Abstract
Introduction
Human malignant melanoma (MM) is a highly malignant tumor of cutaneous melanocytes with a fast progression. We investigated the cellular effects of pyrroline-5- carboxylate reductase 1 (PYCR1) in the MM cell lines, A375 and M14.
Methods
Cell Counting Kit-8 assay, transwell assay, and flow cytometry analysis were performed to evaluate the proliferation, migration and apoptosis of MM cell lines, respectively. To gain more insight into the role of PYCR1 in tumor growth, we analyzed the AKT phosphorylation level in PYCR1-specific siRNA (siPYCR1) and negative control (NC) cells.
Results
Biochemical analysis revealed a clear increase in PYCR1 expression in human MM samples, and its high expression predicted a poor prognosis. Silencing of PYCR1 suppressed the proliferation and migration of A375 and M14 cells. The percentage of apoptosis in cells transfected with siPYCR1 significantly increased in comparison to that of cells transfected with negative control siRNA (NC). The enhanced apoptosis in PYCR1 knockdown cells was consistent with an increased level of markers of apoptosis. siPYCR1 inhibited AKT phosphorylation, as well as the expression of its downstream protein, P70, suggesting that PYCR1 promoted cell growth of the MM cell lines A375 and M14 through stimulation of the AKT pathway. Moreover, forkhead box K2 and regulatory associated protein of MTOR complex 1 shared a similar expression pattern to that of PYCR1 and were significantly downregulated in PYCR1 knockdown cells.
Conclusion
PYCR1 promoted tumor progression through the AKT pathway in human MM in vitro. Our results expand the knowledge of PYCR1 functions in solid tumors and provide a potential target for the clinical treatment of human MM.
Keywords: apoptosis, proliferation, prognosis, AKT pathway
Introduction
Human malignant melanoma (MM) is a highly malignant and rapidly progressing tumor derived from cutaneous melanocytes. MM is one of the leading causes of death from skin cancer, and its incidence is increasing every year.1,2 Currently, the clinical treatment of MM is limited to systemic chemotherapy. Although there is a certain beneficial effect, the 10-year survival rate of MM patients is still less than 10%. With continuous efforts of researchers, the knowledge of the pathogenesis of MM has made notable progress; however, the precise mechanism of the occurrence and development of MM remains unknown.3,4 Therefore, investigating the molecular mechanism of melanoma pathogenesis is the focus of current research.
Pyrroline-5-carboxylate reductase (PYCR) is a key enzyme in proline metabolism in organisms. As an important housekeeping protein in prokaryotes and eukaryotes, PYCR catalyzes the transformation of P5C to proline with the oxidation of NAD(P)H to NAD(P)+.5,6 Currently, three PYCR family members have been identified in humans, namely PYCR1 (17q25.3), PYCR2 (1q42.13) and PYCRL (8q24.3).7 PYCR1 is the member that plays a leading role in humans.8 Previous research has established that not only is proline necessary for cells to maintain normal physiological function, but it also plays a special role in the occurrence of tumors.9 Several enzymes involved in proline metabolism play an important role in tumorigenesis.10,11 In recent years, PYCR1, a key enzyme in proline metabolism, was gradually found to be involved in the progression of tumors, including breast cancer, lung cancer, prostate cancer and lymphoma.12–14 Nonetheless, the research on the role and mechanism of PYCR1 in tumors is still at its early stage.
While studying prophase, a biochemical analysis revealed a clear increase of PYCR1 expression in 468 cases of cutaneous melanoma in comparison to 558 cases of normal skin tissues. A survival analysis showed that the overall survival (OS) of patients with low PYCR1 expression was significantly better than that of patients with high PYCR1 expression. In the present study, we demonstrated that PYCR1 plays an oncogenic role in MM by contributing to the regulation of cell proliferation, migration and apoptosis.
Materials and methods
Cell lines
Human MM cell lines, A375 and M14, were obtained from the Type Culture Collection of the Chinese Academy of Sciences. A375 and M14 cells were cultured in DMEM supplemented with 10% of fetal bovine serum (FBS) and incubated in a humidified atmosphere at 37°C with 5% CO2. When cells reached approximately 70% confluency, they were transfected with PYCR1-specific siRNA (siPYCR1) or negative control siRNA (NC) by using Lipofectamine 2000 according to the manufacturer’s guidelines. Cells were treated with 10 ng/mL IGF-1 (Proteintech) for the verification of signaling pathway.
