0.60 |
| Afternoon (12 p.m.–8 a.m.) | 0.08 | − 0.20 to 0.36 | 0.60 |
*Statistically significant (P < 0.05)
Discussion
The goals of this study were twofold: to aid in the validation of qPCR detection of avian schistosomes, and to utilize the assay in monitoring projects to assess the effects of environmental variables on the distribution of cercariae in the water.
Method Validation
The common merganser has long been assumed to contribute the majority of avian schistosomes to the lakes in northern Michigan (Blankespoor and Reimink 1991), and our survey confirms this observation. We also report Dendritobilharzia spp. miracidia, another schistosome species capable of causing swimmer’s itch in the feces of mallard ducks (Table 1). The qPCR assay in its initial validation by Narayanan et al. (2015) was designed to detect all species of the family Schistosomatidae, which includes Trichobilharzia and Dendritobilharzia spp.
qPCR has become a well-vetted and well-established method for recreational water monitoring for enteric bacteria (USEPA 2012); however, it is still an emerging methodology in parasitology. Molecular methods for the detection of the environmental stages of parasites have become more common in recent years and can be applied to fill crucial knowledge gaps related to the environmental transmission stages. As a review by Bass et al. (2015) points out, environmental sampling for parasites coupled with molecular methods can reduce sampling bias incurred by specimen-based sampling. Additionally, molecular parasitological methods have been demonstrated to be more cost-effective, less labor intensive than specimen-based detection (Huver et al. 2015). However, there are a number of limitations, notably sample inhibition, DNA losses, and difficulties in quantifying to the organismal level, and the inability to distinguish between live and dead organisms. Eukaryotic targets in qPCR can be more challenging to validate because they are multi-cellular, and the 18S rRNA gene target is multi-copy within the genome (Weiss et al. 2017; Bik et al. 2013). Ribosomal gene copy numbers are known to vary by orders of magnitude in bacteria and fungi, and apparent copy numbers quantified by qPCR may also be influenced by downstream methods such as losses due to DNA extraction (Cankar et al. 2006; Adamska et al. 2010; Green and Field 2012). Given this, quantitation to an organismal level for a eukaryotic target must include a conversion between gene copy number and organismal number, which is predicated on an assessment of DNA losses due to extraction. The majority of the Michigan lakes studies were relatively pristine, oligotrophic lakes. Silt and sandy debris in the water samples occasionally presented a challenge, but could be overcome by decanting off the liquid portion of a sample, leaving the debris behind before extraction. Procedures to the DNA extraction protocols may need to be modified if a similar study was replicated in a mesotrophic or eutrophic environment, or in the presence of cyanobacteria or algal blooms.
The average 18S rRNA gene copy number in a single cercaria is 55,851 copies; however, our proposed conversion between gene copy number and cercariae number utilizes the conversion equation and accounts for losses due to DNA extraction. We opted to use a standard curve to predict the number of cercariae rather than using a single number to estimate, as interpolating based on a standard curve better accounts for uncertainty and variability in the target measurement compared to using a single-point measurement. Extraction efficiencies between 0.5 and 4% have been reported for bacteria (Hwang et al. 2012). Therefore, it is not surprising that DNA extraction efficiencies ranged between 1 and 7% (Table 3). These results demonstrate how imperative it is to quantify losses through the extraction procedure in order to avoid underestimations in target concentrations. Other studies that have used microscopic cercariometry to quantify cercariae abundance have found as many as between 200 and 300 cercariae/L in still-water environments (Theron et al. 1978), while typical values in a more dynamic system range typically average 10 cercariae/L (Aoki et al. 2003).
Accuracy of the qPCR methods was tested in a single blinded study. Using the conversion equation and an estimation extraction efficiency of 4.2%, the actual number of cercariae was estimated correctly in each vial, suggesting that under ideal circumstances (i.e., purified cercariae in a clean matrix), this method of estimating cercariae concentrations from qPCR copy numbers is accurate and robust (Fig. 1B). Precision was tested in the field by collecting 3 consecutive 25-L water samples at ten beaches from Crystal Lake in 2016. Variation between samples in this dynamic natural environment is low, with the highest samples exhibiting variation of between 8 and 12 cercariae in between replicate samples. These results demonstrate that the assay provides a precise estimate of cercariae concentration over time in a natural environment (Fig. 1C), but that multiple samples will result in more robust estimation of cercariae concentrations at a particular location.
