Abstract
Background
We critically evaluated the available evidence on genomic tests in breast cancer to define their prognostic ability and likelihood to determine treatment benefit.
Design
Independent evaluation of six genomic tests [Oncotype Dx™, MammaPrint®, Genomic Grade Index, PAM50 (ROR-S), Breast Cancer Index, and EndoPredict] was carried out by a panel of experts in three parameters: analytical validity, clinical validity, and clinical utility based on the principles of the EGAPP criteria.
Panel statements
The majority of the working group members found the available evidence on the analytical and clinical validity of Oncotype Dx™ and MammaPrint® to be convincing. None of the genomic tests demonstrated robust evidence of clinical utility: it was not clear from the current evidence that modifying treatment decisions based on the results of a given genomic test could result in improving clinical outcome.
Conclusions
The IMPAKT 2012 Working Group proposed the following recommendations: (i) a need to develop models that integrate clinicopathologic factors along with genomic tests; (ii) demonstration of clinical utility should be made in the context of a prospective randomized trial; and (iii) the creation of registries for patients who are subjected to genomic testing in the daily practice.
Keywords: breast cancer, genomic signatures, prediction, prognosis
introduction
Over the past decade, it has been made clear that breast cancer (BC) is not a single disease, but rather a group of heterogeneous tumors at the molecular level [1–3]. Gene expression profiling has shed light on the complex molecular background of this disease and holds the potential for more accurate prognostication and patient stratification for therapy. Several genomic tests have been developed with the aim of improving prognostic information beyond that provided by classic clinicopathologic parameters [4]. Some of these tests are currently available in the clinic and are used to determine prognosis and more importantly to assist in determining the need for adjuvant chemotherapy particularly in patients with ER-positive disease. Available data suggest that information generated from genomic tests has resulted in a change in decision making in approximately 25%–30% of cases [5, 6]. Such decision changes mandate a critical evaluation of the quality of available evidence on the different genomic tests and their ability to define prognosis and determine treatment benefit.
Hence, a working group was established in order to evaluate the medical utility of six genomic tests that have been developed in the field of early BC: Oncotype Dx™, MammaPrint®, Genomic Grade Index (GGI), PAM50 (ROR-S), Breast Cancer Index (BCI), and EndoPredict. A critical review of the available evidence was carried out by the panel over the course of 1 year, and the recommendations were presented at the IMPAKT BC Conference in May 2012.
methods
genomic tests
Gene expression profiling allows the evaluation of a wide array of genes by determining the expression of the cellular messenger RNA in tumor tissue. Two main methods are used for gene expression analysis: DNA microarray technology, which is mainly carried out on fresh or frozen tissue, and quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR), which can also be carried out on formalin-fixed paraffin-embedded (FFPE) tissue. Table 1 summarizes the six signatures that were evaluated.
Table 1.
Summary of evaluated genomic tests
| Oncotype Dx™ | MammaPrint® | GGI | PAM50 (ROR-S) | Breast Cancer Index | EndoPredict | |
|---|---|---|---|---|---|---|
| Provider | Genomic Health | Agendia | Ipsogen | – | Biotheranostics | Sividon Diagnostics |
| Type of assay | 21-Gene recurrence score | 70-Gene assay | 97-Gene assay | 50-Gene assay | 2-Gene ratio HOXB13 to IL17R and molecular grade index | 11-Gene assay |
| Tissue sample | FFPE | Fresh or frozen | Fresh or frozen | FFPE | FFPE | FFPE |
| Technique | qRT–PCR | DNA microarray | DNA microarray | qRT–PCR | qRT–PCR | qRT–PCR |
GGI, Genomic Grade Index; FFPE, formalin-fixed paraffin-embedded; qRT–PCR, quantitative reverse transcriptase–polymerase chain reaction.
literature search
On 4 November 2011, we searched the MEDLINE database using the algorithm summarized in supplementary Table S1, available at Annals of Oncology online, which was carried out for each genomic test separately. Initial results were reviewed and cross-referencing was carried out among the identified studies to ensure that all eligible studies were captured. The main eligibility criterion was testing the genomic test of interest in BC patients. We set minimum criteria on the basis of which the studies were deemed not eligible for this evaluation:
Studies evaluating the genomic test in the pre-operative setting. These studies were excluded to avoid the potential impact of tissue sampling (biopsies or fine-needle aspirations versus surgical specimens) on the results of the genomic test. In addition, the primary end point of these studies is pathological complete response, and not long-term outcome.
Studies evaluating the cost–benefit of the genomic test. These studies are largely dependent on the health care system of each country. Such studies could be more valuable in the case the test has demonstrated robust evidence of being medically useful.
Studies using approximate versions of the evaluated genomic tests.
evaluation method and procedure
This project was undertaken by a working group composed of oncologists, pathologists, and scientists with expertise in the field of BC and/or genomic testing. The working group adopted a Delphi process [7], which was coordinated by a medical oncologist and completed over the course of 1 year. The Delphi process is a structured communication technique: joining together an expert panel to answer to a pre-defined question. The experts answer questionnaires blinded to the responses provided by other members of the group in at least two rounds. After each round, the coordinator provides a summary of the experts' forecasts from the previous round as well as the reasons they provided for their judgments. Thus, experts are encouraged to revise their earlier answers in light of the replies of other members. Finally, the process is stopped after a consensus is reached.