Reverse transcription quantitative PCR (qPCR)
Total RNA was isolated from A375 and M14 cells after 48 hours of transfection with PYCR1 or NC siRNA using an Ultrapure RNA Kit (CWBIO, Beijing, China) and was reverse transcribed with a Reverse Transcription Reaction Kit (CWBIO) according to the manufacturer’s manual. qPCR was performed by using gene-specific primer pairs for PYCR1 (5′-TGGCTGCCCACAAGATAATG-3′ and 5′-TCAATGTCGGCGCCTATTTC-3′), RPTOR (5′-CACAGAGGGAGGGAGTTTGAC-3′ and 5′-GCCACCAGATTCCTCTGTCAA-3′), RAC3 (5′-CGAGAATGTTCGTGCCAAGTG-3′ and 5′-GGCTGAGCACTCCAGGTATTT-3′), FOXK2 (5′-GTCTTCAGGGTACAAGGTGGG-3′ and 5′-GCTTTGAATCATCCTTCGGGC-3′) and β-actin as a control (5′-CCCGAGCCGTGTTTCCT-3′ and 5′-GTCCCAGTTGGTGACGATGC-3′) on an H-4800 Real-Time PCR System using Super TaqMan Mixture (CWBIO) per the manufacturer’s instructions.
Western blot
For the immunoblot analysis, A375 and M14 cells were collected and lysed with RIPA buffer after 24 hours of transfection. Forty micrograms of lysates was separated on a 10% SDS-PAGE gel, and then transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% of fat-free milk for 1 hour, and probed with primary (1:1,000) and secondary (1:5,000) antibodies. The subsequent immunoblot analysis was performed using ECL. The primary antibodies used for the immunoblot analysis were PYCR1 (Proteintech), AKT (Cell Signaling Technology), p-AKT (Cell Signaling Technology), Caspase3-p17 (Proteintech), Bax (Proteintech), Bcl2 (Proteintech), p70 (Abcam) and α-Tubulin (Abcam). The densitometry was measured using ImageJ software, and the protein levels were normalized to α-Tubulin.
Cell Counting Kit-8 (CCK8) assay for proliferation detection
The proliferation of A375 and M14 cells was assessed by using CCK8. Cells were transfected with PYCR1 or NC siRNA, and then transferred into 96-well plates at a density of 5,000 cells per well in 200 µL of culture medium. At every 24 hours, 10 µL of CCK8 solution was added to each well and cells were cultured for 2 hours. Then, the OD values were measured on a microplate reader. Each experiment was performed in triplicate.
Transwell assay for migration detection
Transwell assay was performed to evaluate cell migration using a transwell chamber (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. For the migration assay, cells were seeded in a chamber at a density of 1×104 cells/well. After 48 hours of incubation at 37°C, cells that had migrated to the lower chamber were washed with PBS and stained with crystal violet for 10 minutes. The average number of migrating cells was calculated from triplicates.
Flow cytometric analysis for apoptosis detection
Flow cytometric analysis was performed to evaluate the level of apoptosis by staining MM cells with propidium iodide (PI) and annexin V-fluorescein isothiocyanate (FITC) according to the manufacturer’s manual. To determine the number of apoptotic cells, A375 and M14 cells transfected with siRNA were starved for 24 hours and stained with annexin V-FITC/PI for 5 minutes. Cell samples were analyzed using flow cytometric analysis and the Flowjo software. Experiments were performed in triplicate.
Statistical analyses
All results in this study were presented as the mean ± standard deviation (SD). All analyses were evaluated using SPSS v17.0 software (SPSS Inc., Chicago, IL, USA). The statistical significance of differences between NC and siPYCR1 groups was calculated using one-way analysis of variance (ANOVA). Differences with P-values less than 0.05 were considered statistically significant.
