Abstract
Background
Helicobacter pylori (H. pylori) infection has been regarded as a main risk factor for gastric cancer. Nod-like receptor pyrin domain-containing protein 6 (NLRP6), a component of inflammasome, has been linked to colorectal tumorigenesis. Here, we aimed to evaluate NLRP6 expression profile and functions in gastric cancer.
Materials and methods
We examined NLRP6 expression in gastric cancer and adjacent normal gastric tissues. The biological functions and mechanism of NLRP6 overexpression in gastric cancer cells were investigated.
Results
Downregulated NLRP6 expression in human gastric cancer significantly correlated with H. pylori infection, tumor size, TNM stage, lymph node metastasis, and overall survival. NLRP6 overexpression in gastric cancer cells led to a significant decrease in cell proliferation, migration, and invasion, as well as a notable increase in cell apoptosis, whereas NLRP6 knockdown had opposing effects. In addition, NLRP6 overexpression significantly repressed STAT3 phosphorylation and the transcription of its target genes, Bcl-2 and MMP-2. Moreover, forkhead box O3 (FOXO3), a transcription factor regulated by H. pylori, was demonstrated as an upstream regulator of NLRP6 transcription.
Conclusion
Our study may provide insight into the understanding of NLRP6 as a tumor suppressor and implicate the potential application of NLRP6 for gastric cancer treatment.
Keywords: NLRP6, H. pylori, cell proliferation, cell apoptosis, gastric cancer
Introduction
Gastric cancer is one of the most prevalent cancer1 and the third leading cause of cancer death.2 Currently, Helicobacter pylori (H. pylori) infection, living environment, diet, genetic and immune factors, and chronic gastritis are defined as main risk factors for gastric cancer.3 In recent years, remarkable progresses have been made in surgery, chemotherapy, and radiotherapy for gastric cancer. However, the 5-year survival rate for gastric cancer remains less than 25%.4 Therefore, more research is needed to identify more sensitive diagnosis and prognosis markers and to investigate molecular mechanisms of tumorigenesis.
Nod-like receptor pyrin domain-containing protein 6 (NLRP6), belonging to nod-like receptor (NLR) family of pattern recognition receptors, consists of a N-terminal pyrin domain (PYD), a central NOD domain, and C-terminal leucine-rich repeats (LRRs).5 NLRP6 is known as a component of inflammasome and participates in inflammasome signaling.6 NLRP6 plays critical roles in innate immunity and host defense. It negatively regulates MAPK and the canonical NF-κB pathway signaling and impedes clearance of intracellular bacterial pathogens Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli.7 Moreover, experimental evidence has linked NLR6 to tumorigenesis. NLRP6 exhibits enhanced levels in colorectal cancer tissues and has been proposed as a diagnostic marker for this malignancy.8 Mice lacking NLRP6 are highly susceptible to chemically induced colitis and colitis-associated tumorigenesis, accompanied with reduced serum IL-18 level and increased expression of Wnt-target genes.5,9–11 However, the expression pattern and biological functions of NLRP6 in gastric cancer remain poorly understood.
In the current study, we identified downregulated NLRP6 expression in gastric cancer. The protein expression of NLRP6 was associated with malignant tumor size, TNM stage, lymph node metastasis, and overall survival. We assessed NLRP6 function by overexpression and RNA interference, which suggested that NLRP6 played a critical role in the proliferation, apoptosis, migration, and invasion of gastric cancer cells. Further study identified forkhead box O3 (FOXO3) as a H. pylori-regulated transcription factor for NLRP6.
Materials and methods
Tissue samples
A total of 120 gastric cancer patients undergoing surgery at Department of Gastroenterology, Shanghai Sixth People’s Hospital (South), were enrolled in this study. All patients gave written informed consent. The study was approved by the independent ethics committee of Shanghai Sixth People’s Hospital (South, KY-201501). The tumor stage was determined according to the TNM classification. For RNA or protein extraction, 45 pairs of tumor tissue samples and adjacent nontumorous samples were obtained, frozen in liquid nitrogen immediately after surgical resection and stored at –80°C. For immunohistochemistry (IHC) analysis, formalin-fixed, paraffin-embedded gastric cancer specimens were collected from all patients. Tissue collection was conducted in accordance with the Declaration of Helsinki.
Quantitative reverse transcription (qRT)-PCR
Total RNA was extracted from normal/tumor paired tissues and cell lines using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Reverse transcription (RT) reaction was performed with ProSTAR First-Strand RT-PCR kit (Stratagene, La Jolla, CA, USA). cDNA was used as a template for qRT-PCR with SYBR Green PCR kit (Thermo Fisher Scientific) on ABI 7300 (Thermo Fisher Scientific) thermal cycler. The specific primers were designed with Primer 5 software (Premier Biosoft, Palo Alto, CA, USA; Table 1). The optimized PCR conditions were as follows: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 45 seconds. The relative gene expression was normalized to GAPDH expression. All data represent the average of three replicates.
Table 1.
Primers for qRT-PCR
| Gene | Forward | Reverse |
|---|---|---|
| NLRP6 | 5′-CAGTACCTGGTGGGTATG-3′ | 5′-CCTGAAGCTCCTGTAGTG-3′ |
| Bcl-2 | 5′-GGTGGGGTCATGTGTGTGG-3′ | 5′-CGGTTCAGGTACTCAGTCATCC-3′ |
| MMP-2 | 5′-TACAGGATCATTGGCTACACACC-3′ | 5′-GGTCACATCGCTCCAGACT-3′ |
| GAPDH | 5′-CACCCACTCCTCCACCTTTG-3′ | 5′-CCACCACCCTGTTGCTGTAG-3′ |
Abbreviation: qRT, quantitative reverse transcription.
Western blot analysis
To determine protein expression levels, normal/tumor paired tissues and cell lines were lysed in precooled RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) with freshly added protease inhibitor cocktail (Hoffman-La Roche Ltd., Basel, Switzerland). Protein concentration was quantified by bicinchoninic acid assay kit (Thermo Fisher Scientific). Same amount of protein was separated by SDS-PAGE and subjected to Western blot analysis as previously described.12 Antibodies against STAT3, phospho-STAT3, AKT, phosphor-AKT, phosphor-FOXO3, and GAPDH were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against NLRP6 and MMP-2 were from Abcam (Cambridge, MA, USA). Antibodies against Bcl-2 were from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were from Beyotime Biotechnology. The experiments were repeated independently at least three times.
