Abstract
Background
The peroxiredoxin (PRDX) protein family is involved in cancer cell invasion and metastasis, but its prognostic value in lung cancer remain elusive.
Methods
In this report, we accessed the overall survival (OS) of each individual PRDX mRNA expression through the Kaplan–Meier plotter (KM plotter) database, in which updated gene expression data and survival information include a total of 1,926 lung cancer patients.
Results
Our results indicated that PRDX1 and PRDX2 mRNA expressions were associated with improved OS in all lung cancer patients especially in lung adenocarcinoma patients, whereas PRDX5 and PRDX6 mRNA expressions were associated with poor OS in all lung cancer patients. In addition, the prognostic value of PRDXs in the different clinicopathological features according to smoking status, pathological grades, clinical stages, and chemotherapeutic treatment of lung cancer patients was further assessed in the KM plotter database by the multivariate cox regression analysis.
Conclusion
Our finding will elucidate the prognostic role of PRDXs in lung cancer and might promote development of PRDX-targeted inhibitors for the treatment of lung cancer.
Keywords: PRDX, expression, prognostic, KM plotter, smoking status, lung cancer
Introduction
Lung cancer is one of the most common cancers and the leading cause of cancer death worldwide. It is generally classified as small-cell lung cancer and non-small-cell lung cancer (NSCLC), accounting for 15% and 85% of all lung cancers, respectively.1 NSCLC includes adenocarcinoma (Ade), squamous cell carcinoma (SCC) and large cell carcinoma. While standard treatment and better supportive care have improved overall survival (OS) and life quality, the prognosis of lung cancer is still unfavorable with an overall 5-year survival rate <15%.2 Therefore, there is much interest in identifying a more accurate prognostic assessment and molecular biomarkers for lung cancer.
Peroxiredoxins (PRDXs) are a ubiquitous family of small (22–27 kDa) non-seleno peroxidases which catalyze peroxide reduction to balance cellular hydrogen peroxide (H2O2) levels.3,4 They are divided into two classes: 2-Cys PRDXs and 1-Cys PRDX. Six PRDX isoforms have been identified.5 Increased or decreased levels of PRDXs have been demonstrated in many human cancers.6–10 Studies performed in vitro or in vivo models have demonstrated that overexpression of PRDXs may either inhibit cancer development or promote cancer growth, depending on the specific PRDX family member and on the cancer context.11–15 However, the prognostic value of mRNA expression of individual PRDXs in lung cancer patients is still elusive. In this study, we determined the prognostic roles of mRNA expression of six PRDXs in lung cancer patients.
KM plotter database generated data from The European Genome-phenome Archive (EGA), Gene Expression Omnibus (GEO), and The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) cancer data sets,16 which include gene expression data and survival information from a total of 1,926 lung cancer patients. In the present study, we used the KM plotter database and accessed the prognostic roles of individual PRDX mRNA expression in patients with lung cancer.
Materials and methods
The Kaplan–Meier plotter (KM plotter, http://kmplot.com/analysis/) has information of 54,675 genes on survival using 10,461 cancer samples, which include breast,17 ovarian,18 gastric,19 and lung16 cancer patients with a mean follow-up of 69, 40, 49, and 33 months, respectively. Gene expression data and OS information are downloaded from EGA, GEO (Affymetrix microarrays only), and TCGA, which is handled by a PostgreSQL server and integrates clinical data and gene expression information simultaneously. The database collected survival information of 1,926 lung cancer patients, which comprises clinical data such as gender, age, histology, grade, stage, and chemotherapy of all patients in WinStat 2013.16
In our study, the prognostic values of the individual mRNA expression of PRDXs in lung cancer were speculated using the KM plotter online database. In addition, we evaluated the correlation between OS of lung cancer patients and specific PRDXs based on smoking status, pathological grades, clinical stages, and chemotherapeutic treatment. Briefly, the JetSet probe IDs representing the six genes of PRDXs (PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6) individually entered into the database (http://kmplot.com/analysis/index.php?p=service&cancer=lung) to analyze Kaplan–Meier survival curves. With the purpose to determine the prognostic value of a particular gene, certain gene mRNA expression between the lower and upper quartiles were computed, and the best performing threshold was used as the final cutoff in a univariate cox regression analysis. Smoking status, pathological grades, clinical stages, and chemotherapeutic treatment were calculated in the multivariate analysis. Finally, the expression cutoff points of the PRDX genes were determined according to the median expression of the gene from the selected lung cancer samples. PRDX expression were split into “low” and “high” depending on the comparisons between expression values with established cutoffs. The number of cases, median values of mRNA expression levels, HR, 95% CI, and log rank P were determined and presented on the webpage. P-value of <0.05 was deemed to be statistically significant to reduce the false-positive rate.
