Abstract
Purpose
Accumulating studies showed that the expression of microRNAs (miRNAs) was dysregulated in osteosarcoma (OS). In this study, we sought to investigate the effect of let-7a on OS progression and its potential molecular mechanism.
Patients and methods
Quantitative real-time PCR (qRT-PCR) was performed to evaluate the expression level of let-7a and Aurora-B (AURKB) in OS tissues and cells. The OS cells were treated with let-7a mimic, let7a inhibitor, negative mimic and Lv-AURKB combined with let-7a. The ability of cell proliferation, migration and invasion was measured using Cell Counting Kit-8 (CCK-8) and wound-healing and transwell invasion assays. The protein of AURKB, NF-κβp65, MMP2 and MMP9 was measured by Western blot analysis. Xenograft model was performed to investigate the effects of let-7a on tumor growth and metastasis. The lung metastasis was measured by counting the metastatic node using H&E staining.
Results
Let-7a expression was significantly underexpressed in OS cell lines and tissues compared with human osteoblast cell lines, hFOB1.19, and adjacent normal bone tissues. Exogenous let-7a inhibited the viability, migratory and invasive ability of OS cells in vitro. In addition, the overexpression of AURKB in OS cells could partly rescue let-7a-mediated tumor inhibition. Also, the overexpression of let-7a inhibited OS cell growth and lung metastasis in vivo. Furthermore, the results showed that let-7a could decrease the expression of NF-κβp65, MMP2 and MMP9 proteins by targeting AURKB in OS cells.
Conclusion
Let-7a inhibits the malignant phenotype of OS cells by targeting AURKB at least partially. Targeting let-7a and AURKB/NF-κβ may be a novel therapeutic strategy for the treatment of OS.
Keywords: let-7a, Aurora-B, osteosarcoma, malignant phenotype
Introduction
Osteosarcoma (OS) is the most common type of primary malignant tumor of the bone in young adolescents and children.1 The prognosis of patients with localized OS has been improved by the current treatment strategies, including preoperative chemotherapy, surgical resection and postoperative chemotherapy.2 However, the use of adjuvant chemotherapy provides no survival advantage for patients with pulmonary metastases, whereas pulmonary metastasis is the leading cause of mortality in patients with OS.3 Therefore, it is of particular importance to develop new molecular targets for the management of OS.
MicroRNAs (miRNAs) are a class of noncoding single-stranded RNA molecules encoded by endogenous genes with the length of about 22 nucleotides, which are involved in the regulation of posttranscriptional gene expression in plants and animals. Up to now, over 15,000 miRNA gene loci in over 140 species and over 17,000 distinct mature miRNA sequences have been found.4 Deep-sequencing technologies have delivered a sharp rise in the rate of novel miRNA discovery. Substantial studies have revealed that miRNAs are involved in the development and progression of cancers, acting as tumor suppressors or oncogenes by modulating gene expression.5–9 Let-7 was first identified in Caenorhabditis elegans as a heterochronic gene.10 The let-7 cluster was found to be dysregulated in various malignant tumors, and let-7 can prevent the malignant phenotype via the downregulation of oncogenes such as zinc finger-1,11 high mobility group AT-Hook 2 (HMGA2)12 and LIN2.13 Let-7a is one of the members of let-7 family. Previous studies found that let-7a, as a tumor suppressor, plays an important role in the development of tumors by targeting oncogene cancer in several tumors.14–16
Aurora-B (AURKB) is a component of chromosome passenger complex (CPC), which is composed of additional three activation regulators such as INCEP, survivin and borealin. AURKB plays important biological functions in regulating chromosome condensation and spindle assembly checkpoint (SAC), rectifying the faulty attachment between spindle and kinetochore, maintaining the correct chromosome alignment and the faithful chromosomal segregation. Accumulating evidence has shown that AURKB was highly expressed in various malignant tumors and as an important antitumor target.17–20 Our previous study indicated that AURKB showed increased expression in OS and the knockdown of AURKB inhibited proliferation, migration and invasion of OS cells in vitro.21,22 Although several miRNAs have been found to target AURKB, including let-7b23 and miRNA-378a-5p,24 the correlation of AURKB expression and let-7a in OS cells is still unclear.
In the current study, we found that the expression of let-7a was decreased, whereas that of AURKB was increased in OS tissues and cell lines compared with the normal bone tissues and hFOB1.19. Based on the inverse correlation between let-7a and AURKB expression in OS, we speculated that the elevated expression of AURKB in OS may be mediated, at least partially, by let-7a. We aimed to assess the effects of let-7a on OS progress and to determine whether the let-7a regulated AURKB expression in OS.
Patients and methods
Patients and clinical samples
Twenty-one specimens of OS were obtained by excision biopsy from patients with histologically proven OS of the extremities treated at the First Affiliated Hospital of Nanchang University and the Cancer Hospital of Jiangxi Province (Nanchang, People’s Republic of China) between 2009 and 2012. The tissues obtained from an area 5 cm from the tumor margin were accounted as negative controls. No patient had a history of prior therapies with anticancer drugs or radiotherapy. The samples were stored in the –80°C fridge before quantitative PCR (qPCR) detection. The pathological diagnosis was confirmed by two practiced pathologists. All research methods were approved by the medical ethics committee of the First Affiliated Hospital of Nanchang University and followed the Declaration of Helsinki. All subjects were notified about the objectives, contents, latent risks and signed informed consents.