IHC analysis
IHC staining was performed as previously described.9 Briefly, 5-μm-thick sections were deparaffinized and rehydrated. Antigen retrieval was performed by heating the slides in citrate buffer (pH 6.0) for 20 minutes. To block endogenous peroxidase activity, the slides were soaked in 3% hydrogen peroxide for 10 minutes. The slides were incubated with the polyclonal rabbit anti-NLRP6 (Novus Biologicals, Littleton, CO, USA; dilution: 1/50) for 1 hour and then with HRP-labeled secondary antibody for 1 hour. Sections were developed with 3,3′-diaminobenzidine substrate and counterstained with hematoxylin. The expression of NLRP6 was reviewed blindly by two pathologists independently. The specimens were graded into NLRP6 lower expression group and NLRP6 higher expression group using 20% of tumor cells positively stained as a cutoff.
Cell culture, transfection, lentiviral infection, and RNA interference
Seven cell lines derived from human gastric cancer, MKN-28, BGC-823, AGS, MGC-803, HGC-27, SGC-7901 and MKN-45, were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China) and maintained in RPMI 1640 medium (HyClone, Logan, UT, USA) supplemented with 10% FBS and antibiotics at 37°C in a humidified incubator with 5% CO2. All the cell lines were authenticated by DNA fingerprinting analysis in 2015 and passaged in our laboratory for less than 6 months. Cell lines were tested routinely to ensure the absence of mycoplasma contamination.13
Lentiviral constructs of GV348 empty vector (GeneChem, Shanghai, China) or GV348-NLRP6-Flag were cotransfected with the helper virus packaging systems into 293 T cells with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacture’s instruction. At 48 hours posttransfection, viral supernatant was harvested and filtered through 0.45 μm filter. BGC-823 and HGC-27 cells were infected with NLRP6 lentivirus or control vector len-tivirus. Stable cells were obtained in the presence of 0.5 μg/mL puromycin (Sigma-Aldrich Co., St Louis, MO, USA).
NLRP6 siRNA (siNLRP6, GUGUCCGAGUACAAGAAGA) and negative control siRNA (siNC) were synthesized by GenePharma Co., Ltd (Shanghai, China). FOXO3 siRNA (siFOXO3; sc-37887) was obtained from Santa Cruz Biotechnology.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) assay was performed by standard methods. Briefly, 3×103 cells per well were seeded onto 96-well plates and treated as indicated. At indicated time point, CCK-8 solution was added to each well and incubated for 1 hour. Absorbance at wavelength 450 nm was measured using a microplate reader. All conditions were tested in three replicates.
Evaluation of cell apoptosis by flow cytometry
The percentage of cells in early and late apoptosis was determined by Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining. Cells were harvested at 48 hours after treatment and double labeled with Annexin V-FITC and PI apoptosis detection kits (eBioscience, San Diego, CA, USA). Cell apoptosis was analyzed using a FACScan flow cytometry (BD Biosciences, San Jose, CA, USA). Early apoptosis and late apoptosis were defined by Annexin V+/PI– staining and Annexin V+/PI+ staining, respectively. The experiments were performed in triplicate.
In vivo tumorigenicity assay
Animal experiments were approved by Animal Care and Use Committee of Shanghai Sixth People’s Hospital (South) and performed in accordance with the principles and procedures outlined in the National Institute of Health Guide. Four-to five-week-old BALB/c nude mice were provided by SLAC Animal Co Ltd. (Shanghai, China), housed under specific pathogen-free conditions. BGC-823 stable cells (2×106) were subcutaneously injected into the flank of each mouse. Tumor volume (mm3) was estimated using the following formula: 0.5×(the shortest diameter)2×(the longest diameter) every 3 days. At 27 days after cell inoculation, the mice were sacrificed and the tumors were collected. Tumor xenografts were subjected to qRT-PCR analysis, TUNEL assay (Hoffman-La Roche Ltd), and IHC staining with anti-Ki-67 (Abcam).
Transwell assays
To evaluate the cell migration and invasive capacity of cells, Transwell assays were performed using Matrigel noncoated and Matrigel-coated Boyden chamber (BD Biosciences), respectively. Stable cells were serum-starved overnight, harvested, and resuspended in the serum-free medium. Cells (1×105) were then plated to the upper chamber. The medium containing 10% FBS was added to the lower chamber. After incubation at 37°C for 24 hours, the migrant and invasion cells attached to the lower surface of the membrane were fixed with formalin, stained with 0.5% crystal violet, and counted using a microscope. The experiments were performed in triplicate.
Luciferase reporter assay for the activity of Bcl-2, MMP-2, and NLRP6 promoter
The full length of Bcl-2 promoter,14 MMP-2 promoter,15 or NLRP6 promoter16 was inserted into pGL3 luciferase vector (Promega Corporation, Fitchburg, WI, USA) as previously described. Cells were transfected with pGL3-Bcl-2, pGLS-MMP-2, or pGLS-NLRP6. At 48 hours posttransfection, luciferase activity was determined using a luciferase assay kit (Promega Corporation), normalizing to protein concentration and then to a control sample transfected with pGL3.
H. pylori infection and LY294002 treatment
The CagA-positive/VacA-positive H. pylori strain (NCTC11637) was cultured and collected as previously described.17 BGC-823 and HGC-27 cells were infected with H. pylori at a multiplicity of infection (MOI) of 0, 25, or 100. Cells were pretreated with PI3K inhibitor LY294002 (Calbiochem, San Diego, CA, USA) as indicated.
Statistical analyses
GraphPad Prism software Version 6.0 (San Diego, CA, USA) was used for statistical analysis. All data were presented as the mean±SD. Student’s t-test and ANOVA test followed by Tukey’s test were performed to assess the statistical significance between two groups and among more than two groups, respectively. Chi-square test was done to determine the relationship between NLRP6 protein expression and clinicopathological features. Overall survival time was analyzed by the Kaplan–Meier survival curves and log-rank test. P<0.05 was considered as statistically significant.