Results
PYCR1 expression was upregulated and predicted a poor prognosis in human melanoma
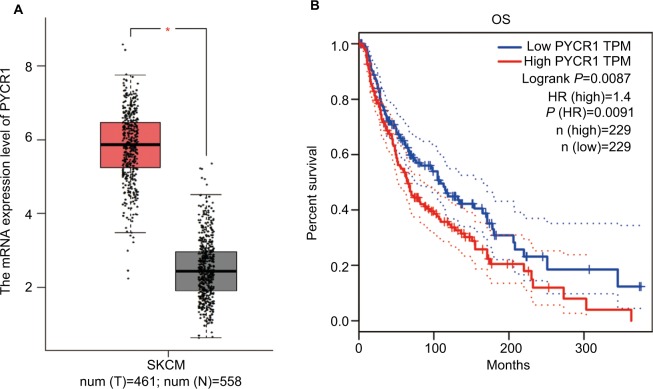
With the development of sequencing technology, an overwhelming amount of cancer genomics data have been generated, and genome-wide individual genes analysis is now possible. Here, we assessed PYCR1 expression and its prognosis role using Gene Expression Profiling Interactive Analysis (GEPIA),15 which was specifically designed to explore gene expression profiles and to perform interactive analyses using the dataset resourced from Genotype-Tissue Expression and the Cancer Genome Atlas.16,17 For the expression analysis, the |Log2FC| cutoff was 1 and the log2(TPM+1) was used for log-scale. For the prognosis analysis, the group cutoff was median, and the hazard ratio (HR) was calculated based on the Cox PH Model. Figure 1 shows the results of the bioinformatics analysis. From the plot, we proved that PYCR1 expression was significantly upregulated in human skin cutaneous melanoma (SKCM) samples in comparison to normal skin tissues. Moreover, the survival curve was plotted for SKCM patients, who were divided into two groups, PYCR1 high expression (n=229) and low expression (n=229) groups. Patients with low PYCR1 level showed better OS compared with patients with high PYCR1 level.
Figure 1.
PYCR1 expression was upregulated and predicted a poor prognosis in human MM.
Notes: The expression and prognosis role of PYCR1 were assessed by using the GEPIA online database. (A) PYCR1 expression was significantly increased in SKCM tumor tissues (n=461, red box) in comparison to normal skin tissue (n=558, gray box). (B) The survival curve was plotted in SKCM patients who were divided into PYCR1 high-expression group (n=229) and PYCR1 low-expression group (n=229). As seen from the curve, SKCM patients with low PYCR1 level showed better OS than those with high PYCR1 level. *P<0.05.
Abbreviations: MM, malignant melanoma; GEPIA, Gene Expression Profiling Interactive Analysis; SKCM, skin cutaneous melanoma; OS, overall survival; TPM, transcripts per million; PYCR1, pyrroline-5-carboxylate reductase 1.
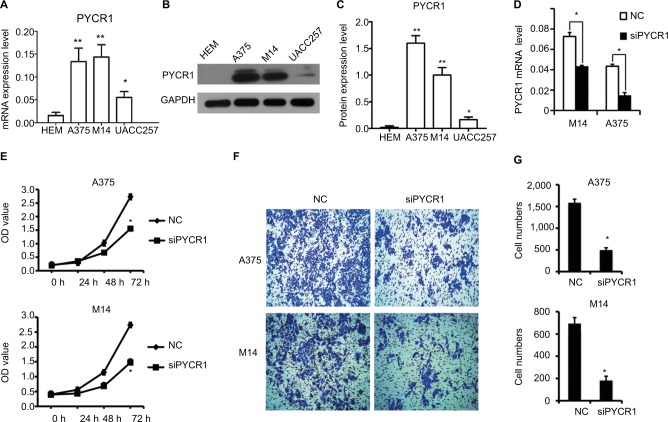
We further analyzed the different expression level of PYCR1 between normal skin and MM cells. One human normal melanocyte cell line, HEM, and three human melanoma cell lines, A375, M14 and UACC257, were used to investigate the difference in PYCR1 expression. From the qPCR (Figure 2A) and Western blot (Figure 2B and C) results, PYCR1 was upregulated in A375, M14 and UACC257 cell lines compared with the HEM cell line. The differences between A375, M14 and HEM were statistically different (P<0.01). The above results demonstrated that PYCR1 might contribute to the regulation of tumor cell activity in human MM.
Figure 2.
Knockdown of PYCR1 inhibited proliferation and migration in human MM cells.