Environmental and Seasonal Drivers of Cercariae Abundance
Laboratory studies have demonstrated that cercariae of Trichobilharzia are positively phototactic and negatively geotactic, and the parasites are typically shed in the morning (Feiler and Haas 1988; Haas et al. 2008). Given these biological determinants of cercariae behavior, it could be expected that avian schistosome cercariae will be more likely to be found during the morning and in the topmost column of the water (Feiler and Haas 1988). Results of the water depth study demonstrated that the majority of cercariae reside in the top 15–20 cm of the water (Fig. 1D), which confirms observations made by others in the laboratory (Feiler and Haas 1988). The time of day experiment shows that on average, more cercariae are found in the water earlier in the day, between 8 a.m. and 12 p.m., and gradually decline throughout the day (Fig. 1E). However, the majority of samples collected on this day were collected during onshore or alongshore winds; therefore, it remained unclear if wind, or the time of sample collection may be responsible for the higher concentrations of cercariae observed between 8 a.m. and 12 p.m.
To better elucidate the biological and environmental drivers of cercariae abundance, a generalized linear model was performed on the entire dataset from 2016 to 2017. Results of the generalized linear model demonstrated the importance of onshore, and alongshore wind in explaining higher cercariae concentrations. Time of sampling (morning or afternoon) did not explain higher cercariae concentrations. There were 2 weeks, corresponding to early July and early August, which also predicted higher numbers of cercariae that could be due to seasonal trends in patent infections among snails (Table 4). Wind conditions likely blow cercariae closer to shore, as such it is intuitive that on- and alongshore winds would increase concentrations of cercariae along the shore. This is intuitive given that our study has also confirmed laboratory experiments that cercariae position themselves in the topmost portion of the water body, and therefore, their movement in the water is likely influenced greatly by surface winds, akin to the movement of surface cyanobacteria scums (Kahru et al. 2007; Kanoshina et al. 2003). Wind direction has also been previously implicated in moving Trichobilharzia cercariae (Leighton et al. 2000). Our model suggests that time of day does not have a strong relationship with cercariae abundance. However, as avian trematodes do leave their snail hosts in the morning, early morning bathers may still be at greater risk of encountering high concentrations of cercariae. There are numerous reports in the literation of strong seasonal trends in snails shedding cercariae, and this likely explains the higher overall cercariae concentrations found in weeks 4 and 8. Numerous studies have reported seasonal trends in patent infections in snails (Crews and Esch 1986; Brown et al. 1988; Gordy et al. 2016). It must be acknowledged that we tested a few factors which might affect cercariae concentrations—however, there are a number of potentially important variables which might influence cercariae concentrations, including water temperature, unpredictable cercariae shedding from snails, and shoreline bird abundance (Lo and Lee 1996; Abrous et al. 1999; Byers et al. 2008; Soldánová et al. 2016). Furthermore, qPCR as a DNA-based molecular method will detect both live and dead cercariae in the water column and therefore may overestimate infection risk in some instances (Bass et al. 2015).
The continued development of molecular testing for environmental stages of parasite development is essential to fully elucidating and understanding transmission and infection risks in parasitic lifecycles (Bass et al. 2015). We hope that our contributions to the avian schistosomes qPCR cercariometry method enable others to continue to elucidate environmental and biological drivers of infection in a variety of host parasite systems. Here we confirm a few previously hypothesized drivers of infection risk, but with this method, a variety of other hypotheses can be tested in the future.
This study demonstrates the utility of these molecular methods more broadly. For instance, analogous methods could be applied to tracking the spatial and temporal changes in Schistosoma mansoni, or Schistosoma japonicum cercariae which cause human schistosomiasis. While qPCR tests have been developed for both of these species, and have been trialed in ambient waters, to our knowledge no comprehensive assessment of cercariae concentrations and conditions that may increase or decrease their concentrations has been undertaken (Hertel et al. 2004; Hung and Remais 2008). Such an assessment is a natural next step for both DNA-based parasitological testing, but also could aid in monitoring the effectiveness of control efforts (Melo et al. 2006).
Electronic supplementary material
Below is the link to the electronic supplementary material.
A diagram of the stacked V trap water column-sampling device used to sample the number of parasites at different depths in the water column. (TIFF 850 kb)
Acknowledgements
The authors would like to thank Dr. Randy DeJong for their insightful comments on the manuscript, and Richard Golonka and Nathan McClure for their statistical critique.