Results
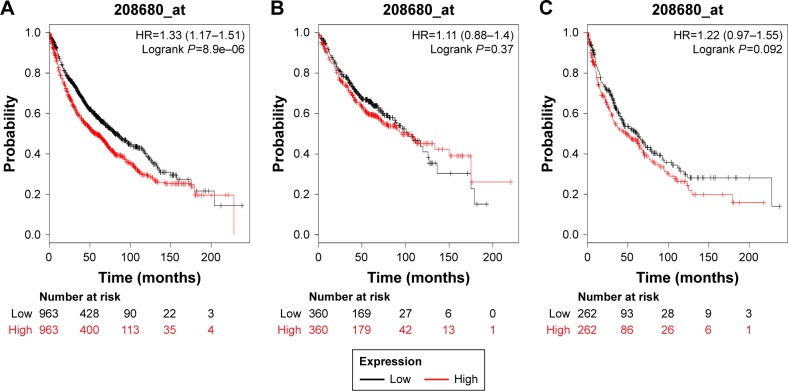
In our present study, all the six PRDX family Kaplan–Meier survival information can be determined on www.kmplot.com. We used KM plotter and determined the prognostic value of PRDX1 in the database. The valid Affymetrix ID is 208680_at (PRDX1). Survival curves are drafted for all lung cancer patients (n=1,926) (Figure 1A), Ade patients (n=720) (Figure 1B), and SCC patients (n=524) (Figure 1C). PRDX1 mRNA high expression was correlated with worse OS for all lung cancer patients who were followed for 20 years (HR=1.33, 95% CI: 1.17–1.51, P=8.9e–6). Nevertheless, with regard to Ade and SCC patients, there was no significant difference in HR estimates between study strata.
Figure 1.
The prognostic value of PRDX1 expression in the database.
Notes: The valid Affymetrix ID is 208680_at (PRDX1). Survival curves are plotted for (A) all patients (n=1,926), HR=1.33 (95% CI: 1.17–1.51); (B) adenocarcinoma (n=720), HR=1.11 (95% CI: 0.88–1.4); and (C) squamous cell carcinoma (n=524), HR=1.22 (95% CI: 0.97–1.55).
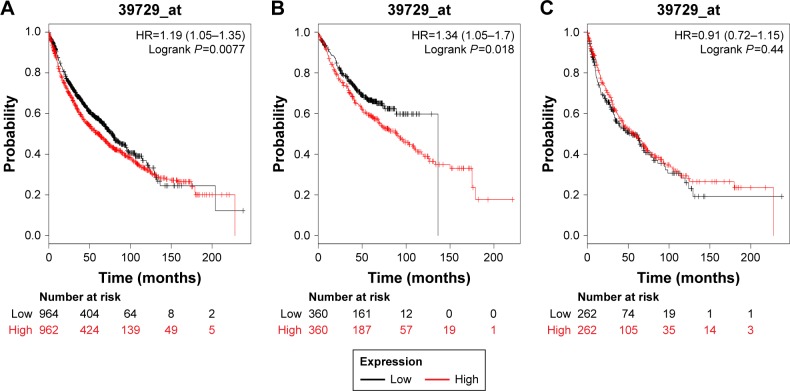
We next determined the prognostic value of PRDX2 in the database. The valid Affymetrix ID is 39729_at (PRDX2). PRDX2 mRNA high expression was significantly correlated with worse OS for all patients (HR=1.19, 95% CI: 1.05–1.35, P=0.0077; Figure 2A). PRDX2 mRNA high expression was also correlated with worse OS in Ade patients (HR=1.34, 95% CI: 1.05–1.7, P=0.018; Figure 2B), but not in SCC patients (HR=0.91, 95% CI: 072–1.15, P=0.44; Figure 2C).