Cell culture and transfection
The U2-OS and 143B cell lines and the human osteoblast cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were routinely cultured in DMEM (Hyclone, Logan, UT, USA) supplemented with 10% FBS (Sigma-Aldrich Co., St Louis, MO, USA). Cells were cultured at 37°C in 5% CO2. The let-7a mimic, let-7a inhibitor and negative mimic were produced by Yingqi Biotechnology Company (Wuhan, People’s Republic of China). Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used to transfect cells with let-7a mimics, let-7a inhibitor and negative mimic. The lentivirus vectors were compound by GeneChem Co. (Shanghai, People’s Republic of China). U2-OS and 143B cells were transfected with lentivirus vectors of upregulating AURKB (Lv-AURKB) and negative lentivirus vectors (NC-AURKB), respectively. Infection/transfection process was operated according to the manufacturer’s instructions.
Biological information prediction
Prediction of the AURKB 3′-UTR as a miRNA-binding target was determined using TargetScan (www.targetscan.org), mirDIP (http://ophid.utoronto.ca) and starBase (http://starbase.sysu.edu.cn).
Luciferase reporter assay
To investigate whether let-7a could target AURKB directly, the luciferase reporter assay was performed as described in our previous study.25
Reverse transcription quantitative real-time PCR (RT-qPCR)
The RNA was extracted from fresh OS tissues, adjacent non-tumor tissues, OS cells and hFOB1.19 cells using the Qiagen RN easy FFPE protocol (Qiagen NV, Venlo, the Netherlands). The expression level of let-7a and AURKB was evaluated by qRT-PCR (StepOne™; Bio-Rad Laboratories Inc., Hercules, CA, USA), and U6 snRNA and GAPDH were used as endogenous references. Total RNA in OS cells or tissues was extracted using TRIzol (Thermo Fisher Scientific). Reverse transcription was performed using Prime Script RT Reagent Kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer’s protocol. Then, each sample was analyzed by qPCR under the conditions described in the manufacturer’s instructions for SYBR Premix Ex Tap II (Thermo Fisher Scientific): 50°C for 2 minutes, 95°C for 2 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 30 seconds. Relative expression was calculated using the 2−ΔΔCt method. All of primer sequences are summarized in Table 1.
Table 1.
Primer information
| Primers | Sequences (5′–3′) |
|---|---|
|
| |
| Let-7a-RT | TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCATAGGAAAGA |
| U6-RT | TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAAAAATATGGA |
| Let-7a-Q-F | CACCCACCACTGGGAGATAAC |
| Let-7a-Q-R | TATGGTTGTTCACGACTCCTTCAC |
| U6-Q-F | CGCAAGGATGACACGCAAAT |
| U6-Q-R | TATGGTTGTTCACGACTCCTTCAC |
| AURKB F | AGAAGGAGAACTCCTACCCCT |
| AURKB R | CGCGTTAAGATGTCGGGTG |
| GAPDH F | CAGGGCTGCTTTTAACTCTGGT |
| GAPDH R | GATTTTGGAGGGATCTCGCT |
Abbreviation: AURKB, Aurora-B.
Western blot analysis
Total protein was extracted using radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology) containing 60 µg/mL PMSF (Solarbio, Beijing, People’s Republic of China). Protein concentration was determined by a Bradford assay (Bio-Rad Laboratories Inc.). Western blot analysis was conducted as described in the study by He et al.26 Three independent experiments were performed over multiple days.
Cell Counting Kit-8 (CCK-8) assays
The proliferation of OS cells was evaluated using CCK-8. OS cells (4,000/well) were seeded per well in a 96-well plate and to grow for 24 hours. Then, 24 hours and 48 hours after transfection or infection, 10 µL CCK-8 solution was added to wells and incubated for 12 hours at 37°C. The absorbance of each well at 450 nm with microplate reader was measured. Three independent experiments were performed over multiple days.
Wound-healing assays
In brief, cells were grown to confluence in six-well tissue culture plastic dishes to a density of approximately 5×106 cells/well. The cells were denuded by dragging a rubber policeman (Thermo Fisher Scientific) through the center of the plate. Cell culture dishes were rinsed with PBS. Then, we added fresh quiescent medium alone or containing 10% FBS into the dishes and put the dishes into the incubator (37°C for 24 hours). Images were captured at 0 hour and 24 hours using an ECLIPSE-TS-100 microscope (magnification, ×200; Nikon Corporation, Tokyo, Japan), and the migrated distance was measured by Image J, version 1.48 (National Center for Biotechnology Information, Bethesda, MD, USA). The migration rates of cells were obtained by counting three fields per area and represented as the average of three independent experiments done over multiple days.
Transwell invasion assays
We used the BD BioCoatTM BD MatrigelTM Invasion Chamber (BD, Franklin Lakes, NJ, USA) to analyze the cell’s invasion ability according to the specification. The medium in the lower chamber contained 5% fetal calf serum as a source of chemoattractants. Cells were suspended in serum-free medium and added to the upper chambers at the same time. Cells that passed through the Matrigel-coated membrane were stained with Diff-Quik (Sysmex, Kobe, Japan) and photographed (magnification, ×400). Images were captured at 24 hours, and cell counting was measured by Image J software. The values for invasion were obtained by counting three fields per membrane and represented as the average of three independent experiments conducted over multiple days.
In vivo tumor-bearing nude mice model
The athymic nude BALB/c mice (weight, 15±2 g; age, 6±2 weeks) were provided by the Department of Laboratory Animal Science of Nanchang University (Nanchang, People’s Republic of China). The experimental model was established by injecting 1×107 cells, suspended in 200 µL PBS, in the proximal tibia bone marrow cavity of nude mice. Following 14 days, mice were randomly divided into three groups (n=12): 1) injected with let-7a mimic; 2) injected with let-7a inhibitor and 3) injected with negative mimic. Using the Entranster™ in vivo kit (Engreen Biosystem Co., Ltd., People’s Republic of China), 0.2 mL of let-7a mimic, let-7a inhibitor and negative mimic was intratumorally injected three times every other day, respectively. The mice were sacrificed at 6 weeks after injection. The volume and weight of the tumors were evaluated. Tumor volumes were calculated using the following formula: V (mm3) = length × width2/2. The lung metastasis was measured by counting the metastatic node of OS cells using H&E staining. All animal experiments were approved by the human ethical review committee of the Nanchang University, Jiangxi, People’s Republic of China, and performed according to the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (1996).