Funding
Funding was provided by Natural Sciences and Engineering Research Council of Canada (Grant No. 418540), Alberta Innovates-Energy and Environment Solutions (Grant Nos. 2078, 2332) and Michigan Swimmer’s Itch Partnership.
References
- Abrous M, Rondelaud D, Dreyfuss G. Influence of low temperatures on the cercarial shedding of Paramphistomum daubneyi from the snail Lymnaea truncatula. Parasite. 1999;6:85–88. doi: 10.1051/parasite/1999061085. [DOI] [PubMed] [Google Scholar]
- Adamska M, Leońska-Duniec A, Maciejewska A, Sawczuk M, Skotarczak B. Comparison of efficiency of various DNA extraction methods from cysts of Giardia intestinalis measured by PCR and TaqMan real time PCR. Parasite. 2010;17:299–305. doi: 10.1051/parasite/2010174299. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Sata K, Muhoho ND, Noda SI, Kimura E. Cercariometry for detection of transmission sites for schistosomiasis. Parasitology International. 2003;52:403–408. doi: 10.1016/S1383-5769(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Baird JK, Wear DJ. 12 Cercarial dermatitis: The swimmer’s itch. Clinics in Dermatology. 1987;5:88–91. doi: 10.1016/S0738-081X(87)80013-5. [DOI] [PubMed] [Google Scholar]
- Bass D, Stentiford GD, Littlewood DTJ, Hartikainen H. Diverse applications of environmental DNA methods in parasitology. Trends in Parasitology. 2015;31:499–513. doi: 10.1016/J.PT.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Batten PJ., Jr The histopathology of swimmers’ itch. I. The skin lesions of Schistosomatium douthitti and Gigantobilharzia huronensis in the unsensitized mouse. American Journal of Pathology. 1956;32:363–377. [PMC free article] [PubMed] [Google Scholar]
- Bik HM, Fournier D, Sung W, Bergeron RD, Thomas WK. Intra-genomic variation in the ribosomal repeats of nematodes. PLoS ONE. 2013 doi: 10.1371/journal.pone.0078230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D, Islam KS. The life-cycle and morphology of Trichobilharzia australis n.sp. (Digenea: Schistosomatidae) from the nasal blood vessels of the black duck (Anas superciliosa) in Australia, with a review of the genus Trichobilharzia. Systematic Parasitology. 1983;5:89–117. doi: 10.1007/BF00049237. [DOI] [Google Scholar]
- Blair D, Ottesen P. Nasal schistosomiasis in Australian Anatids. Journal of Parasitology. 1979;65:982. doi: 10.2307/3280265. [DOI] [Google Scholar]
- Blankespoor HD, Reimink RL. The control of swimmer’s itch in Michigan: past, present, and future. Michigan Academician. 1991;24:7–23. [Google Scholar]
- Brant SV, Loker ES. Schistosomes in the southwest United States and their potential for causing cercarial dermatitis or “swimmer”s itch’. Journal of Helminthology. 2009;83:191. doi: 10.1017/S0022149X09308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant SV, Bochte CA, Loker ES. New Intermediate Host Records for the Avian Schistosomes Dendritobilharzia pulverulenta, Gigantobilharzia huronensis, and Trichobilharzia querquedulae From North America. Journal of Parasitology. 2011;97:946–949. doi: 10.1645/GE-2743.1. [DOI] [PubMed] [Google Scholar]
- Brown KM, Leathers BK, Minchella DJ. Trematode Prevalence and the Population Dynamics of Freshwater Pond Snails. American Midland Naturalist. 1988;120:289. doi: 10.2307/2426001. [DOI] [Google Scholar]
- Byers JE, Blakeslee AMH, Linder E, Cooper AB, Maguire TJ. Controls of spatial variation in the prevalence of trematode parasites infecting a marine snail. Ecology. 2008;89:439–451. doi: 10.1890/06-1036.1. [DOI] [PubMed] [Google Scholar]
- Cankar K, Štebih D, Dreo T, Žel J, Gruden K. Critical points of DNA quantification by real-time PCR—effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. BMC Biotechnology. 2006;6:37. doi: 10.1186/1472-6750-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady NR, Muzzall PM, Burton TM, Snider RJ, Saxton J, Sergeant M, Sommers A. Ubiquitous variability in the prevalence of Trichobilharzia stagnicolae (Schistosomatidae) infecting stagnicola emarginata in three northern michigan lakes. Journal of Parasitology. 2006;92:10–15. doi: 10.1645/GE-3336.1. [DOI] [PubMed] [Google Scholar]
