Abstract
Discoveries made during the 1918 influenza A pandemic and reports of severe disease associated with coinfection during the 2009 hemagglutinin type 1 and neuraminidase type 1 (commonly known as H1N1 or swine flu) pandemic have renewed interest in the role of coinfection in disease pathogenesis. The authors assessed how various timings of coinfection with influenza virus and pneumonia-causing bacteria could affect the severity of illness at multiple levels of interaction, including the biologic and population levels. Animal studies most strongly support a single pathway of coinfection with influenza inoculation occurring approximately 7 days before inoculation with Streptococcus pneumoniae, but less-examined pathways of infection also may be important for human disease. The authors discussed the implications of each pathway for disease prevention and what they would expect to see at the population level if there were sufficient data available. Lastly, the authors identified crucial gaps in the study of timing of coinfection and proposed related research questions.
Keywords: community-acquired infections; influenza, human; pneumococcal infections; pneumonia
Community-acquired pneumonia (CAP) often follows influenza infection. The hypothesized synergistic interaction resulting from coinfection with influenza and agents of CAP is thought to be a major factor in the severity of the 1918 influenza A pandemic (1, 2). Today, up to 20% of persons who have CAP show evidence of recent exposure to the influenza virus (3), and pneumonia is a leading indicator of influenza severity (4). Although Staphylococcus aureus is a relatively uncommon cause of CAP after influenza infection, methicillin-resistant S. aureus has been considered an important pathogen in deaths of coinfected pediatric patients. Data from the Centers for Disease Control and Prevention on the 2004–2007 influenza seasons showed that methicillin-resistant S. aureus was present in 60% of the 20 pediatric patients who died from S. aureus coinfection, with the highest rate during the 2006–2007 season (5), which suggests that the problem of antibiotic resistance among children with CAP in this age group is growing.
Influenza is transmitted via respiratory droplets, through either direct or indirect contact, and is highly infectious. Each year, seasonal influenza infects hundreds of thousands of people worldwide and accounts for an estimated 36,000 deaths in the United States (6), with pandemic strains often resulting in higher mortality rates (7, 8). Complications from influenza are most severe in the young, the elderly, and persons with compromised immune systems (9). Bacteria that cause CAP are transmitted through person-to-person direct contact; however, the rates of infectivity and progression to pneumonia are thought to be lower than those of influenza, and many of the CAP-causing bacteria are found in healthy individuals. The most common causes of CAP are Staphylococcus pneumoniae, S. aureus, Haemophilus influenzae, Mycoplasma pneumoniae, and Chlamydophila pneumoniae, although there are regional variations (10). Although investigators performing studies in which they used animals and experimental systems have begun to address how coinfection may enhance pathogenesis, there is little in the literature on the impact of coinfection on transmission of or susceptibility to bacteria. Perhaps most importantly, although population-level synergy has been observed, the timing of transmission related to the order of infection from each agent has not been definitively established (11, 12). Understanding the order and timing of this synergistic relation and the resulting population-level effects is especially relevant to epidemiologists who are engaged in planning for pandemics.
In the present commentary, we use S. pneumoniae, the most common cause of CAP, as a model organism to explore the relation between influenza A and CAP. With over 90 identified serotypes and multiple licensed vaccines, S. pneumoniae remains one of the best researched of the pneumonia-causing bacteria (13). We present 3 pathways to coinfection and discuss the impact of order and timing of coinfection on what epidemiologic patterns might be observed. We close with a discussion of outstanding research questions and their implications for CAP prevention.