Figure 2.
The prognostic value of PRDX2 expression in the database.
Notes: The valid Affymetrix ID is 39729_at (PRDX2). Survival curves are plotted for (A) all patients (n=1,926), HR=1.19 (95% CI: 1.05–1.35); (B) adenocarcinoma (n=720), HR=1.34 (95% CI:1.05–1.7); and (C) squamous cell carcinoma (n=524), HR=0.91 (95% CI: 072–1.15).
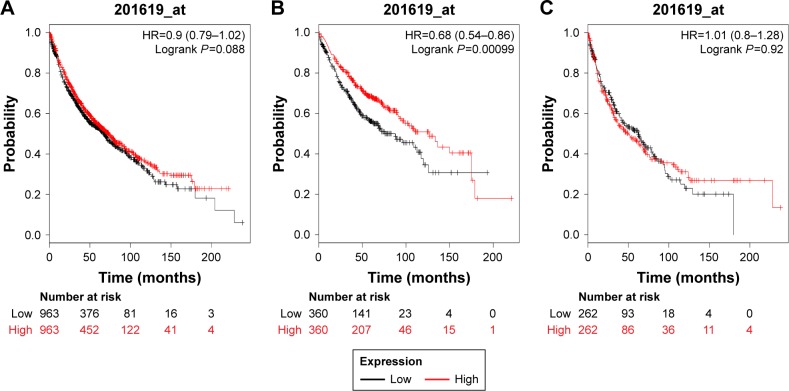
Figure 3 demonstrates the prognostic value of PRDX3 in the database. The valid Affymetrix ID is 201619_at (PRDX3). PRDX3 mRNA high expression was not significantly correlated with OS for all lung cancer patients (HR=0.9, 95% CI: 0.79–1.02, P=0.088; Figure 3A), SCC patients (HR=1.01, 95% CI: 0.8–1.28, P=0.9174; Figure 3B). However, PRDX3 mRNA expression predicted a better OS in Ade patients (HR=0.68, 95% CI: 0.54–0.86, P=0.0011; Figure 3C).
Figure 3.
The prognostic value of PRDX3 expression in the database.
Notes: The valid Affymetrix ID is 201619_at (PRDX3). Survival curves are plotted for (A) all patients (n=1,926), HR=0.88 (95% CI: 0.79–1.02); (B) adenocarcinoma (n=720), HR=0.68 (95% CI: 0.54–0.86); and (C) squamous cell carcinoma (n=524), HR=1.01 (95% CI: 0.8–1.28).
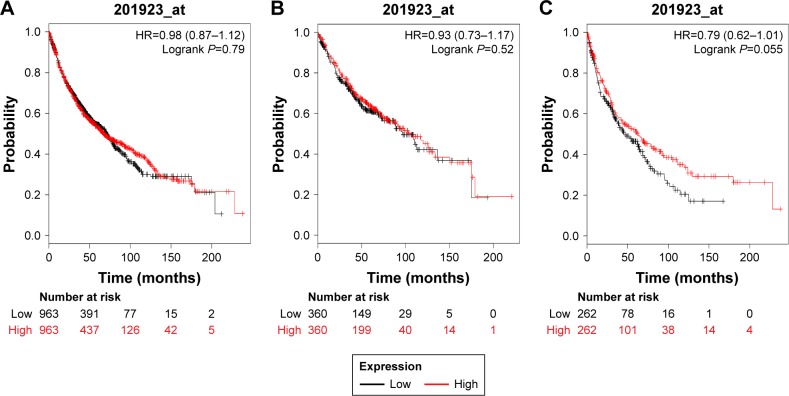
Figure 4 shows the prognostic value of PRDX4 in the database. The valid Affymetrix ID is 201923_at (PRDX4). PRDX4 mRNA high expression was not significantly correlated with OS for all lung cancer patients (HR=0.98, 95% CI: 0.87–1.12, P=0.79; Figure 4A), Ade patients (HR=0.93, 95% CI: 0.73–1.17, P=0.5238; Figure 4B), as well as patients with SCC (HR=0.79, 95% CI: 0.62–1.01, P=0.0556; Figure 4C).