Statistical analyses
All measurement data were presented as mean ± SD. Student’s t-test was used in two-sample analysis, and one-way ANOVA was used in multiple sample analysis. P<0.05 was considered to indicate a statistically significant difference. The Pearson correlation was used to evaluate the relationship between let-7a and AURKB in OS tissues. All analyses were performed using SPSS statistical software, version 13.0 (IBM Corporation, Armonk, NY, USA).
Results
Negative relationship between let-7a and AURKB in OS tissues and cell lines
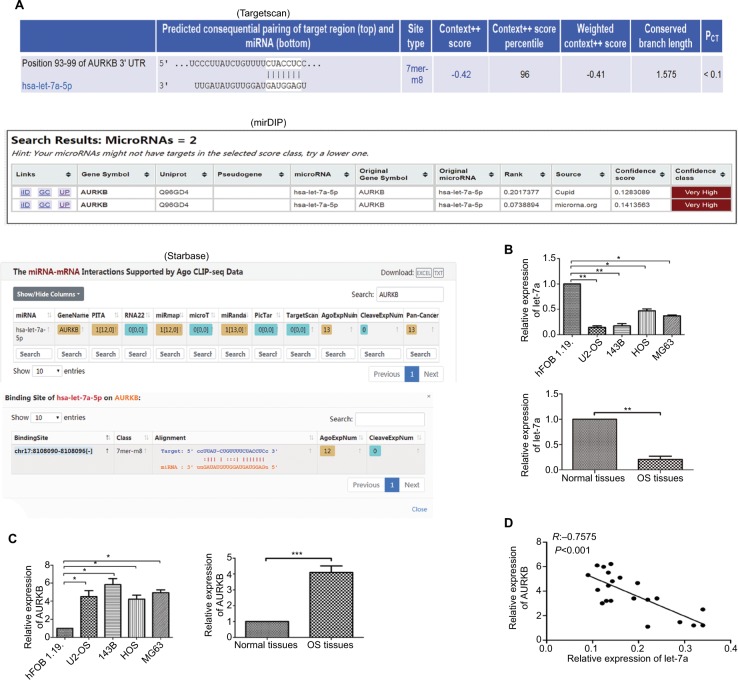
The result of biological information prediction showed that the AURKB 3′-UTR is a let-7a binding target as shown in Figure 1A. To investigate the expression level of let-7a in OS tissues and cell lines (U2-OS, 143B, HOS and MG63), qRT-PCR was performed. The results revealed that the expression level of let-7a was significantly lower in OS tissues and cells than that in normal bone tissues and hFOB1.19 cells (Figure 1B). In addition, the upregulated expression of AURKB was found in OS tissues and cell lines (U2-OS, 143B, HOS and MG63) when compared with normal bone tissues and hFOB1.19 cells (Figure 1C). The Pearson correlation assay showed that there was a negative relationship between let-7a and AURKB in OS tissues (Figure 1D). These results suggested that a potential relationship may exist between let-7a and AURKB in OS.
Figure 1.
Expression levels of let-7a and AURKB.
Notes: (A) Results of bioinformatic research about let-7a by targeting AURKB. (B) The expression level of let-7a in OS tissues and normal tissues and in OS cells and osteoblast cells (hFOB1.19 cells) was evaluated by qRT-PCR. Mean ± SD; *P<0.05, **P<0.01, ***P<0.001. (C) The expression level of AURKB in OS tissues and normal tissues and in OS cells and osteoblast cells was evaluated by qRT-PCR. Mean ± SD; *P<0.05, **P<0.01, ***P<0.001. (D) The negative correlation between let-7a and AURKB levels in the OS tissues (R=-0.7575, P<0.001; OS cells and osteoblast cells: n=6/group. OS tissues: n=21). PITA is a database for prediction of target gene.
Abbreviations: AURKB, Aurora-B; OS, osteosarcoma; PCT, preferentially conserved targeting; qRT-PCR, reverse transcription quantitative real-time PCR.
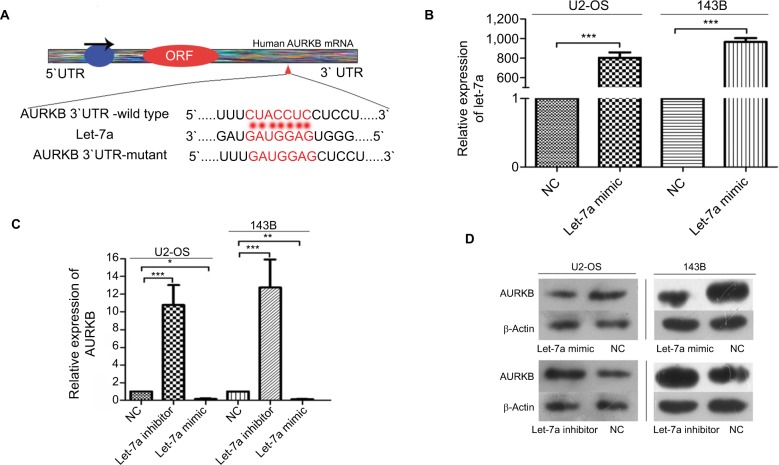
Let-7a negatively regulates AURKB expression in OS cells
We performed the biological information prediction and luciferase reporter assay to investigate whether AURKB is one of the potential targets of let-7a. The results showed that let-7a has predictive binding sites in the 3′-UTR of AURKB and let-7a restrained the relative luciferase activity in the wild-type 3′-UTR of AURKB as shown in our previous study.25 The paired sequence between let-7a and AURKB mRNA 3′-UTR is shown in Figure 2A. qRT-PCR was performed to evaluate the overexpression level of let-7a (Figure 2B). In addition, the protein and mRNA expression of AURKB were significantly lower in cells treated with let-7a mimic than those in cells treated with negative mimic. In addition, the protein and mRNA expression of AURKB were significantly higher in cells managed with let-7a inhibitor when compared with those cells treated with negative inhibitor (Figure 2C and D). These results indicated that let-7a could negatively regulate AURKB expression in OS cells.