Results
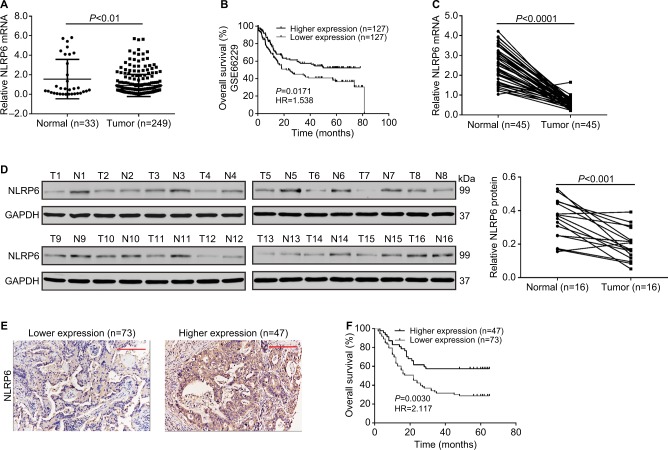
Downregulated expression of NLRP6 in gastric cancer tissues
We analyzed the expression levels of NLR family proteins in gastric cancer tissues, as well as the association between their expression and the overall survival of gastric cancer patients using The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/tcga/) and Gene Expression Omnibus database. The results showed that NLRP6 expression was downregulated in gastric cancer tissues (Figure 1A), and its expression was related with the overall survival time of patients (Figure 1B). To elucidate whether NLRP6 is involved in gastric cancer progression, we performed gene set enrichment analysis (GSEA) on TCGA data set. Enrichment plots of GSEA showed that the genes of cell apoptosis and metastasis down pathways were more correlated with patients with NLRP6 lower expression than those with NLRP6 higher expression (Figure S1), suggesting that NLRP6 may be a key regulator during gastric tumorigenesis.
Figure 1.
NLRP6 is downregulated in gastric cancer tissues.
Notes: (A) NLRP6 expression was significantly decreased in gastric cancer tissues compared with that in normal gastric tissues based on the data from the TCGA data set (P<0.01). (B) Survival analysis of 254 patients from the GSE 66229 data set. The survival time in patients with higher NLRP6 expression patients was significantly longer than that in patients with lower NLRP6 expression. (C) qRT-PCR analysis of NLRP6 expression in 45 pairs of gastric cancer and its corresponding normal tissues (P<0.0001). (D) Western blot analysis of NLRP6 protein expression in 16 pairs of gastric cancer (T1–T16) and its corresponding normal tissues (N1–N16). (E) NLRP6 protein expression was assessed by IHC staining in gastric cancer tissues. Scale bar: 200 µm. (F) Survival analysis of 120 patients with gastric cancer.
Abbreviations: TCGA, The Cancer Genome Atlas; qRT, quantitative reverse transcription; IHC, immunohistochemistry.
We then analyzed NLRP6 mRNA expression by qRT-PCR and showed that NLRP6 mRNA levels in tumor tissues decreased to ~76.9% (0.582±0.04) compared with adjacent normal gastric tissues (2.52±0.13, n=45; Figure 1C). Our data were in line with the public-available data. We also examined the translational expression of NLRP6 in tissues from control and gastric cancer tissues by Western blot analysis and showed that NLRP6 protein levels were markedly reduced in tumor tissues (Figure 1D).
Decreased NLRP6 expression is associated with poor prognosis in gastric cancer patients
IHC staining was then performed and showed that 60.8% (73/120) gastric cancer tissues displayed negative or lower expression of NLRP6 (less than 20% of tumor cells were positively stained) (Figure 1E). Associations between NLRP6 expression and clinicopathologic characteristics were analyzed by chi-squared test. As shown in Table 2, there was a significant correlation between NLRP6 levels and H. pylori infection (P=0.0135), tumor size (P=0.0014), TNM stage (P=0.0179), and lymph node metastasis (P=0.0224). Furthermore, Kaplan–Meier analysis of our patient data (Figure 1F) indicated that the overall survival time of patients with a lower expression of NLRP6 was significantly shorter than that of patients with a higher expression of NLRP6, suggesting that decreased NLRP6 expression may contribute to poor survival.
Table 2.
Correlation of NLRP6 expression with patients’ features in gastric cancer
| Variables | All cases | NLRP6 protein | ||
|---|---|---|---|---|
| Low | High | P-value | ||
| Age at surgery, years | ||||
| <55 | 35 | 18 | 17 | 0.2180 |
| ≥55 | 85 | 55 | 30 | |
| Gender | ||||
| Male | 87 | 50 | 37 | 0.2954 |
| Female | 33 | 23 | 10 | |
| Helicobacter pylori infection | ||||
| Negative | 69 | 35 | 34 | 0.0135* |
| Positive | 51 | 38 | 13 | |
| Tumor location | ||||
| Upper half | 43 | 27 | 16 | 0.7085 |
| Lower half | 56 | 32 | 24 | |
| Whole | 21 | 14 | 7 | |
| TNM | ||||
| I+II | 48 | 23 | 25 | 0.0179* |
| III+IV | 72 | 50 | 22 | |
| Tumor size, cm | ||||
| <5 | 52 | 23 | 29 | 0.0014** |
| ≥5 | 68 | 50 | 18 | |
| Lymph node metastasis | ||||
| Absent | 50 | 24 | 26 | 0.0224* |
| Present | 70 | 49 | 21 | |
Note:
P<0.05,
P<0.01.