Figure 4.
The prognostic value of PRDX4 expression in the database.
Notes: The valid Affymetrix ID is 201923_at (PRDX4). Survival curves are plotted for (A) all patients (n=1,926), HR=0.95 (95% CI: 0.87–1.12); (B) adenocarcinoma (n=720), HR=0.93 (95% CI: 0.73–1.17); (C) squamous cell carcinoma (n=524), HR=0.79 (95% CI: 0.62–1.01).
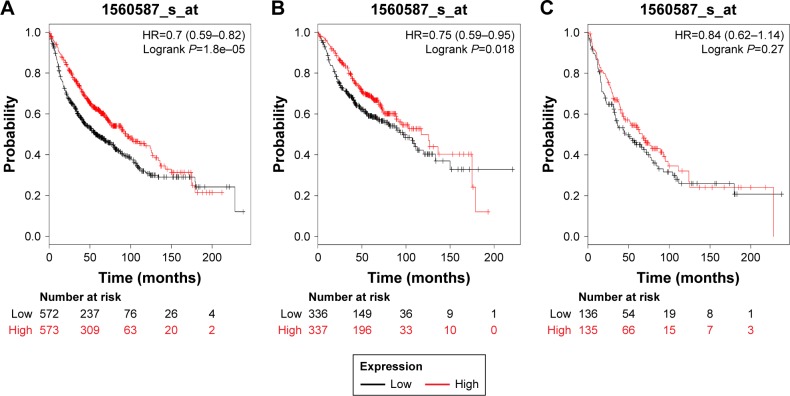
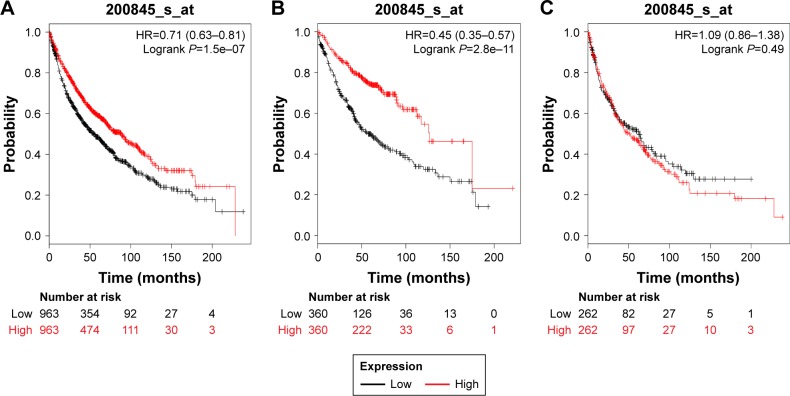
Figures 5 and 6 present the prognostic significance of PRDX5 and PRDX6, respectively. Affymetrix IDs were as follows: 1560587_s_at (PRDX5) and 200845_s_at (PRDX6). Both elevated PRDX5 and PRDX6 mRNA expressions are associated with a favorable OS for all the lung cancer patients (PRDX5: HR=0.7, 95% CI: 0.59–0.82, P=1.8e–5, Figure 5A; PRDX6: HR=0.71, 95% CI: 0.63–0.81, P=1.5e–7, Figure 6A). In addition, only PRDX6 mRNA high expression was significantly associated with worse OS in Ade patients (HR=0.45, 95% CI: 0.35–0.57, P=2.8e–11).
Figure 5.
The prognostic value of PRDX5 expression in the database.
Notes: The valid Affymetrix ID is 1560587_s_at (PRDX5). Survival curves are plotted for (A) all patients (n=1,926), HR=0.7 (95% CI: 0.59–0.82); (B) adenocarcinoma (n=720), HR=0.75 (95% CI: 0.59–0.95); and (C) squamous cell carcinoma (n=524), HR=0.84 (95% CI: 0.62–1.14).
Figure 6.
The prognostic value of PRDX6 expression in the database.