Figure 2.
Let-7a inhibits AURKB expression in OS cells.
Notes: (A) The paired sequence between let-7a and AURKB mRNA 3′-UTR. The complementary nucleotides are highlighted in red. (B) The expression of let-7a was examined by qRT-PCR in OS cells transfected with let-7a mimic or negative mimic. The results confirmed the transfection efficiency. GAPDH was used as the endogenous control. Mean ± SD; *P<0.05, **P<0.01, ***P<0.001. (C) AURKB mRNA levels in OS cells transfected with let-7a mimic, let-7a inhibitor or NC group. Mean ± SD; *P<0.05, **P<0.01, ***P<0.001 (D) AURKB protein levels in OS cells transfected with let-7a mimic, let-7a inhibitor or NC group.
Abbreviations: AURKB, Aurora-B; NC, negative control; OS, osteosarcoma.
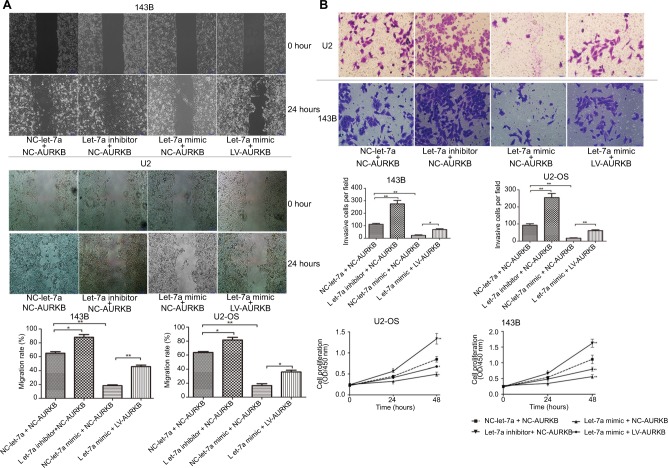
Ectogenous let-7a inhibits the malignant phenotype of OS cells by targeting AURKB
To explore the effect of let-7a on the proliferation, migration and invasion, OS cells were treated with let-7a mimic, let7a inhibitor and negative mimic, respectively. The ability of cell proliferation, migration and invasion was measured using CCK-8 and wound-healing and transwell invasion assays. The results showed that the viability, migratory and invasive ability was significantly lower in cells treated with let-7a mimic than those cells treated with let-7a inhibitor or negative mimic (Figure 3). It indicated that let-7a has anti-malignant phenotype effects in OS cells.
Figure 3.
Ectogenous let-7a inhibits the malignant phenotype of OS cells by targeting AURKB.
Notes: (A) Wound-healing assay (×200); mean ± SD; *P<0.05, **P<0.01, ***P<0.001. (B) Transwell assay (×200); mean ± SD; *P<0.05, **P<0.01, ***P<0.001. (C) The ability of proliferation was determined by CCK-8 assays. Mean ± SD; *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: AURKB, Aurora-B; CCK-8, Cell Counting Kit-8; NC, negative control; OS, osteosarcoma.
In addition, to investigate whether let-7a inhibits the malignant phenotype of OS cells by targeting AURKB, the OS cells were treated with let-7a mimic, let-7a inhibitor and Lv-AURKB combined with let-7a (coinfected). In Western blot assays, the results revealed that the AURKB protein level was significantly decreased in cells treated with let-7a mimic. However, partially downregulated AURKB protein level in co-transfected cells was observed (Figure 4). The malignant phenotype of cells was investigated by measuring the proliferation, migration and invasion abilities. The data showed that the tumor inhibition of let-7a was partly rescued by upregulation of AURKB in OS cells (Figure 3). It suggested that let-7a inhibits the malignant phenotype of OS cells at least partly by targeting AURKB in vitro.
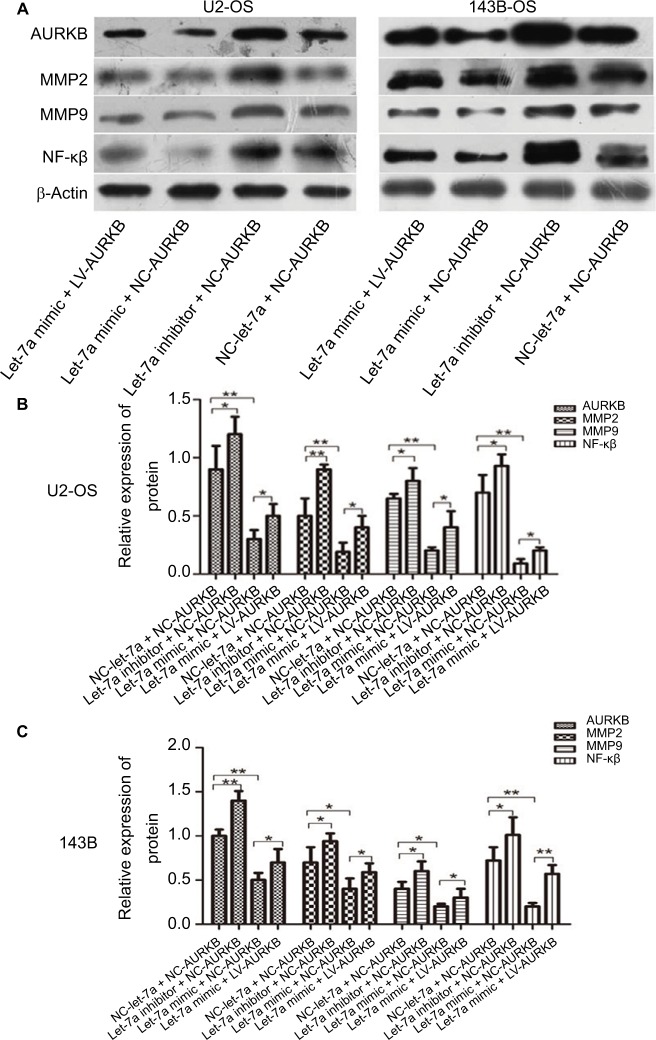
Figure 4.