Notes: PYCR1 expression was analyzed by using (A) qPCR and (B and C) Western blotting in three human melanoma cell lines, A375, M14 and UACC257, and in one human normal melanocyte cell line, HEM. PYCR1 level was upregulated in A375, M14 and UACC257 cell lines compared with HEM. (D) PYCR1 expression was significantly suppressed by PYCR1 siRNA in A375 and M14 cells. (E) CCK8 assay was performed to assess the proliferation of siPYCR1 and NC cells. After 72 hours of transfection, the OD value of siPYCR1 cells decreased significantly. (F and G) Cell migration was determined by transwell assay. The number of migrating cells significantly declined in the siPYCR1 group compared to the NC group. *P<0.05 and **P<0.01.
Abbreviations: MM, malignant melanoma; qPCR, quantitative PCR; CCK8, Cell Counting Kit-8; PYCR1, pyrroline-5-carboxylate reductase 1; siPYCR1, PYCR1-specific siRNA; HEM, human epidermal melanocyte cell line; NC, negative control.
Knockdown of PYCR1 blocked cell proliferation in human MM
PYCR1 expression was suppressed by siPYCR1 to elucidate its biological function in A375 and M14 cells. As seen in Figure 2D, PYCR1 expression was significantly inhibited in A375 and M14 cells transfected with siPYCR1 compared with cells transfected with NC. Next, we investigated the effect of PYCR1 knockdown on the proliferation of human MM cell lines using CCK8 assay (Figure 2E). At 72 hours after transfection of the A375 cell line, the OD value of siPYCR1 cells (1.55±0.04) was statistically and significantly decreased compared with that of NC cells (2.72±0.1, P<0.05). A significant difference was also found between siPYCR1 (1.47±0.03) and NC (2.74±0.1, P<0.05) groups in the M14 cell line. These in vitro results proved that silencing of PYCR1 inhibited the proliferation of MM cells.
Knockdown of PYCR1 inhibited cell migration in human MM
To gain more insight into the role of PYCR1 in human MM, a transwell assay was performed to estimate the migration ability of A375 and M14 cells transfected with control siRNA or siPYCR1. After transfection, cells were seeded at a density of 1×104 cells/well into the upper chamber and incubated for 48 hours. Then, migrating cells were stained and imaged with a microscope. Representative images and statistical analyses are shown in Figure 2F and G, respectively. From the results, the number of migrating cells in the siPYCR1 group (495±55) significantly declined compared to that in the NC group (1,586±83) when using the A375 cell line. Meanwhile, the number of migrating cells in the siPYCR1 group (190±21) also significantly declined compared to that in the NC group (682±59) when using the M14 cell line. These in vitro results confirmed that silencing of PYCR1 inhibited the migration of MM cells.
Knockdown of PYCR1 induced apoptosis in human MM cell lines
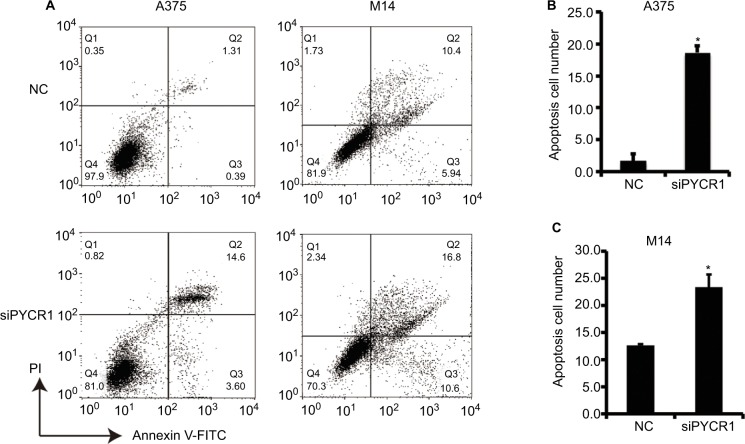
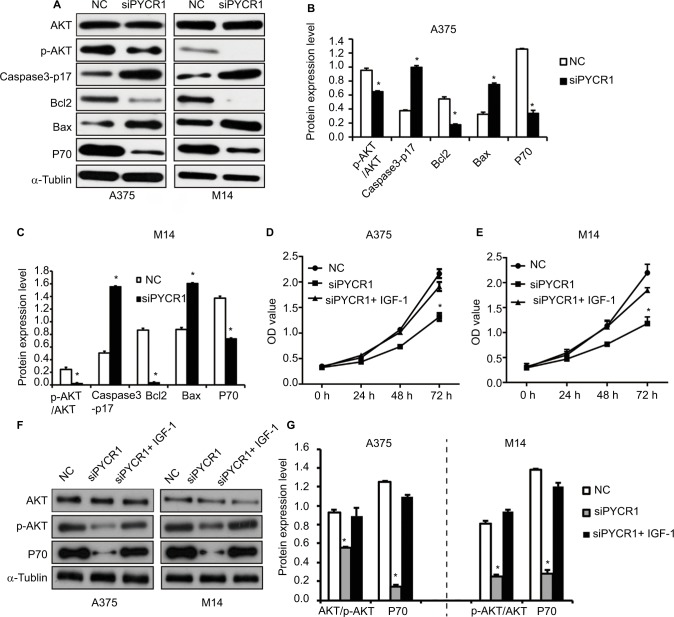