PATHWAYS TO COINFECTION
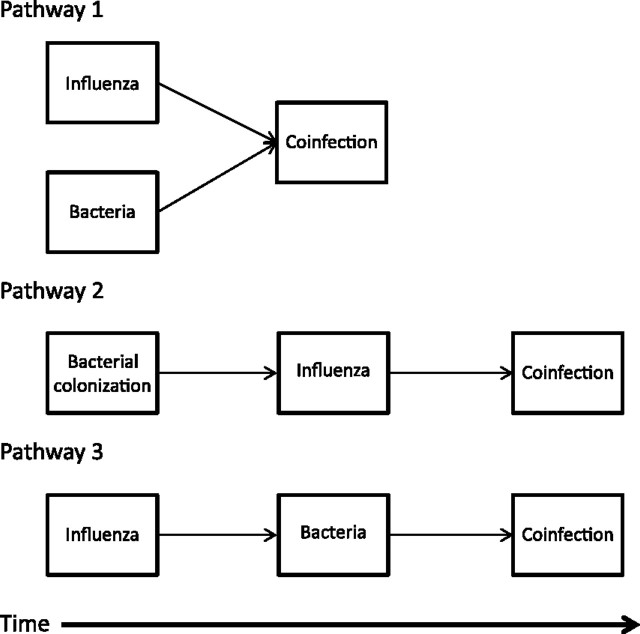
There are 3 possible pathways to coinfection: 1) infections with both etiologic agents occur essentially simultaneously; 2) S. pneumoniae colonization precedes influenza infection; and 3) influenza infection precedes S. pneumoniae colonization (Figure 1). Each of these pathways is possible (although they likely occur at different frequencies); however, we expect the resulting population patterns to differ depending on the relative frequency of each pathway, as will the implications for control measures (discussed below).
Figure 1.
Schematic of 3 pathways of influenza and bacterial pneumonia exposure that lead to coinfection.
Simultaneous acquisition of influenza and S. pneumoniae
Viruses and bacteria can be transmitted together through coughing or sneezing (11, 14–16). However, the extent to which simultaneous infections with influenza and S. pneumoniae lead to CAP is uncertain. In one of the few animal studies in which simultaneous infections were studied, investigators found no evidence of lethal synergy in mice infected with sublethal titers of both influenza and S. pneumoniae (17). In humans, influenza infection alone increases cough, potentially facilitating transmission of both viruses and bacteria (4). Additionally, influenza replicates more rapidly than does bacteria, so even if the infections are acquired simultaneously, the clinical manifestations in an individual would appear sequentially. At the population level, we might expect little or no lag between peaks in influenza and pneumonia occurrence after simultaneous infection. Potential course-of-infection and research questions related to concurrent infection are presented in Table 1.
Table 1.
Examples of Relevant Research Questions for Each Pathway of Influenza and Bacterial Pneumonia Coinfection
| Pathway | Relevant Research Questions | Population Implications |
| Cotransmission | Is a cough or sneeze sufficient for simultaneous infection, or must fomites play a role? | Short or no lag observed between peak of influenza infection and peak of bacterial pneumonia infection |
| Does bacterial survival depend upon droplet size? | ||
| Bacterial colonization followed by influenza infection | Does the body exert more effort to keep certain bacteria in check? If so, is an individual with those bacteria more likely to become infected after influenza infection? | Short lag between peak of influenza infection and peak of bacterial pneumonia infection |
| Can probiotics or prebiotics be used to prevent colonization with pneumonia-causing bacteria? | ||
| Influenza infection followed by bacterial colonization | What role do children play in transmitting bacteria to influenza-infected adults? | Long lag between peak of influenza infection and peak of bacterial pneumonia infection |
| How can we prime the immune system to prevent secondary infection from bacterial pathogens? |
S. pneumoniae colonization preceding influenza
If S. pneumoniae colonization precedes influenza infection, changes in the upper and lower respiratory tracts resulting from the influenza infection would enable the colonizing strain of S. pneumoniae to successfully avoid a host immune response, invade the lung, and cause pneumonia. Although it is assumed that bacteria that colonize the nasopharynx frequently enter the lungs, the host immune response is usually sufficient to prevent infection. The introduction of influenza primes the lungs for bacterial colonization and adversely affects the host response, leading to secondary pneumonia (4, 11, 18). In this case, the endogenous bacteria exploit an opportunity that may not have presented itself in the absence of an influenza infection. The prevalence of S. pneumoniae colonization ranges from 19% in children (19) to approximately 11% in adults in the United States (10). Thus, for pneumonia caused by S. pneumoniae, the relative contribution of coinfection would be age-dependent and rely upon rates of colonization among the elderly or adult interaction with children who had an S. pneumoniae infection.