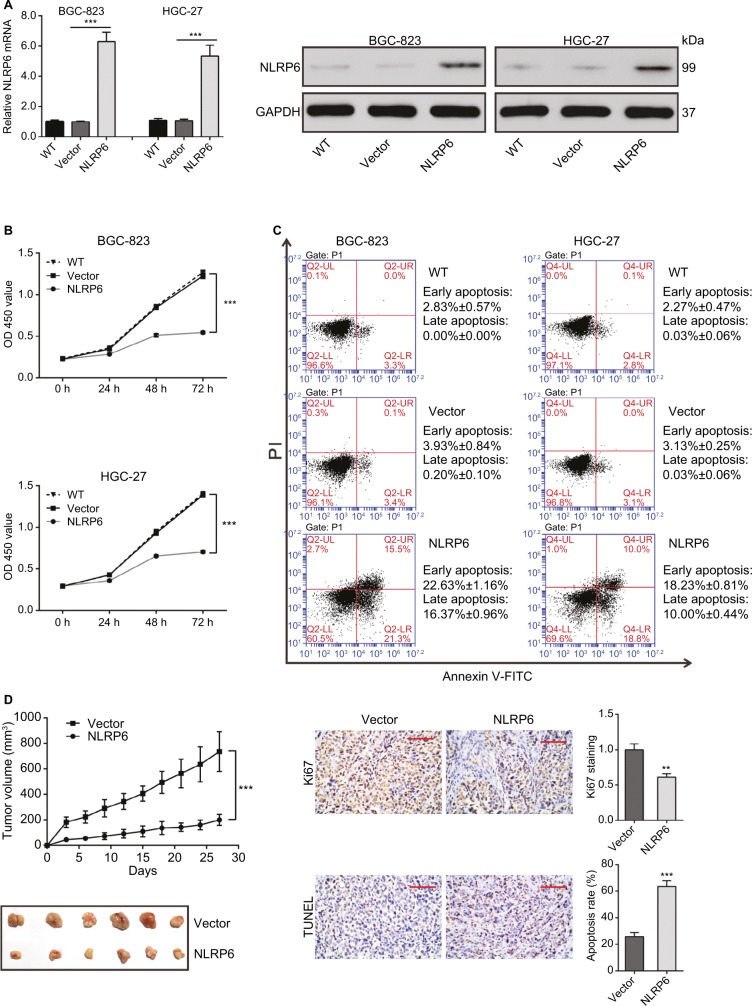
NLRP6 overexpression inhibits gastric cancer cell proliferation in vitro and in vivo
To explore the role of NLRP6 in gastric tumorigenesis, we stably overexpressed NLRP6 in BGC-823 and HGC-27 cells, which had relative lower protein and mRNA levels of NLRP6 (Figure S2). For both cell lines, NLRP6 expression in cells transduced with NLRP6 overexpressing virus was significantly elevated compared to that in wild-type (WT) cells or cells with control vector virus (Figure 2A). The proliferation of BGC-823 and HGC-27 cells overexpressing NLRP6 was significantly suppressed at 24, 48, and 72 hours compared to that of control cells as measured by the CCK-8 assay (Figure 2B). Comparable expression levels of NLRP6 and cell proliferation rates were observed in the WT and vector cells. We then examined whether the proliferation inhibition is associated with cell apoptosis by flow cytometry analysis. Ectopic expression of NLRP6 markedly increased the early and late apoptotic ratios of cells compared to cells transducted with control vector virus (Figure 2C). We also knockdown NLRP6 expression in AGS (Figure S3A), a cell line with a higher NLRP6 expression (Figure S2). Complementary to the results from NLRP6 overexpression, NLRP6 knockdown significantly promoted cell proliferation (Figure S3B) and inhibited cell apoptosis (Figure S3C).
Figure 2.
NLRP6 inhibits cell proliferation of gastric cancer cells.
Notes: (A) Overexpression of NLRP6 in BGC-823 and HGC-27 cells was confirmed by Western blot. Blots are representative of three separate experiments. (B) Cell proliferation was detected at 0, 24, 48, and 72 hours in BGC-823 and HGC-27 stable cells by CCK-8 assay (n=3). (C) Annexin V/PI assay showed that NLRP6 overexpression increased cell early and late apoptosis rates (n=3). (D) NLRP6 inhibits gastric cancer cell proliferation in vivo. BGC-823 cells stably expressed NLRP6 or control vector were subcutaneously inoculated into nude mice (six per group). Tumor volume was measured for 27 days. NLRP6-overexpressed BGC-823 cells grew much slower in nude mice than the control cells (left panels). At 27 days, mice were sacrificed and the tumors were isolated. IHC staining of Ki-67 and TUNEL assay were performed on the xenograft tumors (n=3, right panels). WT, wild-type cells; vector, cells stably expressed control vector; NLR6, cells stably expressed NLRP6. **P<0.01 and ***P<0.001.
Abbreviations: CCK-8, Cell Counting Kit-8; PI, propidium iodide; IHC, immunohistochemistry; FITC, fluorescein isothiocyanate.
Next, we investigated the effect of NLRP6 on tumor growth in vivo. Tumor formation was examined after inoculation of BGC-823 cells stably expressed NLRP6 or control vector into the flank of nude mice. Tumor growth was much slower in mice with NLRP6 overexpressed cells compared to that in mice with cells expressing control vector (Figure 2D). Moreover, Ki-67-positive cells were significantly reduced, whereas apoptotic cells were notably increased in NLRP6 overexpression xenografts. These data indicated that NLRP6 may be a potential regulator of tumor growth in gastric cancer.
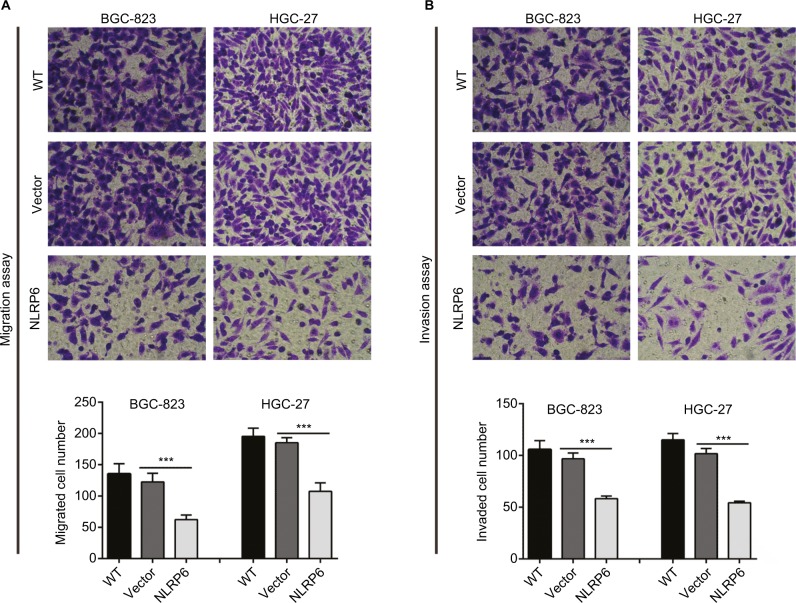
NLRP6 overexpression inhibits the migration and invasion of gastric cancer cells
To determine whether NLRP6 affects migration and invasion of gastric cancer cells, transwell assay was performed. NLRP6 overexpression caused a significant reduction in cell migration and invasion compared to control cells (Figure 3A and B). Decreased migration and invasion of AGS cells were observed in AGS cells with NLRP6 knockdown (Figure S3D).
Figure 3.
NLRP6 overexpression inhibits the migration and invasion of gastric cancer cells.
Notes: Transwell assays were performed to evaluate cell migration (A) and invasive abilities (B) (n=3). For invasion, the upper chamber was precoated with Matrigel. WT, wild-type cells; vector, cells stably expressed control vector; NLR6, cells stably expressed NLRP6. ***P<0.001.