Previous studies have proved that proline metabolism is involved in the regulation of cell apoptosis, but there is no direct research on the role of PYCR1 in cell apoptosis.18 In the present study, the effect of PYCR1 on cellular apoptosis was estimated using flow cytometric analysis. As shown in Figure 3, the percentage of apoptosis in A375 cells transfected with siPYCR1 significantly increased to 27.96±1.25 when compared with the NC group, 12.37±1.28. Meanwhile, the apoptotic percentage of M14 cells transfected with siPYCR1 also significantly increased (23.79±2.52) in comparison to the NC group (12.61±0.11, P<0.05). Furthermore, we assessed the expressions of apoptosis-related key genes by Western blot analysis, and the results are shown in Figure 4A. Silencing of PYCR1 increased the level of the apoptosis markers, Caspase3-p17 and Bax, and suppressed Bcl2 level in both A375 and M14 cell lines (P<0.05), which further confirmed the negative role of PYCR1 in cell apoptosis (Figure 4B and C).
Figure 3.
Knockdown of PYCR1 induced apoptosis in human MM cells.
Notes: (A) Flow cytometric analysis was performed to estimate cellular apoptosis. (B) The percentage of apoptosis in A375 cells transfected with siPYCR1 significantly increased to 18.71±1.01 in comparison to that of the NC group, 1.71±1.02. (C) The percentage of apoptosis in M14 cells transfected with siPYCR1 significantly increased to 23.79±2.52 in comparison to that of the NC group, 12.61±0.11. *P<0.05.
Abbreviations: MM, malignant melanoma; FITC, fluorescein isothiocyanate; PI, propidium iodide; PYCR1, pyrroline-5-carboxylate reductase; siPYCR1, PYCR1-specific siRNA; NC, negative control.
Figure 4.
PYCR1 played a role in tumor progression of human MM through the stimulation of the AKT pathway.
Notes: (A) The expression of AKT signaling pathway-related genes was analyzed by Western blot. (B and C) Statistical analysis of Western blot results was performed using ImageJ software. siPYCR1 inhibited AKT phosphorylation and P70 level, and promoted the expression of the apoptosis factors, Caspase3-p17 and Bax in A375 and M14 cell lines. (D and E) Rescue experiments were performed by activating the AKT pathway with IGF-1 in PYCR1 interference cells (siPYCR1+ IGF-1). CCK8 was used to detect cell proliferation in three cell groups. (F) The expression of AKT signaling pathway-related genes was analyzed by Western blot. (G) Statistical analysis of Western blot results was performed using ImageJ software. *P<0.05.
Abbreviations: MM, malignant melanoma; CCK8, Cell Counting Kit-8; PYCR1, pyrroline-5-carboxylate reductase; siPYCR1, PYCR1-specific siRNA; NC, negative control.
Analysis of the signaling pathway by which PYCR1 promoted cell growth in MM
We analyzed the phosphorylation level of AKT in siPYCR1 and NC cells, a key protein in the PI3K/AKT pathway, which has a vital role in tumorigenesis. The level of AKT phosphorylation was determined using Western blot analysis and analyzed using ImageJ software. The phosphorylated site of AKT we detected was T308. As shown in Figure 4A–C, siPYCR1 inhibited the ratio of p-AKT/AKT in A375 and M14 cells, as well as its downstream protein, P70. To prove the effect of PYCR1 on the AKT pathway, we performed rescue experiments by activating the AKT pathway in PYCR1 interference cells with insulin-like growth factor 1 (IGF-1), which activated T308 phosphorylated sites of AKT, same with siPYCR1. After the treatment with IGF-1, the proliferation ability of PYCR1 knockdown cells was rescued to the control level (Figure 4D and E), as well as AKT phosphorylation and P70 expression (Figure 4F and G). This result demonstrated that PYCR1 promoted cell proliferation by activating the AKT pathway.