There have been few animal models in which this pathway was studied directly. In one mouse model, the introduction of influenza A 3 days after inoculation with S. pneumoniae led to an increase in the presence of pneumonia and facilitated influenza transmission to littermates (12). However, in a second mouse model, investigators found that S. pneumoniae colonization before influenza infection was protective compared with influenza infection before S. pneumoniae colonization (17). These studies seem to suggest that S. pneumoniae can facilitate transmission of influenza, but they provide insufficient evidence for any synergy in terms of coinfection.
At the population level, we would expect a limited time lag before the onset of bacterial pneumonia (Table 1). Because the bacterial agent has had a chance to proliferate in the host, we would expect more rapid proliferation once the lungs were no longer able to clear the bacteria. However, because acquisition of a new bacterial isolate is required, we could anticipate that the onset of coinfection would be much more variable and would be associated with individuals with high risk of exposure, such as those who had contact with young children.
Influenza infection preceding S. pneumoniae colonization
Clinically, it is assumed that bacterial pneumonia associated with influenza results from a bacterial infection that follows the influenza infection. Within an individual, the typical course of secondary bacterial pneumonia involves initial recovery from influenza followed by secondary symptoms, such as cough or fever, 4–14 days later (20). Results from animal models support this clinical observation. When mice are exposed to both wild-type and laboratory influenza strains and various strains of S. pneumoniae and S. aureus, there is an increased number of pneumonia infections, as well as an increased likelihood of death, compared with when any agent is given alone (11, 17, 21–23). In the aggregate, these experiments strongly support the idea of a synergistic interaction between influenza and S. pneumoniae (and S. aureus) when infection occurs sequentially.
At the population level, we would expect to see a relatively long lag time between influenza infection and the presence of CAP because of the time needed for the influenza to reduce the ability of the lungs to clear a bacterial infection. This lag time would enable partial recovery before the bacteria could infect the individual. The bacterial agent would then have to proliferate within the host before the host began to show symptoms of CAP. This lag was observed when data from the 1918 pandemic were reanalyzed. A graph of the timing from influenza infection to death found a lag period of 7–21 days (24), which suggests that affected individuals recovered before developing a more severe bacterial infection. Other research on data from the 1918 influenza A pandemic that suggests the role that secondary bacteria may have played has been supported by similar death curves created from data from infection with bacterial pneumonia alone through the 1920s and 1930s (25).
RESEARCH QUESTIONS AND IMPLICATIONS
Specifying the pathways to coinfection may help to identify new research questions about the interaction between influenza and CAP (Table 1) that can only be answered through basic science, clinical, and population studies. Basic laboratory science studies of the interactions within animal models have provided the most information to date for understanding the interactions between influenza and S. pneumoniae because unlike in human populations, experiments in laboratories can control the timing of exposure to influenza and secondary bacterial infection. Animal research has also shed light on specific cytokine pathways, such as mediation of infection with influenza by interleukin-10 (26) and inhibition of the pulmonary system’s ability to fight infection by interferon-gamma (27, 28), that provide testable hypotheses for an inflammatory response that might occur in humans. Further, in animal studies, investigators can explore what combinations of influenza strains and S. pneumoniae serotypes may result in greater risks to human populations (11, 13, 29, 30).