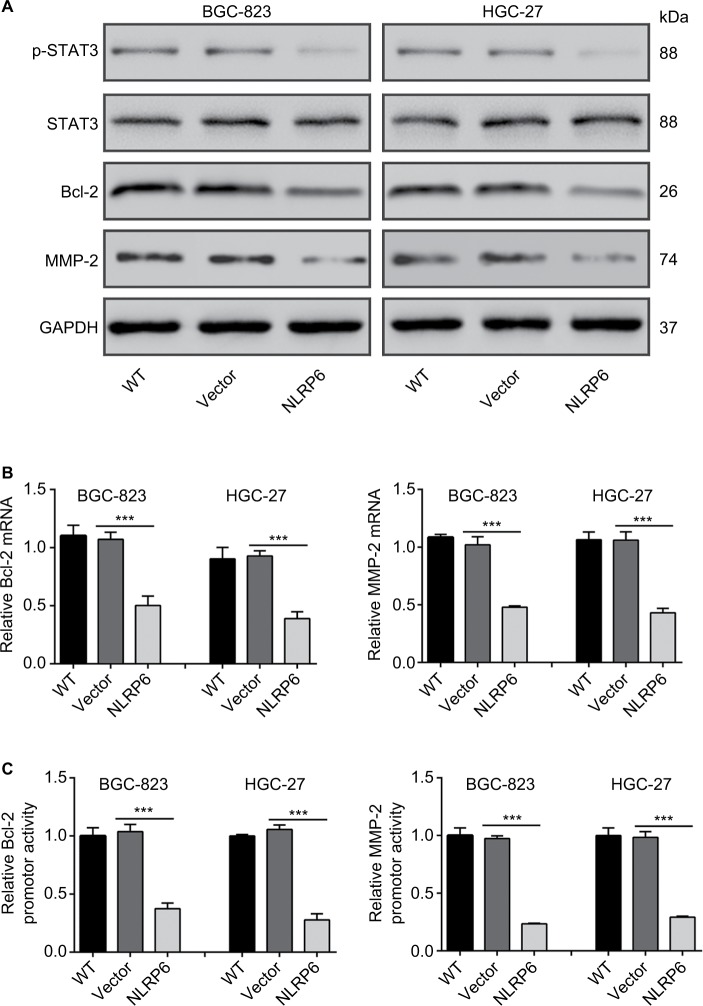
Effects of NLRP6 on STAT3 signaling
Constitutively activated STAT3 is observed in various human cancers.18 It is involved in cell survival, proliferation, and invasion of gastric cancer cell lines.19,20 Then, we tried to explore the effects of NLRP6 on STAT3 signaling. As shown in Figure 4A, NLRP6 overexpression in BGC-823 and HGC-27 cells significantly repressed the phosphoryla tion of STAT3. On the contrary, NLRP6 knockdown in AGS cells remarkably increased STAT3 phosphorylation (Figure S4). The protein levels of Bcl-221 and MMP-2,22,23 two well-known target genes of STAT3, were also reduced by ectopic expression of NLRP6, which was consistent with the results of the biological function analyses.
Figure 4.
Effects of NLRP6 on STAT3 signaling.
Notes: (A) Immunoblot of phosphorylated STAT3, STAT3, Bcl-2, and MMP-2. Blots are representative of three separate experiments. (B) mRNA levels of Bcl-2 and MMP-2 were assessed by qRT-PCR. (C) BGC-823 and HGC-27 cells were transfected with a Bcl-2 or an MMP-2 luciferase reporter plasmid. The cells were then cultured for 48 hours before determination of normalized luciferase activity. WT, wild-type cells; vector, cells stably expressed control vector; NLR6, cells stably expressed NLRP6. ***P<0.001.
Abbreviation: qRT, quantitative reverse transcription.
Furthermore, NLRP6 overexpression resulted in a significant downregulation of the amount of mRNA (Figure 4B) and the promoter activity (Figure 4C) of both Bcl-2 and MMP-2 genes. These data suggested that NLRP6 overexpression in gastric cancer cell lines reduced the STAT3 activity and repressed the transcription of its target gene.
H. pylori reduces NLRP6 expression
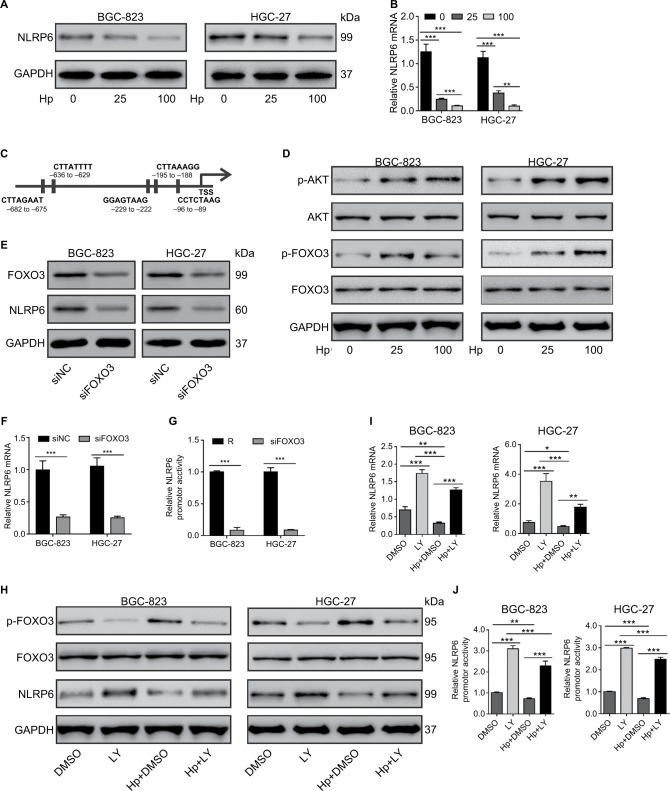
Considering that NLRP6 levels were strongly correlated with H. pylori infection (P=0.0135), we then determined whether NLRP6 expression was modulated by H. pylori infection. As illustrated in Figure 5A and B, H. pylori infection significantly decreased the protein and mRNA levels of NLRP6 in both BC-823 and HGC-27 cells at an MOI of 25 and 100.
Figure 5.
Effect of H. pylori on NLRP6 expression.