Detection of PYCR1 similar genes in human MM
To gain more insight into the mechanism by which PYCR1 was involved in tumor growth, we analyzed PYCR1 coexpression genes in human MM using GEPIA to screen for similar genes. The top 10 genes, generated by GEPIA, that shared a similar expression pattern to that of PYCR1 in melanoma are shown in Table 1. Among the top five genes, forkhead box K2 (FOXK2), regulatory associated protein of MTOR complex 1 (RPTOR) and Rac family small GTPase 3 (RAC3) have been reported to play a role in tumor progression, so we tested if there was a correlation between PYCR1 and their expression levels in MM cell lines. As shown in Figure 5, FOXK2, RPTOR and RAC3 showed a similar expression pattern to that of PYCR1 in SKCM, so their mRNA levels were then determined by qPCR in A375 cell line. From the qPCR results, FOXK2 and RPTOR expression levels significantly declined in cells transfected with siPYCR1 compared to NC cells. However, there was no significant difference in RAC3 expression levels between siPYCR1 cells and NC cells. The above results confirmed that PYCR1 could affect the expression of FOXK2 and RPTOR in MM cells.
Table 1.
Top 10 genes generated by GEPIA that show a similar expression pattern to that of PYCR1 in melanoma
| Gene symbol | Gene ID | PCC |
|---|---|---|
| FOXK2 | ENSG00000141568.19 | 0.54 |
| YARS | ENSG00000134684.10 | 0.5 |
| RAC3 | ENSG00000169750.8 | 0.5 |
| RPTOR | ENSG00000141564.13 | 0.49 |
| MAFG-AS1 | ENSG00000265688.1 | 0.49 |
| GPS1 | ENSG00000169727.12 | 0.48 |
| FAAP100 | ENSG00000185504.16 | 0.48 |
| MRPL12 | ENSG00000262814.6 | 0.47 |
| TMEM214 | ENSG00000119777.18 | 0.46 |
| TMEM104 | ENSG00000109066.13 | 0.45 |
Abbreviations: GEPIA, Gene Expression Profiling Interactive Analysis; PCC, Pearson’s correlation coefficient; PYCR1, pyrroline-5-carboxylate reductase.
Figure 5.
Analysis and detection of PYCR1 coexpression genes in human MM.
Notes: (A–C) PYCR1 coexpression genes were identified using the GEPIA dataset. FOXK2, RPTOR and RAC3 showed a similar expression pattern to that of PYCR1 in SKCM. (D–F) siPYCR1 inhibited FOXK2 and RPTOR expression, but not RAC3 expression in A375 cells. *P<0.05.
Abbreviations: MM, malignant melanoma; NC, negative control; GEPIA, Gene Expression Profiling Interactive Analysis; SKCM, skin cutaneous melanoma; FOXK2, forkhead box K2; PYCR1, pyrroline-5-carboxylate reductase; RPTOR, regulatory associated protein of MTOR complex 1.