Seasonal influenza and pneumococcal vaccinations can protect elderly populations against hospitalization for either influenza or pneumonia, and the effects are additive. We see the fewest hospitalizations among individuals who have had both vaccinations, but persons with either the influenza vaccine or the Streptococcus vaccine have lower rates of hospitalization than do unvaccinated individuals (31). The novel 2009 hemagglutinin type 1 and neuraminidase type 1 influenza (commonly known as H1N1 or swine flu) pandemic provided a natural experiment in a naive population not previously exposed to the influenza strain. Specifically, the lethality of the strain in an unexposed population and potential immunity among the elderly who had been previously exposed to an H1N1 strain provided information about risk of coinfection (8, 32, 33). The pandemic also started a robust debate about when it is appropriate to provide antibiotics to prevent secondary infection and how to ration them (34–36). Because etiology is determined in only 30%–50% of patients with CAP who are tested (16) and antibiotics are often given without identifying the bacterial agent, clinical research studies will need to include bacterial surveillance. Additionally, determining the effectiveness of neuraminidase inhibitors and specific age groups to target to prevent a secondary pneumonia infection is needed to provide evidence of which populations will benefit the most from limited resources.
Historical research into the role that bacterial infections played in the deaths from the 1918 influenza A pandemic provides a baseline for our understanding of coinfection at the population level (24, 25, 37) but little insight into the role of order and timing of the infections. Although current recommendations to stay home after influenza infection have been suggested to prevent secondary influenza infections, following the same advice may also prevent subsequent exposure to novel bacteria (pathway 3 in Figure 1) that can cause secondary pneumonia—a major cause of death in 1918. More recent data on the timing between influenza infection and bacterial pneumonia are needed to determine whether antibiotic and antiviral treatments have changed the basic timing and relation at the population level.
We described 3 pathways to CAP associated with influenza and the biologic evidence that supports the potential for each of these pathways to lead to coinfection. By exploring the implications of each pathway and examining coinfection at the biologic, clinical, and population levels, we should be able to identify key signals for predicting and preventing CAP. Although most animal models are currently used to test and support a sequential infection with influenza followed by exposure to bacteria, translating that research to a human model is not straightforward. The interaction between colonization with bacteria and infection, as well as real-life variability in the timing to transmission, means that much further research needs to be conducted for a true understanding of how this coinfection occurs in humans. However, by thinking about the interaction between influenza and pneumonia-causing bacteria in terms of timing of transmission, we are able to raise questions about where to target future human research. Creation of public health recommendations to reduce transmission among at-risk populations and determination of the effectiveness of vaccinations and treatments can be guided by using results from current animal models with a goal of shaping population-level studies. With the majority of historical deaths from influenza pandemics attributable to bacterial infection (2), sorting out the relative contribution of each pathway to disease is a project too important to ignore.
Acknowledgments
Author affiliations: Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan (Brian M. Davis, Allison E. Aiello, Betsy Foxman); Center for Molecular and Clinical Epidemiology of Infectious Diseases, School of Public Health, University of Michigan, Ann Arbor, Michigan (Betsy Foxman, Allison E. Aiello, Pejman Rohani); Center for Social Epidemiology and Population Health, School of Public Health, University of Michigan, Ann Arbor, Michigan (Brian M. Davis, Allison E. Aiello); Department of Pediatrics and Communicable Diseases, University of Michigan Medical School, Ann Arbor, Michigan (Suzanne Dawid); Department of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, Michigan (Suzanne Dawid); Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, Michigan (Pejman Rohani, Sourya Shrestha); Center for the Study of Complex Systems, College of Literature, Science and the Arts, University of Michigan, Ann Arbor, Michigan (Pejman Rohani, Sourya Shrestha); and Fogarty International Center, National Institutes of Health, Bethesda, Maryland (Pejman Rohani).
This work was supported by a Research Partnership Grant from the Rackham Graduate School at the University of Michigan.
The Rackham Graduate School had no involvement in the ideas, discussion, or writing of the manuscript.
Conflict of interest: none declared.