Western blot analysis was carried out to determine the levels of AURKB, NF-κβ p65, MMP2 and MMP9 in transfected/infected U2-OS (left) and 143B (right) cells. (A) the strip of Western blot. (B) The result of relative expression of protein in U2-OS cells. (C) The result of relative expression of protein in 143B cells.
Note: Mean ± SD; n=6/group. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: AURKB, Aurora-B; NC, negative control; OS, osteosarcoma.
Let-7a decreased NF-κβ signaling pathway by targeting AURKB in OS cells
For investigating the downstream of AURKB activation, the AURKB was inhibited or restored in OS cells by let-7a mimic or inhibitor of let-7a. The protein of NF-κβ, MMP2 and MMP9 was measured using Western blot analysis. The results revealed that the expression levels of NF-κβp65, MMP2 and MMP9 proteins were significantly lower in cells treated with let-7a mimic than those in cells treated with negative mimic. While the NF-κβp65, MMP2 and MMP9 proteins were significantly higher in cells treated with let-7a inhibitor than those cells treated with negative mimic. It suggested that the upregulation of let-7a inhibits NF-κβ signaling pathway in OS cells, while the inhibiting effect was restored by the overexpression of AURKB (Figure 4). All the results indicated that let-7a decreased NF-κβ signaling pathway at least partly by targeting AURKB in OS cells.
Let-7a expression inhibits OS cell growth and metastasis in vivo
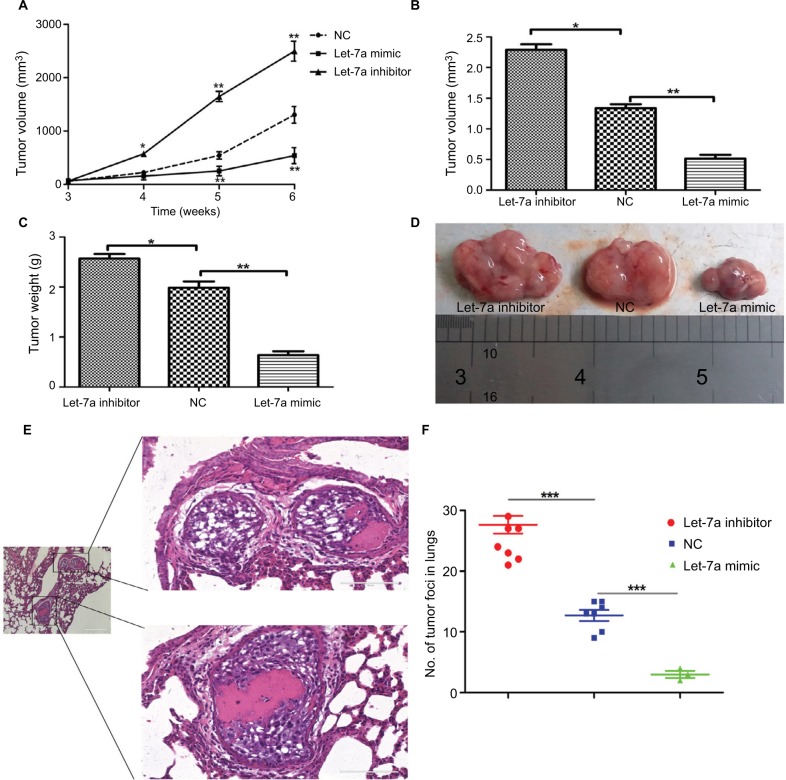
To demonstrate the tumor inhibition of let-7a in OS, we established a xenograft model to investigate the effects of let-7a on tumor growth and metastasis. The results showed that growth and lung metastasis were significantly inhibited in the group treated with let-7a mimic than those treated with negative mimic, while the growth and lung metastasis were significantly promoted in the group treated with let-7a inhibitor when compared with the group treated with negative mimic (Table 2; Figure 5). These results indicated that let-7a has an effective activity to inhibit OS cell growth and lung metastasis in vivo.
Table 2.
The effects of let-7a on tumor metastasis
| Groups (n=12) | Lung metastasis | Rate (%) |
|---|---|---|
| Let-7a mimic | 3 | 25.00 |
| Let-7a inhibitor | 11 | 91.67 |
| Negative mimic | 7 | 58.33 |
Figure 5.
let-7a inhibits OS cell growth in nude mice.
Notes: (A) Growth curve of xenograft tumors. Volume (B) and weight (C) of xenograft tumors in nude mice. (D) A representative image of xenograft tumors from the let-7a mimic, let-7a inhibitor and NC group. (E) Representative H&E stained images. (F) Numbers of lung metastatic foci were counted in each group. Mean ± SD; *P<0.05, **P<0.01, ***P<0.001 vs the NC.
Abbreviations: NC, negative control; OS, osteosarcoma.
Discussion
In the current study, we provided encouraging evidence in support of let-7a as a tumor suppressor via inhibiting NF-κβ/MMP signaling by targeting AURKB in OS.