Notes: The valid Affymetrix ID is 200845_s_at (PRDX6). Survival curves are plotted for (A) all patients (n=1,926), HR=0.71 (95% CI: 0.63–0.81); (B) adenocarcinoma (n=720), HR=0.45 (95% CI: 0.35–0.57); (C) squamous cell carcinoma (n=524), HR=1.09 (95% CI: 0.86–1.38).
To further assess the association of PRDXs with other clinicopathological profiles, we determined the correlation with the patients’ smoking status (Table 1), clinical different stages (Table 2), and chemotherapeutic treatment. As from Table 1, PRDX1 and PRDX6 are significantly correlated with the smoking status. PRDX3 and PRDX6 are significantly associated with lung cancer patients without smoking history. From Table 2, PRDX1, PRDX2, PRDX3, and PRDX6 are significantly correlated with clinical stage I of the patients. Only PRDX6 is significantly associated with clinical stage II of lung cancer patients. However, none of the PRDXs was associated with chemotherapy of lung cancer patients.
Table 1.
Correlation of PRDX mRNA expression with smoking status of lung cancer patients
| Gene | Smoking history | HR (95% CI) | Log rank P |
|---|---|---|---|
| PRDX1 | Smoke | 1.24 (1.01–1.53) | 0.039* |
| Never smoke | 1.17 (0.67–2.05) | 0.5783 | |
| PRDX2 | Smoke | 1.08 (0.87–1.32) | 0.4917 |
| Never smoke | 1.3 (0.75–2.28) | 0.35 | |
| PRDX3 | Smoke | 1.14 (0.92–1.4) | 0.2282 |
| Never smoke | 0.34 (0.18–0.63) | 0.00035* | |
| PRDX4 | Smoke | 0.89 (0.72–1.09) | 0.26 |
| Never smoke | 1.08 (0.62–1.87) | 0.7966 | |
| PRDX5 | Smoke | 0.91 (0.61–1.37) | 0.6625 |
| Never smoke | 2.17 (0.93–5.06) | 0.067 | |
| PRDX6 | Smoke | 0.72 (0.58–0.89) | 0.002* |
| Never smoke | 0.33 (0.18–0.62) | 0.00028* |
Note:
P<0.05.
Abbreviation: PRDX, peroxiredoxin.
Table 2.
Correlation of PRDX mRNA expression with clinical stages of lung cancer patients
| Gene | Clinical stages | HR (95% CI) | Log rank P |
|---|---|---|---|
| PRDX1 | I | 1.7 (1.29–2.24) | 0.0002* |
| II | 1.39 (0.96–2) | 0.0812 | |
| III | 1.04 (0.6–1.79) | 0.8903 | |
| PRDX2 | I | 2.03 (1.51–2.71) | 1.3e–06* |
| II | 1.06 (0.73–1.53) | 0.753 | |
| III | 1.04 (0.6–1.8) | 0.8863 | |
| PRDX3 | I | 0.73 (0.56–0.95) | 0.0218* |
| II | 0.72 (0.5–1.04) | 0.0782 | |
| III | 1.16 (0.67–1.99) | 0.6013 | |
| PRDX4 | I | 1.09 (0.83–1.43) | 0.5287 |
| II | 0.81 (0.56–1.17) | 0.2608 | |
| III | 0.95 (0.55–1.65) | 0.8551 | |
| PRDX5 | I | 0.77 (0.56–1.05) | 0.0975 |
| II | 0.97 (0.62–1.52) | 0.8819 | |
| III | 1.04 (0.5–2.17) | 0.9078 | |
| PRDX6 | I | 0.59 (0.45–0.77) | 0.00013* |
| II | 0.6 (0.42–0.87) | 0.0063* | |
| III | 1.01 (0.59–1.73) | 0.9748 |
Note:
P<0.05.
Abbreviation: PRDX, peroxiredoxin.