Notes: (A and B) BGC-823 and HGC-27 cells were infected with H. pylori at an MOI of 0, 25, or 100 for 24 hours. The protein (A) and mRNA (B) levels of NLRP6 were detected. (C) The potential binding sites in the promoter of the NLRP6 genes for FOXO3 were shown based on the online program ALGGEN – PROMO. (D) The levels of p-AKT, AKT, p-FOXO3, and FOXO3 in gastric cancer cells with H. pylori infection were determined by Western blot. (E–G) BGC-823 and HGC-27 cells were transfected with siFOXO3 and siNC for 48 hours. Western blot (E), real-time PCR (F), and luciferase reporter assays (H) were performed. (H–J) Gastric cancer cells were treated with 20 µM of LY294002 of DMSO followed by infection with H. pylori (MOI of 100) for 24 hours. Western blot (H), real-time PCR (I), and luciferase reporter assays (J) were performed. *P<0.05, **P<0.01, and ***P<0.001.
Abbreviations: H. pylori, Helicobacter pylori; TSS, transcription start site; siFOXO3, FOXO3 siRNA; siNC, negative control siRNA; DMSO, dimethyl sulfoxide; MOI, multiplicity of infection; LY, LY294002.
Role of AKT/FOXO3 on H. pylori-inhibited NLRP6 expression
We then tried to investigate which transcription factors were involved in H. pylori-mediated NLRP6 expression. A previ ous study has shown that H. pylori infection in gastric cells induces AKT activation, which phosphorylates and inactivates FOXO3,24 a forkhead transcription factor and potential tumor suppressor.25–28 By utilizing the online program ALG-GEN – PROMO (based on the 8.3 version of TRANSFAC),29 we found that FOXO3 was a potential transcription factor for NLRP6 gene (Figure 5C). As expected, H. pylori infection in both BC-823 and HGC-27 cells for 1 hour significantly induced the phosphorylation of AKT and FOXO3 but had no obvious effects on the total protein of AKT and FOXO3 (Figure 5D).
To further verify whether FOXO3 regulated the NLRP6 gene expression, gastric cancer cells were transfected with FOXO3 siRNA. FOXO3 knockdown led to a significant downregulation in the protein expression (Figure 5E), mRNA expression (Figure 5F), and promoter activity (Figure 5G) of NLRP6. Furthermore, a PI3K inhibitor LY294002 was applied to gastric cancer cells exposed to H. pylori. As illustrated in Figure 5H, LY294002 treatment in gastric cancer cells remarkably suppressed the effects of H. pylori infection on the phosphorylation of FOXO3 and the protein levels of NLRP6. Furthermore, H. pylori led to a significant downregulation of the amount of mRNA (Figure 5I) and the promoter activity (Figure 5J) of NLRP6 gene, while LY294002 treatment displayed reversed effects. LY294002 treatment can rescue the effects of H. pylori infection. These data suggested that downregulation of NLRP6 expression in gastric cancers may be associated with H. pylori/AKT/FOXO3 signaling.
Discussion
NLRP6 has been proposed as a diagnostic marker for colorectal cancer.8 NLRP6-deficient mice are highly susceptible to experimental colitis and colitis-induced tumorigenesis.5,9,10 Here, we provided the first evidence of NLRP6 expression in gastric cancer, showing that NLRP6 was significantly downregulated in gastric cancer tissues compared to that in adjacent noncancerous tissues. In addition, a lower NLRP6 expression in gastric cancer tissues was associated with H. pylori infection, higher TNM grade, larger tumor size, higher incidence of lymph node metastasis, and poorer overall survival of patients (Tables 2 and S1), although multivariate analysis did not reveal NLRP6 expression to be an independent prognostic factor (Table S2). These data suggest that NLRP6 may be a novel prognostic marker for gastric cancer.
GSEA on TCGA data set showed that cell apoptosis and metastasis down pathways were negatively correlated with NLRP6 expression in gastric cancer patients, which was further validated by a series of functional experiments. The ectopic overexpression of NLRP6 in gastric cancer cells significantly suppressed cell growth, cell migration, and cell invasion but enhanced cell apoptosis, whereas siRNA-mediated knockdown displayed the reverse effects. The proliferation-suppressing and apoptosis-promoting effects of NLRP6 were also observed in xenograft tumors. These findings indicate that NLRP6 may work as a tumor suppressor in gastric cancer.
STAT3 is activated by growth factors and cytokines via the Janus kinase (JAK). Upon activation, it dimerizes and translocates to the nucleus to activate the transcription of target genes, such as Bcl-2 and MMP-2.18,21–23 Multiple lines of evidence suggest a crucial role for STAT3 in the development and progression of various human cancers,18 including gastric cancer.19,30 STAT3 expression in gastric cancer indicates a poor prognosis.30 Constitutively, STAT3 signaling supports gastric cancer cell survival.19 Suppression of Bcl-2 may contribute to cell apoptosis in gastric carcinoma31 and gastric cancer cells.32 MMP-2, which degrades the extracellular matrix, has been related to lymph node metastasis of gastric cancer.33,34 Here, NLRP6 overexpression in gastric cancer cells significantly repressed the phosphorylation of STAT3 as well as the transcription of Bcl-2 and MMP-2, suggesting that the apoptosis-promoting, anti-migration, and anti-invasion effects of NLRP6 were mediated by STAT3 signaling.
H. pylori-induced gastritis is a main risk factor for gastric cancer.3 H. pylori activates the PI3K/AKT signaling pathway, which activates β-catenin signaling and upregulates the transcriptional activation of target genes that affect carcinogenesis.35 Here, we found that NLRP6 levels were strongly correlated with H. pylori infection in patients with gastric cancer. H. pylori infection significantly decreased the levels of NLRP6 in gastric cancer cells. Furthermore, we tried to investigate the transcription factor for NLRP6. FOXO3 was phosphorylated by H. pylori infection in a PI3K/AKT-dependent manner in gastric cells as previously reported.24 Knockdown of FOXO3 causes a significant decrease in the protein expression, mRNA expression, and promoter activity of NLRP6. H. pylori infection had the same effects as FOXO3 knockdown, and such inhibitory effects were also PI3K/AKT dependent. Overall, these results inferred that H. pylori infection downregulated the transcription of NLRP6 via AKT/FOXO3 signaling.