- Cort WW. Schistosome dermatitis in the United States (Michigan) JAMA, the Journal of the American Medical Association. 1928;90:1027. doi: 10.1001/jama.1928.02690400023010. [DOI] [Google Scholar]
- Crews AE, Esch GW. Seasonal dynamics of Halipegus occidualis (Trematoda: Hemiuridae) in Helisoma anceps and its impact on fecundity of the snail host. Journal of Parasitology. 1986;72:646. doi: 10.2307/3281451. [DOI] [Google Scholar]
- Feiler W, Haas W. Host-finding in Trichobilharzia ocellata cercariae: swimming and attachment to the host. Parasitology. 1988;96:493–505. doi: 10.1017/S0031182000080136. [DOI] [PubMed] [Google Scholar]
- Gordy MA, Pila EA, Hanington PC. The role of fibrinogen-related proteins in the gastropod immune response. Fish & Shellfish Immunology. 2015;46:39–49. doi: 10.1016/j.fsi.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Gordy MA, Kish L, Tarrabain M, Hanington PC. A comprehensive survey of larval digenean trematodes and their snail hosts in central Alberta, Canada. Parasitology Research. 2016;115:3867–3880. doi: 10.1007/s00436-016-5152-9. [DOI] [PubMed] [Google Scholar]
- Green HC, Field KG. Sensitive detection of sample interference in environmental qPCR. Water Research. 2012;46:3251–3260. doi: 10.1016/j.watres.2012.03.041. [DOI] [PubMed] [Google Scholar]
- Haas W. Physiological analyses of host-finding behaviour in trematode cercariae: adaptations for transmission success. Parasitology. 1994;109:S15–S29. doi: 10.1017/S003118200008505X. [DOI] [PubMed] [Google Scholar]
- Haas W, Beran B, Loy C. Selection of the host’s habitat by cercariae: from laboratory experiments to the field. Journal of Parasitology. 2008;94:1233–1238. doi: 10.1645/GE-1192.1. [DOI] [PubMed] [Google Scholar]
- Hertel J, Kedves K, Hassan AH, Haberl B, Haas W. Detection of Schistosoma mansoni cercariae in plankton samples by PCR. Acta Tropica. 2004;91:43–46. doi: 10.1016/J.ACTATROPICA.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Hung YW, Remais J. Quantitative Detection of Schistosoma japonicum Cercariae in Water by Real-Time PCR. PLoS Neglected Tropical Diseases. 2008;2:e337. doi: 10.1371/journal.pntd.0000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huver JR, Koprivnikar J, Johnson PTJ, Whyard S. Development and application of an eDNA method to detect and quantify a pathogenic parasite in aquatic ecosystems. Ecological Applications. 2015;25:991–1002. doi: 10.1890/14-1530.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu W-T. Evaluation of methods for the extraction of dna from drinking water distribution system biofilms. Microbes and Environments. 2012;27:9–18. doi: 10.1264/jsme2.ME11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahru M, Savchuk OP, Elmgren R. Satellite measurements of cyanobacterial bloom frequency in the Baltic Sea: interannual and spatial variability. Marine Ecology Progress Series. 2007;343:15–23. doi: 10.3354/meps06943. [DOI] [Google Scholar]
- Kanoshina I, Lips U, Leppänen JM. The influence of weather conditions (temperature and wind) on cyanobacterial bloom development in the Gulf of Finland (Baltic Sea) Harmful Algae. 2003;2:29–41. doi: 10.1016/S1568-9883(02)00085-9. [DOI] [Google Scholar]
- Keas BE, Blankespoor HD. The prevalence of cercariae from Stagnicola emarginata (Lymnaeidae) over 50 years in northern Michigan. Journal of Parasitology. 1997;83:536–540. doi: 10.2307/3284427. [DOI] [PubMed] [Google Scholar]
- Kolářová L, Skirnisson K, Horák P. Schistosome cercariae as the causative agent of swimmer’s itch in Iceland. Journal of Helminthology. 1999;73:215–220. doi: 10.1017/S0022149X99000335. [DOI] [PubMed] [Google Scholar]
- Leighton BJ, Zervos S, Webster JM. Ecological factors in schistosome transmission, and an environmentally benign method for controlling snails in a recreational lake with a record of schistosome dermatitis. Parasitology International. 2000;49:9–17. doi: 10.1016/S1383-5769(99)00034-3. [DOI] [PubMed] [Google Scholar]