Discussion
PRDX1 was first reported as an antioxidant enzyme.20 Recent studies have shown that abnormal expression of PRDX1 has been observed in several human cancers, including breast, esophageal, liver, lung, and prostate cancers.21–24 PRDX1 is expressed at significantly higher levels in lung cancer tissues compared with normal lung tissues, and elevated PRDX1 associated with shorter survival in lung cancer.25,26 Furthermore, PRDX1 is upregulated in NSCLC tissue interstitial fluid, and high level of PRDX1 expression is related to lymph node metastasis and tumor differentiation.27 Knockdown of PRDX1 in lung cancer cells significantly inhibits TGF-β1-induced epithelial-mesenchymal transition (EMT) and cell migration, whereas PRDX1 overexpression enhances TGF-β1-induced EMT and cell migration.28 In addition, in vivo studies have shown that silencing of PRDX1 leads to tumor suppression of growth and metastases by reducing the activation of c-Jun.12 Consistent with these findings, our data suggested that high mRNA expression of PRDX1 was associated with worse OS in patients with stage I lung cancer. Furthermore, PRDX1 is significantly correlated with the smoking status of patient with lung cancer.
PRDX2, also named as NKEF-B, was the most homologous molecule of PRDX1 and showed similar cellular localization.29 Lu et al demonstrated that PRDX2 overexpression in colorectal cancer tissues was strongly correlated with a more aggressive cancer behavior, tumor metastasis, and the tumor–node–metastasis (TNM) stage.30,31 PRDX2 overexpression was an independent prognostic indicator for stage I–III colorectal cancer patients.32 However, the biologic role and the prognostic effect of PRDX2 in lung cancer are poorly understood. According to our results, increased mRNA expression of PRDX2 was correlated with worse OS in all lung cancer patients as well as Ade patients, but not in SCC patients. Furthermore, high mRNA expression of PRDX2 was associated with worse OS in stage I lung cancer patients. Additional studies are needed to explore the mechanism of PRDX2 in lung cancer.
PRDX3 was mainly located in mitochondria. Several studies have indicated that PRDX3 is overexpressed in several human cancers, including lung cancer.33 But its role in prognosis of lung cancer is barely explored. In the present study, overexpressed PRDX3 mRNA only predicted a better OS in Ade patients, but not in SCC patients.
Wei et al demonstrated that Srx-PRDX4 axis plays a very important role in tumor progression and metastasis in lung cancer through the modulation of specific phosphokinase signaling.34 However, our results showed that PRDX4 was not significantly related to OS in patients with lung cancer.
Increased levels of PRDX5 have been reported in breast cancer and malignant mesothelioma.35,36 PRDX5 overexpression was associated with larger tumor size, positive lymph node status, poor differentiation, and a shorter patient survival in breast cancer patients.37 PRDX5 was involved in the regulation of several mitochondrial processes, which is essential in the protection of lung cancer cells against apoptosis induced by anticancer drugs.38 In this report, we observed that PRDX5’s high mRNA expression was found to be significantly correlated with worse OS for all lung cancer patients.
PRDX6 is a bifunctional enzyme with both GSH peroxidase and phospholipase A2 activities. PRDX6 is the only mammalian 1-Cys member of the PRDX family and is expressed in all major organs with a particularly high level in lung.39 Schremmer et al showed that PRDX6 expression was significantly associated with high-grade dysplasia and tumor progression in lung cancer cells.40 The invasion-promoting action of PRDX6 has also been confirmed using lung cancer cells, in which upregulation of PRDX6 results in the activation of Akt via PI3K and p38 kinase, which, in turn, promotes cell invasion by inducing uPA.41 Yun et al found that PRDX6 promotes lung tumor progression via its GPx and iPLA2 activities and promotes lung tumor development via activation of the JAK2/STAT3 pathway in a carcinogen-induced mouse lung tumor model.42,43 Our recent study demonstrated that high mRNA expression of PRDX6 was associated with worse OS in lung cancer patients, Ade patients, and those with early stage I–II.