Discussion
The human PYCR1 gene originally identified in mitochondria codes for a key enzyme of the proline synthesis pathway, which catalyzes the conversion of P5C into proline. PYCR1 can protect cells from mitochondrial damage caused by oxidative stress, and play an important role in the normal physiological function of cells and in the occurrence of many diseases.19,20 Several studies have proved that proline has an antiapoptotic effect, while P5C has a positive role in apoptosis. Thus, PYCR1 is considered to be involved in the initiation of cell apoptosis. In addition, a recent study has confirmed that PYCR1 was regulated by the oncogenic transcription factor c-MYC (MYC) and was overexpressed in renal carcinoma, which in turn activated the proline metabolism and proliferation of tumor cells. Moreover, MYC can also increase the expression of PYCR1 in human lymphoma and prostate cancer, which can lead to the increase of proline synthesis and to the survival of tumor cells.21 Metabolic disorder of proline catabolism is observed in various cancers, and is closely related to the extracellular matrix degradation mediated by metalloproteinase (MMP); however, its specific pathway remains unclear.22 The latest research has shown that PYCR1 is overexpressed in breast cancer. Silencing of PYCR1 inhibited the proliferation and invasion ability of the breast cancer cell lines MCF-7 (ER positive) and MDA-MB-231 (ER negative) by regulating the AKT/ERK pathway.13
In the present study, we first confirmed that PYCR1 promoted tumor progression in human MM cells in vitro. Knockdown of PYCR1 blocked cell proliferation and migration, and induced apoptosis in the human MM cell lines A375 and M14. Kardos et al confirmed that aldehyde dehydrogenase 18 family, member A1 (ALDH18A1) plays a vital role in the regulation of MM cell growth via proline biosynthesis. The silencing of ALDH18A1 activated general control nonderepressible 2 (GCN2) and suppressed protein synthesis, which was reversed with proline supplementation.23 De Ingeniis et al have tracked the fate of 113C-labeled precursors to assess the role of three isozymic versions of PYCR in 10 human melanoma cell lines. PYCR1 and PYCR2 were localized in the mitochondria, and primarily linked to the transformation of glutamate to proline. In contrast, PYCRL was localized in the cytosol, and exclusively involved in the transformation of ornithine to proline. Moreover, De Ingeniis et al have demonstrated that PYCR1 is the leading member of PYCR that contributes to the proline metabolism in melanoma cells.7 All these research studies proved a significant role for PYCR1 in human MM.
However, the specific function and related mechanism of PYCR1 in tumorigenesis of human MM have not been investigated. In the present study, we found that knockdown of PYCR1 inhibited the AKT signaling pathway in A375 and M14 cells, which was confirmed by the rescue experiment. Our results were consistent with the latest research on the role of PYCR1 in breast cancer, suggesting that the AKT signaling pathway may be a specific pathway for PYCR1 to participate in tumor progression.13 To gain more insight into the mechanism by which PYCR1 promoted tumor growth, we analyzed and verified the expression of PYCR1 coexpression genes in human MM. siPYCR1 suppressed the expression of two PYCR1 coexpression genes, FOXK2 and RPTOR, which declined significantly in MM cells transfected with siPYCR1. As a transcription factor, FOXK2 has been found to play a role in a variety of tumors including liver cancer, lung cancer and breast cancer, but its specific mechanism remains to be further studied. RPTOR is an important component of mTORC1, and mTORC1 is an important member of the mTOR signaling pathway. However, according to the present knowledge, the function of these two proteins remains to be further studied.24–26 Our results proved that PYCR1 may affect the expression of FOXK2 and RPTOR. We predict that PYCR1, FOXK2 and RPTOR belong to the same regulatory network, either by direct or indirect protein interactions; however, this will require more experimental research, which is the goal of our future research direction.
Conclusion
Our biochemical analysis revealed a clear increase of PYCR1 expression in human MM samples, and PYCR1 low expression predicted a better prognosis. Silencing of PYCR1 blocked cell proliferation and migration, and promoted cell apoptosis in human MM cells by inhibiting the AKT signaling pathway. Our results contribute to expand the knowledge of PYCR1 functions in promoting tumor progression, and provide a potential target for the clinical treatment of human MM.