- Lo CT, Lee KM. Pattern of Emergence and the Effects of Temperature and Light on the Emergence and Survival of Heterophyid Cercariae (Centrocestus formosanus and Haplorchis pumilio) Journal of Parasitology. 1996;82:347. doi: 10.2307/3284178. [DOI] [PubMed] [Google Scholar]
- McCarthy AM. The influence of temperature on the survival and infectivity of the cercariae of Echinoparyphium recurvatum (Digenea: Echinostomatidae) Parasitology. 1999;118:383–388. doi: 10.1017/S003118209900400X. [DOI] [PubMed] [Google Scholar]
- Melo FL, Gomes ALdoV, Barbosa CS, Werkhauser RP, Abath FGC. Development of molecular approaches for the identification of transmission sites of schistosomiasis. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2006;100:1049–1055. doi: 10.1016/j.trstmh.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Narayanan J, Mull BJ, Brant SV, Loker ES, Collinson J, Secor WE, Hill VR. Real-time pcr and sequencing assays for rapid detection and identification of avian schistosomes in environmental samples. Applied and Environment Microbiology. 2015 doi: 10.1128/AEM.00750-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pila EA, Tarrabain M, Kabore AL, Hanington PC, Cheung M, Sturrock S. A novel toll-like receptor (tlr) influences compatibility between the gastropod biomphalaria glabrata, and the digenean trematode schistosoma mansoni. PLoS Pathogens. 2016;12:e1005513. doi: 10.1371/journal.ppat.1005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudko SP, Ruecker NJ, Ashbolt NJ, Neumann NF, Hanington PC (2017) Investigating Enterobius vermicularis as a novel surrogate of helminth ova presence in tertiary wastewater treatment plants. Applied and Environmental Microbiology AEM.00547-17. 10.1128/aem.00547-17 [DOI] [PMC free article] [PubMed]
- Sluiters JF, Brussaard-Wüst CM, Meuleman EA. The relationship between miracidial dose, production of cercariae, and reproductive activity of the host in the combination Trichobilharzia ocellata and Lymnaea stagnalis. Parasitology Research. 1980;63:13–26. doi: 10.1007/BF00927722. [DOI] [PubMed] [Google Scholar]
- Soldánová M, Selbach C, Kalbe M, Kostadinova A, Sures B. Swimmer’s itch: etiology, impact, and risk factors in Europe. Trends in Parasitology. 2013;29:65–74. doi: 10.1016/j.pt.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Soldánová M, Selbach C, Sures B. The early worm catches the bird? productivity and patterns of Trichobilharzia szidati cercarial emission from Lymnaea stagnalis. PLoS ONE. 2016;11:e0149678. doi: 10.1371/journal.pone.0149678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron A, Pointier JP, Combes C. An ecological study of the involvement of man and rat in the maintenance of transmission of Schistosoma mansoni in Guadeloupe. Ann Parasitol Hum Comp. 1978;53:223–234. doi: 10.1017/S0031182000061734. [DOI] [PubMed] [Google Scholar]
- USEPA Method 1611: Enterococci in Water by TaqMan Quantitative Polymerase Chain Reaction (qPCR) Assay. Environmental Protection. 2012 doi: 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- Verbrugge LM, Rainey JJ, Reimink RL, Blankespoor HD. Prospective study of swimmer’s itch incidence and severity. Journal of Parasitology. 2004;90:697–704. doi: 10.1645/GE-237R. [DOI] [PubMed] [Google Scholar]
- Webster BL. Isolation and preservation of schistosome eggs and larvae in RNAlater(R) facilitates genetic profiling of individuals. Parasites & Vectors. 2009;2:50. doi: 10.1186/1756-3305-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, Knight R. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilrich C, Wilrich PT. Estimation of the POD Function and the LOD of a Qualitative Microbiological Measurement Method. Journal of AOAC International. 2009;92:1763–1772. [PubMed] [Google Scholar]
- Zbikowska E. Infection of snails with bird schistosomes and the threat of swimmer’s itch in selected Polish lakes. Parasitology Research. 2004;92:30–35. doi: 10.1007/s00436-003-0997-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A diagram of the stacked V trap water column-sampling device used to sample the number of parasites at different depths in the water column. (TIFF 850 kb)