Conclusion
In our present study, we comprehensively investigated the prognostic value of six PRDX members in patients with lung cancer by using the KM plotter database. In which, we revealed that PRDX1 and PRDX2 mRNA expressions were associated with improved OS in all lung cancer patients especially with Ade patients, whereas PRDX5 and PRDX6 mRNA expressions were associated with poor OS in all lung cancer patients. The prognostic value of the PRDX protein should be further evaluated in clinical studies. Our finding provides new insights into the prognostic functions of PRDX proteins in lung cancer and might promote the development of PRDX-targeted inhibitors for the treatment of lung cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ali A, Goffin JR, Arnold A, Ellis PM. Survival of patients with non-small-cell lung cancer after a diagnosis of brain metastases. Curr Oncol. 2013;20(4):e300–e306. doi: 10.3747/co.20.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B, Wang Y, Su Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer Lett. 2009;286(2):154–160. doi: 10.1016/j.canlet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- 4.Park MH, Jo M, Kim YR, Lee CK, Hong JT. Roles of peroxiredoxins in cancer, neurodegenerative diseases and inflammatory diseases. Pharmacol Ther. 2016;163:1–23. doi: 10.1016/j.pharmthera.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chae HZ, Oubrahim H, Park JW, Rhee SG, Chock PB. Protein glutathionylation in the regulation of peroxiredoxins: a family of thiol-specific peroxidases that function as antioxidants, molecular chaperones, and signal modulators. Antioxid Redox Signal. 2012;16(6):506–523. doi: 10.1089/ars.2011.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Chae HZ, Kim YJ, et al. Preferential elevation of Prx I and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell Biol Toxicol. 2003;19(5):285–298. doi: 10.1023/b:cbto.0000004952.07979.3d. [DOI] [PubMed] [Google Scholar]
- 7.Quan C, Cha EJ, Lee HL, Han KH, Lee KM, Kim WJ. Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J Urol. 2006;175(4):1512–1516. doi: 10.1016/S0022-5347(05)00659-2. [DOI] [PubMed] [Google Scholar]
- 8.Kinnula VL, Lehtonen S, Sormunen R, et al. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196(3):316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 9.Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, Chae HZ. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 2001;21(3B):2085–2090. [PubMed] [Google Scholar]
- 10.Neumann CA, Cao J, Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8(24):4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J, Schulte J, Knight A, et al. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28(10):1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang H, Wu L, Mishra M, Chawsheen HA, Wei Q. Expression of peroxiredoxin 1 and 4 promotes human lung cancer malignancy. Am J Cancer Res. 2014;4(5):445–460. [PMC free article] [PubMed] [Google Scholar]
- 13.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300(5619):650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 14.Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid Redox Signal. 2011;15(3):781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 15.Rostila A, Puustinen A, Toljamo T, et al. Peroxiredoxins and tropomyosins as plasma biomarkers for lung cancer and asbestos exposure. Lung Cancer. 2012;77(2):450–459. doi: 10.1016/j.lungcan.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 16.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 18.Győrffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 19.Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding C, Fan X, Wu G. Peroxiredoxin 1 – an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21(1):193–202. doi: 10.1111/jcmm.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren P, Ye H, Dai L, et al. Peroxiredoxin 1 is a tumor-associated antigen in esophageal squamous cell carcinoma. Oncol Rep. 2013;30(5):2297–2303. doi: 10.3892/or.2013.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai CY, Zhai LL, Wu Y, Tang ZG. Expression and clinical value of peroxiredoxin-1 in patients with pancreatic cancer. Eur J Surg Oncol. 2015;41(2):228–235. doi: 10.1016/j.ejso.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 23.Sun YL, Cai JQ, Liu F, Bi XY, Zhou LP, Zhao XH. Aberrant expression of peroxiredoxin 1 and its clinical implications in liver cancer. World J Gastroenterol. 