Glossary
Abbreviation
- CAP
community-acquired pneumonia
References
- 1.McCullers JA, English BK. Improving therapeutic strategies for secondary bacterial pneumonia following influenza. Future Microbiol. 2008;3(4):397–404. doi: 10.2217/17460913.3.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz M, Ewig S, Marcos MA, et al. Etiology of community-acquired pneumonia: impact of age, comorbidity, and severity. Am J Respir Crit Care Med. 1999;160(2):397–405. doi: 10.1164/ajrccm.160.2.9808045. [DOI] [PubMed] [Google Scholar]
- 4.McCullers JA. Insights into the interaction between influenza virus and Pneumococcus. Clin Microbiol Rev. 2006;19(3):571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finelli L, Fiore A, Dhara R, et al. Influenza-associated pediatric mortality in the United States: increase of Staphylococcus aureus coinfection. Pediatrics. 2008;122(4):805–811. doi: 10.1542/peds.2008-1336. [DOI] [PubMed] [Google Scholar]
- 6.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 7.Thompson WW, Comanor L, Shay DK. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis. 2006;194(suppl 2):S82–S91. doi: 10.1086/507558. [DOI] [PubMed] [Google Scholar]
- 8.Palacios G, Hornig M, Cisterna D, et al. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS One. 2009;4(12):e8540. doi: 10.1371/journal.pone.0008540. (doi:10.1371/journal.pone.0008540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilleman MR. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine. 2002;20(25-26):3068–3087. doi: 10.1016/s0264-410x(02)00254-2. [DOI] [PubMed] [Google Scholar]
- 10.File TM. Community-acquired pneumonia. Lancet. 2003;362(9400):1991–2001. doi: 10.1016/S0140-6736(03)15021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullers JA, McAuley JL, Browall S, et al. Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J Infect Dis. 2010;202(8):1287–1295. doi: 10.1086/656333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diavatopoulos DA, Short KR, Price JT, et al. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 2010;24(6):1789–1798. doi: 10.1096/fj.09-146779. [DOI] [PubMed] [Google Scholar]
- 13.Weinberger DM, Trzciński K, Lu YJ, et al. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 2009;5(6):e1000476. doi: 10.1371/journal.ppat.1000476. (doi:10.1371/journal.ppat.1000476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wadowsky RM, Mietzner SM, Skoner DP, et al. Effect of experimental influenza A virus infection on isolation of Streptococcus pneumoniae and other aerobic bacteria from the oropharynges of allergic and nonallergic adult subjects. Infect Immun. 1995;63(4):1153–1157. doi: 10.1128/iai.63.4.1153-1157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaal KP. Medical and microbiological problems arising from airborne infection in hospitals. J Hosp Infect. 1991;18(suppl A):451–459. doi: 10.1016/0195-6701(91)90056-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morawska L. Droplet fate in indoor environments, or can we prevent the spread of infection? Indoor Air. 2006;16(5):335–347. doi: 10.1111/j.1600-0668.2006.00432.x. [DOI] [PubMed] [Google Scholar]
- 17.McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis. 2002;186(3):341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- 18.Marston BJ, Plouffe JF, File TM, Jr, et al. Incidence of community-acquired pneumonia requiring hospitalization: results of a population-based active surveillance study in Ohio. The Community-Based Pneumonia Incidence Study Group. Arch Intern Med. 1997;157(15):1709–1718. [PubMed] [Google Scholar]
- 19.Bogaert D, van Belkum A, Sluijter M, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363(9424):1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 20.Rothberg MB, Haessler SD. Complications of seasonal and pandemic influenza. Crit Care Med. 2010;38(4 suppl):e91–e97. doi: 10.1097/CCM.0b013e3181c92eeb. [DOI] [PubMed] [Google Scholar]