Collectively, our study suggests that NLRP6 acts as a tumor suppressor in gastric cancer, and the downregulation of NLRP6 expression is closely associated with patients’ poor prognosis. We provided key evidence that NLRP6 suppresses the proliferation, migration, and invasion of gastric cancer cells through regulating STAT3 signaling. H. pylori regulated the expression of NLRP6 via AKT/FOXO3 signaling.
Supplementary materials
NLRP6-associated pathways in gastric cancer.
Notes: GSEA was performed on TCGA data set by using GSEA version 2.0 from the Broad Institute at MIT as previously described.1 Gene set permutations were performed 1,000 times, and the pathway set list was sorted by the NES. GSEA was performed using TCGA data set. The cell apoptosis (A) and metastasis (B) pathways were strongly associated with NLRP6 lower expression.
Abbreviations: GSEA, gene set enrichment analysis; TCGA, The Cancer Genome Atlas; NES, normalized enrichment score; ES, enrichment score.
NLRP6 protein expression and mRNA expression in seven gastric cancer cell lines were determined by qRT-PCR (A) and Western blot (B) analysis, respectively.
Abbreviation: qRT, quantitative reverse transcription.
NLRP6 knockdown in AGS cells promoted cell proliferation, migration, and invasion but inhibited cell apoptosis. (A) qRT-PCR (upper panel) and Western blot (lower panel) analysis of knockdown efficiency in AGS cells. (B) CCK-8 assay showed that NLRP6 knockdown promoted cell proliferation. (C) Annexin V/PI assay showed that NLRP6 knockdown decreased cell early and late apoptosis. (D) Transwell assays showed that NLRP6 knockdown enhanced migration and invasive capacity. WT, wild-type cells. **P<0.01 and ***P<0.001.
Abbreviations: qRT, quantitative reverse transcription; CCK-8, Cell Counting Kit-8; PI, propidium iodide; siNC, negative control siRNA.
Effects of NLRP6 knockdown on STAT3 phosphorylation.
Notes: Blots are representative of three separate experiments. WT, wild-type cells; siNC, negative control siRNA.
Table S1.
Univariate Cox regression of prognostic parameters for survival in patients with gastric cancer
| Prognostic parameter | Univariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P-value | |
| Expression of NLRP6 (low vs. high) | 0.470 | 0.280–0.788 | 0.004** |
| Helicobacter pylori infection (negative vs. positive) | 0.221 | 0.135–0.362 | <0.001 |
| TNM (I+II vs. III+IV) | 4.457 | 2.473–8.034 | <0.001 |
| Tumor size (<5 cm vs. ³5 cm) | 4.275 | 2.486–7.352 | <0.001 |
| Lymph node metastasis (absent vs. present) | 22.304 | 9.414–52.846 | <0.001 |
Note:
P<0.01.
Table S2.
Multivariate Cox regression of prognostic parameters for survival in patients with gastric cancer
| Prognostic parameter | Multivariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P-value | |
| Expression of NLRP6 (low vs. high) | 0.825 | 0.483–1.409 | 0.482 |
| Helicobacter pylori infection (negative vs. positive) | 0.518 | 0.306–0.876 | 0.014* |
| TNM (I+II vs. III+IV) | 2.383 | 1.249–4.549 | 0.008** |
| Tumor size (<5 cm vs. ≥5 cm) | 2.689 | 1.492–4.848 | 0.001** |
| Lymph node metastasis (absent vs. present) | 13.465 | 5.398–33.589 | 0.000*** |
Note:
P<0.05,
P<0.01, and
P<0.001.
Reference
- 1.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This study was supported by the Natural Science Foundation of Shanghai (16ZR1429100).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cancer IAfRo . World Cancer Report. Geneva: WHO; 2014. [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14(4):302–308. [PubMed] [Google Scholar]
- 4.Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20(7):1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen GY, Liu M, Wang F, Bertin J, Núñez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186(12):7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grenier JM, Wang L, Manji GA, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530(1-3):73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 7.Anand PK, Malireddi RK, Lukens JR, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488(7411):389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed FE, Vos P. Molecular markers for human colon cancer in stool and blood identified by RT-PCR. Anticancer Res. 2004;24(6):4127–4134. [PubMed] [Google Scholar]
- 9.Normand S, Delanoye-Crespin A, Bressenot A, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A. 2011;108(23):9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu B, Elinav E, Huber S, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A. 2013;110(24):9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YH, Wang Y, Yusufali AH, et al. Cytotoxic genes from traditional Chinese medicine inhibit tumor growth both in vitro and in vivo. J Integr Med. 2014;12(6):483–494. doi: 10.1016/s2095-4964(14)60057-1. [DOI] [PubMed] [Google Scholar]
- 13.Toji LH, Lenchitz TC, Kwiatkowski VA, Sarama JA, Mulivor RA. Validation of routine mycoplasma testing by PCR. In Vitro Cell Dev Biol Anim. 1998;34(5):356–358. doi: 10.1007/s11626-998-0014-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Cheville JC, Pan Y, Roche PC, Schmidt LJ, Tindall DJ. PTEN induces chemosensitivity in PTEN-mutated prostate cancer cells by suppression of Bcl-2 expression. J Biol Chem. 2001;276(42):38830–38836. doi: 10.1074/jbc.M103632200. [DOI] [PubMed] [Google Scholar]
- 15.Bian J, Sun Y. Transcriptional activation by p53 of the human type IV collagenase (gelatinase A or matrix metalloproteinase 2) promoter. Mol Cell Biol. 1997;17(11):6330–6338. doi: 10.1128/mcb.17.11.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kempster SL, Belteki G, Forhead AJ, et al. Developmental control of the Nlrp6 inflammasome and a substrate, IL-18, in mammalian intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300(2):G253–G263. doi: 10.1152/ajpgi.00397.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong H, du W, Sun TT, et al. A positive feedback loop between STAT3 and cyclooxygenase-2 gene may contribute to Helicobacter pylori-associated human gastric tumorigenesis. Int J Cancer. 2014;134(9):2030–2040. doi: 10.1002/ijc.28539. [DOI] [PubMed] [Google Scholar]