AURKB is a component of CPC, which is composed of INCEP, survivin and borealin. AURKB mediates chromosome condensation by phosphorylating histone H3 on Ser10 and variant centrosome protein A (CENP-A) on Ser7. It is also involved in the regulation of SAC, rectifying the faulty attachment between spindle and kinetochore and maintaining the correct chromosome alignment and the faithful chromosomal segregation. Accumulated studies demonstrated that the activation of AURKB mediated phosphorylation of histone H2AX on Ser121, which in turn facilitates the auto-phosphorylation of AURKB.27 During anaphase, AURKB via phosphorylating a series of downstream substrates, including mitotic kinesin-like protein 1 (MKLP1) and RacGAP1,28 accelerated their deposition at mid-body and maintained the stabilization of central spindle. In addition, AURKB could lead to shortening of central spindle and promote cytokinesis via phosphorylation of Kif2A, the microtubule depolymerase.29 Increasing studies revealed that AURKB was involved in malignant tumor cell growth and metastasis.17–20 In our previous studies, we found that AURKB was overexpressed in OS tissues and cells and the knockdown of AURKB by shRNA and small molecular inhibitor could inhibit the proliferation, migration and invasion abilities of OS cells in vitro.21,22 However, the molecular mechanism mediated AURKB overexpression in OS remains to be explored.
Several studies have shown that miRNAs act as cancer suppressors or oncogenes in OS.30,31 miRNAs regulate cell growth, migration and metastasis via the regulation of its target gene.32,33 Considering the regulation of AURKB expression by miRNAs, we searched public databases described earlier for candidate miRNAs that targeted AURKB. Let-7 cluster was found in the databases. Numerous studies have identified members of the let-7 family to be suppressors of tumor cell proliferation.11–13 There are 12 miRNAs in the let-7 cluster in human beings, including let-7a which has been confirmed to be decreased expression and closely related to the prognosis of patients with malignant tumor.14–16 A previous study showed that let-7a was decreased expression in OS cell lines, and the upregulation of let-7a inhibits OS cell proliferation and metastasis.34,35 Accordingly, we investigated the expression of let-7a in OS tissues and cell lines. Consistent with the former study, the expression of let-7a was attenuated in tissues and OS cell lines when compared with adjacent normal bone tissues and hFOB1.19 cells. However, the accurate biological function and relative molecular mechanisms in OS are largely unknown. Target genes of let-7a include c-Myc, HMGA2, STAT3 and E2F2.34–36 Although Liu et al37 reported that let-7a regulates AURKB in endometrial carcinoma cells, the role of let-7a on AURKB expression in OS is still unclear. In addition to bioinformatical prediction, we performed a series of studies to investigate the target role of AURKB in OS, as transfection of let-7a caused a substantial reduction in luciferase activity by the luciferase expression constructs that carry the target AURKB fragment compared with the mutant constructs that lack the binding site to let-7a. Further, ectopic expression of let-7a significantly decreased the transcription and translation of the AURKB gene, namely, these results indicated that AURKB is a target of let-7a in OS cells.
In addition, we analyzed the effects of let-7a on the biological behaviors of OS cells. Similar to the study by Iwasaki et al,35 we found that exogenous let-7a prevented OS cell proliferation, migration and invasion in vitro. To further elucidate whether let-7a alters malignant phenotype of OS cells via directly targeting AURKB in OS, the cells, treated with let-7a mimic, were transfected with lentivirus plasmid of overexpression of AURKB, and the malignant phenotype of cells was measured. The results showed that the tumor inhibition mediated by let-7a was partly restored by increasing AURKB. In addition, in this study, the upregu-lation of let-7a could suppress OS cell growth and lung metastasis in nude mice. These data suggested that let-7a alters the malignant phenotype of OS cells at least partly by targeting AURKB.
Looking downstream of AURKB activation, we investigated the effect of altering AURKB by let-7a mimic and inhibitor on NF-κβ signaling pathway. Most importantly, our previous studies have confirmed the role of the AURKB/NF-κβ signaling pathway in OS, and the knockdown of AURKB inhibits NF-κβ signaling pathway.26 NF-κβ is a member of the Rel family proteins, which regulates protein expression mediating cell cycle/proliferation, anti-apoptosis and cytokine secretion. NF-κβ is composed of DNA-binding subunits (p50 and p52) and subunits with transcriptional activity (p65 and RelB or c-Rel), which dimerizes in various combinations. The primary form of NF-κβ is a heterodimer of the p50 and p65 subunits and is localized mainly to the cytoplasm in an inactive form bound to I-κB. Several studies showed that the activation of NF-κβ could promote MMP2 and MMP9 proteins.38 Decreased expression of MMP-2 and MMP-9 promotes cell invasion and metastasis by degrading components of the basement membrane and epimatrix in malignant tumor.39–42 In this study, we found that let-7a inhibits the growth and metastasis of OS cells and decreases the expression of NF-κβ p65, MMP2 and MMP9 proteins.
The abovementioned results indicated that the expression of let-7a was downregulated in OS patients and negatively correlated with that of AURKB, and let-7a inhibits OS cells and lung metastasis by targeting AURKB in vivo and in vitro. It is suggested that targeting let-7a/AURKB axis may be a therapeutic strategy for OS management. However, in view of the functions of AURKB, such as regulating chromosome condensation and SAC, rectifying the faulty attachment between spindle and kinetochore, maintaining the correct chromosome alignment and the faithful chromosomal segregation, clinical practice of targeting let-7a/AURKB axis may lead to side effects that are worthy of attention, such as dysregulation of mitotic mechanisms. In addition, whether let-7a/AURKB axis differs between patients with and without lung metastasis remains unexplored. Therefore, more further clinical studies are particularly important now.