Acknowledgments
The present study was supported by the Key Discipline Project of Ningbo No.2 Hospital (grant no. 2016013).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Niezgoda A, Niezgoda P, Czajkowski R. Novel Approaches to Treatment of Advanced Melanoma: A Review on Targeted Therapy and Immunotherapy. Biomed Res Int. 2015;2015:1–16. doi: 10.1155/2015/851387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garbe C, Eigentler TK, Keilholz U, Hauschild A, Kirkwood JM. Systematic review of medical treatment in melanoma: current status and future prospects. Oncologist. 2011;16(1):5–24. doi: 10.1634/theoncologist.2010-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millet A, Martin AR, Ronco C, Rocchi S, Benhida R. Metastatic Melanoma: Insights Into the Evolution of the Treatments and Future Challenges. Med Res Rev. 2017;37(1):98–148. doi: 10.1002/med.21404. [DOI] [PubMed] [Google Scholar]
- 4.Lázár-Molnár E, Hegyesi H, Pállinger E, et al. Inhibition of human primary melanoma cell proliferation by histamine is enhanced by interleukin-6. Eur J Clin Invest. 2002;32(10):743–749. doi: 10.1046/j.1365-2362.2002.01020.x. [DOI] [PubMed] [Google Scholar]
- 5.Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15(2):89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Kukavica-Ibrulj I, Sanschagrin F, Peterson A, et al. Functional genomics of PycR, a LysR family transcriptional regulator essential for maintenance of Pseudomonas aeruginosa in the rat lung. Microbiology. 2008;154(Pt 7):2106–2118. doi: 10.1099/mic.0.2007/011239-0. [DOI] [PubMed] [Google Scholar]
- 7.De Ingeniis J, Ratnikov B, Richardson AD, et al. Functional specialization in proline biosynthesis of melanoma. PLoS One. 2012;7(9):e45190. doi: 10.1371/journal.pone.0045190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kretz R, Bozorgmehr B, Kariminejad MH, et al. Defect in proline synthesis: pyrroline-5-carboxylate reductase 1 deficiency leads to a complex clinical phenotype with collagen and elastin abnormalities. J Inherit Metab Dis. 2011;34(3):731–739. doi: 10.1007/s10545-011-9319-3. [DOI] [PubMed] [Google Scholar]
- 9.Phang JM, Liu W, Hancock CN, Fischer JW. Proline metabolism and cancer: emerging links to glutamine and collagen. Curr Opin Clin Nutr Metab Care. 2015;18(1):71. doi: 10.1097/MCO.0000000000000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phang JM, Liu W. Proline metabolism and cancer. Front Biosci. 2012;17:1835. doi: 10.2741/4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu CA, Bart Williams D, Zhaorigetu S, Khalil S, Wan G, Valle D. Functional genomics and SNP analysis of human genes encoding proline metabolic enzymes. Amino Acids. 2008;35(4):655. doi: 10.1007/s00726-008-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng T, Zhu L, Liao M, et al. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol. 2017;34(2):27. doi: 10.1007/s12032-016-0870-5. [DOI] [PubMed] [Google Scholar]
- 13.Ding J, Kuo ML, Su L, et al. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis. 2017;38(5):519–531. doi: 10.1093/carcin/bgx022. [DOI] [PubMed] [Google Scholar]
- 14.Cai F, Miao Y, Liu C, et al. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol Lett. 2018;15(1):731–740. doi: 10.3892/ol.2017.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mclendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 2008 Sep 4;455(7216):1061–1068. doi: 10.1038/nature07385. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arentson BW, Sanyal N, Becker DF. Substrate channeling in proline metabolism. Front Biosci. 2012;17:375. doi: 10.2741/3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashraf M, Foolad MR. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot. 2007;59(2):206–216. [Google Scholar]
- 20.Yasuda T, Kaji Y, Agatsuma T, et al. DJ-1 cooperates with PYCR1 in cell protection against oxidative stress. Biochem Biophys Res Commun. 2013;436(2):289–294. doi: 10.1016/j.bbrc.2013.05.095. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Le A, Hancock C, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A. 2012;109(23):8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kardos GR, Wastyk HC, Robertson GP. Disruption of Proline Synthesis in Melanoma Inhibits Protein Production Mediated by the GCN2 Pathway. Mol Cancer Res. 2015;13(10):1408–1420. doi: 10.1158/1541-7786.MCR-15-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Ao X, Jia Z, et al. FOXK2 transcription factor suppresses ERα-positive breast cancer cell growth through down-regulating the stability of ERα via mechanism involving BRCA1/BARD1. Sci Rep. 2015;5:8796. doi: 10.1038/srep08796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Y, Xia S, Feng Z. Sox9 mediated transcriptional activation of FOXK2 is critical for colorectal cancer cells proliferation. Biochem Biophys Res Commun. 2017;483(1):475–481. doi: 10.1016/j.bbrc.2016.12.119. [DOI] [PubMed] [Google Scholar]
- 26.Dugué PA, Milne RL, Southey MC. A prospective study of peripheral blood DNA methylation at RPTOR, MGRN1 and RAPSN and risk of breast cancer. Breast Cancer Res Treat. 2017;161(1):181–183. doi: 10.1007/s10549-016-4032-4. [DOI] [PubMed] [Google Scholar]