- 21.LeVine AM, Koeningsknecht V, Stark JM. Decreased pulmonary clearance of S. pneumoniae following influenza A infection in mice. J Virol Methods. 2001;94(1-2):173–186. doi: 10.1016/s0166-0934(01)00287-7. [DOI] [PubMed] [Google Scholar]
- 22.Tashiro M, Ciborowski P, Reinacher M, et al. Synergistic role of staphylococcal proteases in the induction of influenza virus pathogenicity. Virology. 1987;157(2):421–430. doi: 10.1016/0042-6822(87)90284-4. [DOI] [PubMed] [Google Scholar]
- 23.Margolis E, Yates A, Levin BR. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host’s immune response. BMC Microbiol. 2010;10(1):59. doi: 10.1186/1471-2180-10-59. (doi:10.1186/1471-2180-10-59) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klugman KP, Astley CM, Lipsitch M. Time from illness onset to death, 1918 influenza and pneumococcal pneumonia. Emerg Infect Dis. 2009;15(2):346–347. doi: 10.3201/eid1502.081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brundage JF, Shanks GD. Deaths from bacterial pneumonia during 1918–19 influenza pandemic. Emerg Infect Dis. 2008;14(8):1193–1199. doi: 10.3201/eid1408.071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahangian A, Chow EK, Tian X, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119(7):1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MH, Arrecubieta C, Martin FJ, et al. A postinfluenza model of Staphylococcus aureus pneumonia. J Infect Dis. 2010;201(4):508–515. doi: 10.1086/650204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat Med. 2008;14(5):558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 29.Giebink GS, Wright PF. Different virulence of influenza A virus strains and susceptibility to pneumococcal otitis media in chinchillas. Infect Immun. 1983;41(3):913–920. doi: 10.1128/iai.41.3.913-920.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harboe ZB, Thomsen RW, Riis A, et al. Pneumococcal serotypes and mortality following invasive pneumococcal disease: a population-based cohort study. PLoS Med. 2009;6(5):e1000081. doi: 10.1371/journal.pmed.1000081. (doi:10.1371/journal.pmed.1000081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christenson B, Hedlund J, Lundbergh P, et al. Additive preventive effect of influenza and pneumococcal vaccines in elderly persons. Eur Respir J. 2004;23(3):363–368. doi: 10.1183/09031936.04.00063504. [DOI] [PubMed] [Google Scholar]
- 32.Soto-Abraham MV, Soriano-Rosas J, Díaz-Quiñónez A, et al. Pathological changes associated with the 2009 H1N1 virus. N Engl J Med. 2009;361(20):2001–2003. doi: 10.1056/NEJMc0907171. [DOI] [PubMed] [Google Scholar]
- 33.Cunha B, Syed U, Strollo S. During the “herald wave” of the pandemic bacterial pneumonia relatively rare with fatal swine influenza (H1N1) pneumonia: if chest films have no focal segmental/lobar infiltrates, antibiotic therapy is unnecessary. J Chemother. 2009;21(5):584–589. doi: 10.1179/joc.2009.21.5.584. [DOI] [PubMed] [Google Scholar]
- 34.Wright PF, Kirkland KB, Modlin JF. When to consider the use of antibiotics in the treatment of 2009 H1N1 influenza-associated pneumonia. N Engl J Med. 2009;361(24):e112. doi: 10.1056/NEJMopv0910749. (doi:10.1056/NEJMopr0910749) [DOI] [PubMed] [Google Scholar]
- 35.Cinti SK, Barnosky AR, Gay SE, et al. Bacterial pneumonias during an influenza pandemic: how will we allocate antibiotics? Biosecur Bioterror. 2009;7(3):311–316. doi: 10.1089/bsp.2009.0019. [DOI] [PubMed] [Google Scholar]
- 36.Cunha BA. Swine influenza (H1N1) pneumonia: clinical considerations. Infect Dis Clin North Am. 2010;24(1):203–228. doi: 10.1016/j.idc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedson DS. Was bacterial pneumonia the predominant cause of death in the 1918–1919 influenza pandemic? J Infect Dis. 2009;199(9):1408–1409. doi: 10.1086/597621. [DOI] [PubMed] [Google Scholar]