- 18.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanda N, Seno H, Konda Y, et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23(28):4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 20.Wei Z, Jiang X, Qiao H, et al. STAT3 interacts with Skp2/p27/p21 pathway to regulate the motility and invasion of gastric cancer cells. Cell Signal. 2013;25(4):931–938. doi: 10.1016/j.cellsig.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya S, Ray RM, Johnson LR. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. Biochem J. 2005;392(Pt 2):335–344. doi: 10.1042/BJ20050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo JM, Park S, Kim JH. Leukotriene B4 receptor-2 promotes invasiveness and metastasis of ovarian cancer cells through signal transducer and activator of transcription 3 (STAT3)-dependent up-regulation of matrix metalloproteinase 2. J Biol Chem. 2012;287(17):13840–13849. doi: 10.1074/jbc.M111.317131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie TX, Wei D, Liu M, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23(20):3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 24.Tabassam FH, Graham DY, Yamaoka Y. Helicobacter pylori-associated regulation of forkhead transcription factors FoxO1/3a in human gastric cells. Helicobacter. 2012;17(3):193–202. doi: 10.1111/j.1523-5378.2012.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bullock MD, Bruce A, Sreekumar R, et al. FOXO3 expression during colorectal cancer progression: biomarker potential reflects a tumour suppressor role. Br J Cancer. 2013;109(2):387–394. doi: 10.1038/bjc.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu MC, Lee DF, Xia W, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117(2):225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Li P, Chen T, et al. Expression of microRNA-96 and its potential functions by targeting FOXO3 in non-small cell lung cancer. Tumour Biol. 2015;36(2):685–692. doi: 10.1007/s13277-014-2698-y. [DOI] [PubMed] [Google Scholar]
- 28.Park SH, Lee JH, Berek JS, Hu MC, Mc-T H. Auranofin displays anticancer activity against ovarian cancer cells through FOXO3 activation independent of p53. Int J Oncol. 2014;45(4):1691–1698. doi: 10.3892/ijo.2014.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 30.Kim DY, Cha ST, Ahn DH, et al. STAT3 expression in gastric cancer indicates a poor prognosis. J Gastroenterol Hepatol. 2009;24(4):646–651. doi: 10.1111/j.1440-1746.2008.05671.x. [DOI] [PubMed] [Google Scholar]
- 31.Koshida Y, Saegusa M, Okayasu I. Apoptosis, cell proliferation and expression of Bcl-2 and Bax in gastric carcinomas: immunohistochemical and clinicopathological study. Br J Cancer. 1997;75(3):367–373. doi: 10.1038/bjc.1997.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Guo QL, You QD, Wu ZQ, Gu HY. Gambogic acid induces apoptosis and regulates expressions of Bax and Bcl-2 protein in human gastric carcinoma MGC-803 cells. Biol Pharm Bull. 2004;27(7):998–1003. doi: 10.1248/bpb.27.998. [DOI] [PubMed] [Google Scholar]
- 33.Mönig SP, Baldus SE, Hennecken JK, et al. Expression of MMP-2 is associated with progression and lymph node metastasis of gastric carcinoma. Histopathology. 2001;39(6):597–602. doi: 10.1046/j.1365-2559.2001.01306.x. [DOI] [PubMed] [Google Scholar]
- 34.Matsumura S, Oue N, Nakayama H, et al. A single nucleotide polymorphism in the MMP-9 promoter affects tumor progression and invasive phenotype of gastric cancer. J Cancer Res Clin Oncol. 2005;131(1):19–25. doi: 10.1007/s00432-004-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama M, Hisatsune J, Yamasaki E, et al. Helicobacter pylori VacA-induced inhibition of GSK3 through the PI3K/Akt signaling pathway. J Biol Chem. 2009;284(3):1612–1619. doi: 10.1074/jbc.M806981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NLRP6-associated pathways in gastric cancer.
Notes: GSEA was performed on TCGA data set by using GSEA version 2.0 from the Broad Institute at MIT as previously described.1 Gene set permutations were performed 1,000 times, and the pathway set list was sorted by the NES. GSEA was performed using TCGA data set. The cell apoptosis (A) and metastasis (B) pathways were strongly associated with NLRP6 lower expression.
Abbreviations: GSEA, gene set enrichment analysis; TCGA, The Cancer Genome Atlas; NES, normalized enrichment score; ES, enrichment score.
NLRP6 protein expression and mRNA expression in seven gastric cancer cell lines were determined by qRT-PCR (A) and Western blot (B) analysis, respectively.
Abbreviation: qRT, quantitative reverse transcription.
NLRP6 knockdown in AGS cells promoted cell proliferation, migration, and invasion but inhibited cell apoptosis. (A) qRT-PCR (upper panel) and Western blot (lower panel) analysis of knockdown efficiency in AGS cells. (B) CCK-8 assay showed that NLRP6 knockdown promoted cell proliferation. (C) Annexin V/PI assay showed that NLRP6 knockdown decreased cell early and late apoptosis. (D) Transwell assays showed that NLRP6 knockdown enhanced migration and invasive capacity. WT, wild-type cells. **P<0.01 and ***P<0.001.
Abbreviations: qRT, quantitative reverse transcription; CCK-8, Cell Counting Kit-8; PI, propidium iodide; siNC, negative control siRNA.
Effects of NLRP6 knockdown on STAT3 phosphorylation.
Notes: Blots are representative of three separate experiments. WT, wild-type cells; siNC, negative control siRNA.
Table S1.
Univariate Cox regression of prognostic parameters for survival in patients with gastric cancer
| Prognostic parameter | Univariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P-value | |
| Expression of NLRP6 (low vs. high) | 0.470 | 0.280–0.788 | 0.004** |
| Helicobacter pylori infection (negative vs. positive) | 0.221 | 0.135–0.362 | <0.001 |
| TNM (I+II vs. III+IV) | 4.457 | 2.473–8.034 | <0.001 |
| Tumor size (<5 cm vs. ³5 cm) | 4.275 | 2.486–7.352 | <0.001 |
| Lymph node metastasis (absent vs. present) | 22.304 | 9.414–52.846 | <0.001 |
Note:
P<0.01.
Table S2.
Multivariate Cox regression of prognostic parameters for survival in patients with gastric cancer
| Prognostic parameter | Multivariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P-value | |
| Expression of NLRP6 (low vs. high) | 0.825 | 0.483–1.409 | 0.482 |
| Helicobacter pylori infection (negative vs. positive) | 0.518 | 0.306–0.876 | 0.014* |
| TNM (I+II vs. III+IV) | 2.383 | 1.249–4.549 | 0.008** |
| Tumor size (<5 cm vs. ≥5 cm) | 2.689 | 1.492–4.848 | 0.001** |
| Lymph node metastasis (absent vs. present) | 13.465 | 5.398–33.589 | 0.000*** |
Note:
P<0.05,
P<0.01, and
P<0.001.