2015;21(38):10840–10852. doi: 10.3748/wjg.v21.i38.10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Leary PC, Terrile M, Bajor M, et al. Peroxiredoxin-1 protects estrogen receptor α from oxidative stress-induced suppression and is a protein biomarker of favorable prognosis in breast cancer. Breast Cancer Res. 2014;16(4):R79. doi: 10.1186/bcr3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH, Bogner PN, Ramnath N, Park Y, Yu J, Park YM. Elevated peroxiredoxin 1, but not NF-E2-related factor 2, is an independent prognostic factor for disease recurrence and reduced survival in stage I non-small cell lung cancer. Clin Cancer Res. 2007;13(13):3875–3882. doi: 10.1158/1078-0432.CCR-06-2893. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Bogner PN, Baek SH, et al. Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clin Cancer Res. 2008;14(8):2326–2333. doi: 10.1158/1078-0432.CCR-07-4457. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Wang R, Zhang M, Wang L, Cheng S. Proteomic analysis of non-small cell lung cancer tissue interstitial fluids. World J Surg Oncol. 2013;11(1):173. doi: 10.1186/1477-7819-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ha B, Kim EK, Kim JH, et al. Human peroxiredoxin 1 modulates TGF-β1-induced epithelial-mesenchymal transition through its peroxidase activity. Biochem Biophys Res Commun. 2012;421(1):33–37. doi: 10.1016/j.bbrc.2012.03.103. [DOI] [PubMed] [Google Scholar]
- 29.Lee W, Choi KS, Riddell J, et al. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J Biol Chem. 2007;282(30):22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 30.Lu W, Fu Z, Wang H, Feng J, Wei J, Guo J. Peroxiredoxin 2 is upregulated in colorectal cancer and contributes to colorectal cancer cells’ survival by protecting cells from oxidative stress. Mol Cell Biochem. 2014;387(1–2):261–270. doi: 10.1007/s11010-013-1891-4. [DOI] [PubMed] [Google Scholar]
- 31.Lu W, Fu Z, Wang H, Feng J, Wei J, Guo J. Peroxiredoxin 2 knockdown by RNA interference inhibits the growth of colorectal cancer cells by downregulating Wnt/β-catenin signaling. Cancer Lett. 2014;343(2):190–199. doi: 10.1016/j.canlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Peng L, Wang R, Shang J, Xiong Y, Fu Z. Peroxiredoxin 2 is associated with colorectal cancer progression and poor survival of patients. Oncotarget. 2017;8(9):15057–15070. doi: 10.18632/oncotarget.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JH, Kim YS, Lee HL, et al. Expression of peroxiredoxin and thioredoxin in human lung cancer and paired normal lung. Respirology. 2006;11(3):269–275. doi: 10.1111/j.1440-1843.2006.00849.x. [DOI] [PubMed] [Google Scholar]
- 34.Wei Q, Jiang H, Xiao Z, et al. Sulfiredoxin-Peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proc Natl Acad Sci U S A. 2011;108(17):7004–7009. doi: 10.1073/pnas.1013012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinnula VL, Lehtonen S, Sormunen R, et al. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196(3):316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 36.Karihtala P, Mäntyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9(9):3418–3424. [PubMed] [Google Scholar]
- 37.Karihtala P, Mäntyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9(9):3418–3424. [PubMed] [Google Scholar]
- 38.Kropotov A, Gogvadze V, Shupliakov O, et al. Peroxiredoxin V is essential for protection against apoptosis in human lung carcinoma cells. Exp Cell Res. 2006;312(15):2806–2815. doi: 10.1016/j.yexcr.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Chang XZ, Li DQ, Hou YF, et al. Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res. 2007;9(6):R76. doi: 10.1186/bcr1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schremmer B, Manevich Y, Feinstein SI, Fisher AB. Peroxiredoxins in the lung with emphasis on peroxiredoxin VI. Subcell Biochem. 2007;44:317–344. doi: 10.1007/978-1-4020-6051-9_15. [DOI] [PubMed] [Google Scholar]
- 41.Lee SB, Ho JN, Yoon SH, Kang GY, Hwang SG, Um HD. Peroxiredoxin 6 promotes lung cancer cell invasion by inducing urokinase-type plasminogen activator via p38 kinase, phosphoinositide 3-kinase, and Akt. Mol Cells. 2009;28(6):583–588. doi: 10.1007/s10059-009-0152-6. [DOI] [PubMed] [Google Scholar]
- 42.Yun HM, Park KR, Park MH, et al. PRDX6 promotes tumor development via the JAK2/STAT3 pathway in a urethane-induced lung tumor model. Free Radic Biol Med. 2015;80:136–144. doi: 10.1016/j.freeradbiomed.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 43.Yun HM, Park KR, Lee HP, et al. PRDX6 promotes lung tumor progression via its GPx and iPLA2 activities. Free Radic Biol Med. 2014;69:367–376. doi: 10.1016/j.freeradbiomed.2014.02.001. [DOI] [PubMed] [Google Scholar]