Conclusion
Our findings demonstrate that let-7a is underexpressed in OS and acts as a tumor suppressor which inhibits the malignant phenotype of cells via decreased NF-κβ signaling pathway by targeting AURKB partially. Targeting let-7a and AURKB/NF-κβ may be a therapeutic strategy for OS management.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81560435), the Department of Science and Technology Program of Jiangxi Province, China (No. 20162BCB23057), the Natural Science Foundation of Jiangxi Province, China (No.20161ACB20011), and the Department of Science and Technology Program of Jiangxi Province, China (No. 20162BCB22022).
Abbreviations
- OSm
osteosarcoma
- miRNA
microRNA
- AURKB
Aurora-B
- CPC
chromosome passenger complex
- CENP-A
centrosome protein A
- MKLP1
mitotic kinesin-like protein 1
- HMGA2
high mobility group AT-Hook 2
Footnotes
Author contributions
S-HH, Z-LL and WZ contributed to research design. W-SP contributed to acquisition, analysis and interpretation of the data. J-JY and W-SP contributed to drafting of the article. YC, A-FP, Z-YC and J-ML contributed to critical revision of the article. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hegyi M, Semsei AF, Jakab Z, et al. Good prognosis of localized osteosarcoma in young patients treated with limb-salvage surgery and chemotherapy. Pediatr Blood Cancer. 2011;57(3):415–422. doi: 10.1002/pbc.23172. [DOI] [PubMed] [Google Scholar]
- 2.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J Clin Oncol. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Li B, Ren Y, Ye Z. T-Cell-Based Immunotherapy for Osteosarcoma: Challenges and Opportunities. Front Immunol. 2016;7:353. doi: 10.3389/fimmu.2016.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Zhou ZQ, Yang ZR, et al. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66(1):136–151. doi: 10.1002/hep.29116. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Zhang X, Li N, Liu Q, Chen D. miR-30b inhibits cancer cell growth, migration, and invasion by targeting homeobox A1 in esophageal cancer. Biochem Biophys Res Commun. 2017;485(2):506–512. doi: 10.1016/j.bbrc.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Lu G, Shao Y, Xu D. microRNA-802 inhibits epithelial-mesenchymal transition through targeting flotillin-2 in human prostate cancer. Biosci Rep. 2017;37(2):BSR20160521. doi: 10.1042/BSR20160521. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Mao JH, Zhou RP, Peng AF, et al. microRNA-195 suppresses osteosarcoma cell invasion and migration in vitro by targeting FASN. Oncol Lett. 2012;4(5):1125–1129. doi: 10.3892/ol.2012.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long XH, Mao JH, Peng AF, Zhou Y, Huang SH, Liu ZL. Tumor suppressive microRNA-424 inhibits osteosarcoma cell migration and invasion via targeting fatty acid synthase. Exp Ther Med. 2013;5(4):1048–1052. doi: 10.3892/etm.2013.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 11.Tvingsholm SA, Hansen MB, Clemmensen KKB, et al. Let-7 microRNA controls invasion-promoting lysosomal changes via the oncogenic transcription factor myeloid zinc finger-1. Oncogenesis. 2018;7(2):14. doi: 10.1038/s41389-017-0014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Fazio P, Maass M, Roth S, et al. Expression of hsa-let-7b-5p, hsa-let-7f-5p, and hsa-miR-222-3p and their putative targets HMGA2 and CDKN1B in typical and atypical carcinoid tumors of the lung. Tumour Biol. 2017;39(10):1010428317728417. doi: 10.1177/1010428317728417. [DOI] [PubMed] [Google Scholar]
- 13.Zamora-Contreras AM, Alvarez-Salas LM. Let-7 miRNA Precursors Co-express with LIN28B in Cervical Cells. Microrna. 2018;7(1):62–71. doi: 10.2174/2211536607666180115143014. [DOI] [PubMed] [Google Scholar]
- 14.Ling R, Zhou Y, Zhou L, et al. Lin28/microRNA-let-7a promotes metastasis under circumstances of hyperactive Wnt signaling in esophageal squamous cell carcinoma. Mol Med Rep. 2018;17(4):5265–5271. doi: 10.3892/mmr.2018.8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang G, Du R, Tang Z, Kuang Y. MiRNALet-7a mediates prostate cancer PC-3 cell invasion, migration by inducing epithelial-mesenchymal transition through CCR7/MAPK pathway. J Cell Biochem. 2018;119(4):3725–3731. doi: 10.1002/jcb.26595. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Ding R, Han Z, Ma Z, Wang Y. Targeting of cell cycle and let-7a/STAT3 pathway by niclosamide inhibits proliferation, migration and invasion in oral squamous cell carcinoma cells. Biomed Pharmacother. 2017;96:434–442. doi: 10.1016/j.biopha.2017.09.149. [DOI] [PubMed] [Google Scholar]
- 17.Al-Khafaji AS, Davies MP, Risk JM, et al. Aurora B expression modulates paclitaxel response in non-small cell lung cancer. Br J Cancer. 2017;116(5):592–599. doi: 10.1038/bjc.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogen D, Wei JS, Azorsa DO, et al. Aurora B kinase is a potent and selective target in MYCN-driven neuroblastoma. Oncotarget. 2015;6(34):35247–35262. doi: 10.18632/oncotarget.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Loyola A, Fernández-Miranda G, Trakala M, et al. Aurora B Overexpression Causes Aneuploidy and p21Cip1 Repression during Tumor Development. Mol Cell Biol. 2015;35(20):3566–3578. doi: 10.1128/MCB.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang PY, Li Y, Luo DH, et al. Expression of Aurora-B and FOXM1 predict poor survival in patients with nasopharyngeal carcinoma. Strahlenther Onkol. 2015;191(8):649–655. doi: 10.1007/s00066-015-0840-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhu LB, Jiang J, Zhu XP, et al. Knockdown of Aurora-B inhibits osteosarcoma cell invasion and migration via modulating PI3K/Akt/NF-κB signaling pathway. Int J Clin Exp Pathol. 2014;7(7):3984–3991. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu XP, Liu ZL, Peng AF, et al. Inhibition of Aurora-B suppresses osteosarcoma cell migration and invasion. Exp Ther Med. 2014;7(3):560–564. doi: 10.3892/etm.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mäki-Jouppila JH, Pruikkonen S, Tambe MB, et al. MicroRNA let-7b regulates genomic balance by targeting Aurora B kinase. Mol Oncol. 2015;9(6):1056–1070. doi: 10.1016/j.molonc.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winsel S, Mäki-Jouppila J, Tambe M, et al. Excess of miRNA-378a-5p perturbs mitotic fidelity and correlates with breast cancer tumourigenesis in vivo. Br J Cancer. 2014;111(11):2142–2151. doi: 10.1038/bjc.2014.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JM, Long XH, Zhang GM, et al. Let-7g reverses malignant phenotype of osteosarcoma cells by targeting Aurora-B. Int J Clin Exp Pathol. 2014;7(8):4596–4606. [PMC free article] [PubMed] [Google Scholar]
- 26.He JY, Xi WH, Zhu LB, et al. Knockdown of Aurora-B alters osteosarcoma cell malignant phenotype via decreasing phosphorylation of VCP and NF-κB signaling. Tumour Biol. 2015;36(5):3895–3902. doi: 10.1007/s13277-014-3032-4. [DOI] [PubMed] [Google Scholar]
- 27.Shimada M, Goshima T, Matsuo H, et al. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat Commun. 2016;7:12059. doi: 10.1038/ncomms12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minoshima Y, Kawashima T, Hirose K, et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4(4):549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 29.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13(12):789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng N, Miao Z, Wang L, Liu B, Wang G, Guo X. MiR-378 promotes the cell proliferation of osteosarcoma through down-regulating the expression of Kruppel-like factor 9. Biochem Cell Biol. 2018;96(5):515–521. doi: 10.1139/bcb-2017-0186. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Li M, Liu X, Liu F, Zhu J. miR-27a-3p promotes the malignant phenotypes of osteosarcoma by targeting ten-eleven translocation 1. Int J Oncol. 2018;52(4):1295–1304. doi: 10.3892/ijo.2018.4275. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Xu B, Miu X, Deng Z, Liao H, Hao L. Inhibition of miRNA-21 attenuates the proliferation and metastasis of human osteosarcoma by upregulating PTEN. Exp Ther Med. 2018;15(1):1036–1040. doi: 10.3892/etm.2017.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Wang L, Wang Q, Li L, Fu Y, Sun J. MiR-183 inhibits osteosarcoma cell growth and invasion by regulating LRP6-Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2018;496(4):1197–1203. doi: 10.1016/j.bbrc.2018.01.170. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S, Hou C, Li G, et al. A single nucleotide polymorphism in the 3’-untranslated region of the KRAS gene disrupts the interaction with let-7a and enhances the metastatic potential of osteosarcoma cells. Int J Mol Med. 2016;38(3):919–926. doi: 10.3892/ijmm.2016.2661. [DOI] [PubMed] [Google Scholar]
- 35.Iwasaki T, Tanaka K, Kawano M, Itonaga I, Tsumura H. Tumor-suppressive microRNA-let-7a inhibits cell proliferation via targeting of E2F2 in osteosarcoma cells. Int J Oncol. 2015;46(4):1543–1550. doi: 10.3892/ijo.2015.2867. [DOI] [PubMed] [Google Scholar]
- 36.Kawano M, Tanaka K, Itonaga I, Iwasaki T, Tsumura H. c-Myc Represses Tumor-Suppressive microRNAs, let-7a, miR-16 and miR-29b, and Induces Cyclin D2-Mediated Cell Proliferation in Ewing’s Sarcoma Cell Line. PLoS One. 2015;10(9):e138560. doi: 10.1371/journal.pone.0138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, Qi M, Ma C, et al. Let7a inhibits the growth of endometrial carcinoma cells by targeting Aurora-B. FEBS Lett. 2013;587(16):2523–2529. doi: 10.1016/j.febslet.2013.05.065. [DOI] [PubMed] [Google Scholar]
- 38.Zhang XX, Fu Z, Zhang Z, et al. Microcystin-LR promotes melanoma cell invasion and enhances matrix metalloproteinase-2/-9 expression mediated by NF-κB activation. Environ Sci Technol. 2012;46(20):11319–11326. doi: 10.1021/es3024989. [DOI] [PubMed] [Google Scholar]
- 39.Bai XY, Li S, Wang M, et al. Krüppel-like factor 9 down-regulates matrix metalloproteinase 9 transcription and suppresses human breast cancer invasion. Cancer Lett. 2018;412:224–235. doi: 10.1016/j.canlet.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 40.Chen QJ, Shi Y, Shi JF, et al. Liver X receptors agonist T0901317 down-regulates matrix metalloproteinase-9 expression in non-small-cell lung cancer by repressing nuclear factor-κB. Anticancer Drugs. 2017;28(9):952–958. doi: 10.1097/CAD.0000000000000532. [DOI] [PubMed] [Google Scholar]
- 41.Yang CJ, Liu YP, Dai HY, et al. Nuclear HDAC6 inhibits invasion by suppressing NF-κB/MMP2 and is inversely correlated with metastasis of non-small cell lung cancer. Oncotarget. 2015;6(30):30263–30276. doi: 10.18632/oncotarget.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu ZL, Mao JH, Peng AF, et al. Inhibition of fatty acid synthase suppresses osteosarcoma cell invasion and migration via downregulation of the PI3K/Akt signaling pathway in vitro. Mol Med Rep. 2013;7(2):608–612. doi: 10.3892/mmr.2012.1220. [DOI] [PubMed] [Google Scholar]