To assess the quality of the studies, evaluation was carried out based on the general principles of the EGAPP (Evaluation of Genomic Applications in Practice and Prevention) initiative [8]. This initiative provided rigorous evidence-based criteria for evaluating genomic tests for clinical and public health practice and has been used previously to evaluate genomic tests in BC [9].
Each study (where appropriate) was evaluated for the test's ability to accurately and reliably measure the genotype of interest (analytical validity), the test's ability to accurately and reliably identify or predict a relevant BC survival end point 5–10 years after surgery (clinical validity) and the balance of benefit and harms associated with the use of the test in practice, including improvement in measurable clinical outcomes and usefulness/added value in clinical management and decision making compared with not using the test (clinical utility).
In evaluating the ability of the test to accurately predict recurrence risk, we evaluated univariate and multivariate models that were reported for each genomic test. If more than one model was carried out in the same publication on the same dataset but to test different end points, we considered the end point that was specified as the primary end point. If more than one multivariate model was reported in the same paper but on different datasets, the two models were considered.
Each panel member was asked to evaluate the quality of evidence of each genomic test in the three parameters (analytical validity, clinical validity, and clinical utility) as convincing, adequate, or inadequate according to the principles of the EGAPP criteria (Table 2). Evaluation was carried out independently. Each panel member provided his/her review to the working group coordinator. After completing the evaluation process, a summary of all evaluations was shared with all the working group members for discussion to reach a final consensus.
Table 2.
Different parameters that were used to evaluate the eligible studies
| Analytical validity | Clinical validity | Clinical utility |
|---|---|---|
Data source
|
Data source
|
Data source
|
Reproducibility
|
Eligibility criteria | End points
|
| Blinded testing | Sample size and demographics | Data collection
|
Specimen
|
Point estimates of prognostic value
|
Treatment used |
| Report of test failures | Sample size | Randomization |
| Report of indeterminate results | Power calculation | Independence of the test
|
results
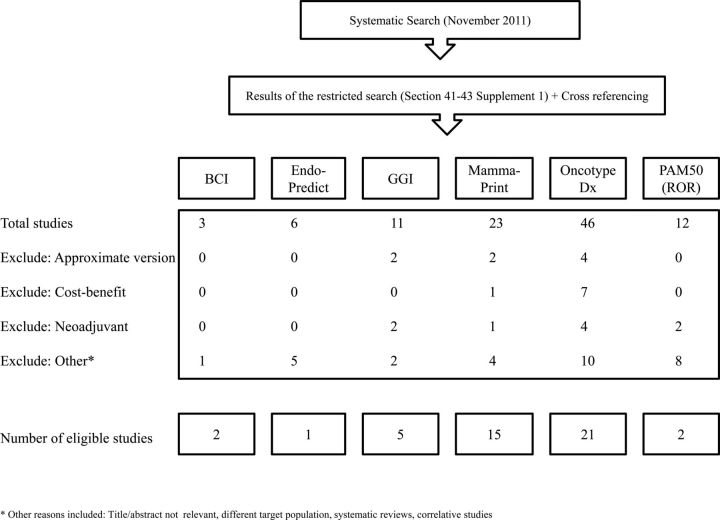
Figure 1 shows the flowchart of eligible studies. The final set of eligible publications included 21, 15, 5, 2, 2, 1 studies for Oncotype Dx™, MammaPrint®, GGI, PAM50 (ROR-S), BCI, and EndoPredict, respectively (supplementary Table S2, available at Annals of Oncology online). A summary of the publications that were evaluated is provided in supplementary Table S3, available at Annals of Oncology online.
Figure 1.
A flow chart summarizing eligible articles.
analytical validity
Supplementary Table S4, available at Annals of Oncology online, summarizes the number of publications that provided information on different analytical validity parameters. The source of tissue samples was mainly from retrospective studies, which were variable in size and quality. On considering all signatures, information on inter- and intra-laboratory reproducibility, blinded testing, and test failures were not reported in 100% of the evaluated studies.
A total of 14 studies evaluated analytical validity parameters of Oncotype Dx™. Habel et al. [10] examined the reproducibility between two and five FFPE blocks/patient in 20 patients to examine the variability of Oncotype Dx™ score between blocks from the same patient, all being conducted in the same laboratory. They found a high concordance (Pearson's r = 0.86) with a standard deviation of 3 recurrence scores between blocks on a continuous scale. Similar results were obtained by Cronin et al. [11]. Drury et al. [12] further showed high correlation between results obtained from whole FFPE section (10 µm) and 0.6 mm cores (P = 0.005), although greater variability was seen between cores than between sections. Of note, Genomic Health carried out all analytic validity experiments of Oncotype Dx™ and hence data are lacking on inter-laboratory validation, which remains a limitation.
Twelve studies provided information on the analytical validity of MammaPrint®. Buyse et al. [13] have reported 100% concordance between the risk classification (high versus low) produced by the manufacturer, Agendia, and results generated by the Swiss Institute of Bioinformatics on a blinded evaluation of a large series of 326 patients. The same study reported a test failure rate of 19%, which was mainly due to poor RNA quality. Ach et al. [14] reported a high intra-laboratory and inter-laboratory reproducibility on tumor sample duplicates across three laboratories in Amsterdam, California, and Paris with a Pearson correlation of >0.98.
Very limited information was available on the analytic validity of GGI, BCI, PAM50 (ROR-S), and EndoPredict, particularly intra-laboratory and inter-laboratory validation, the latter was not reported in any of the studies. At the time of undertaking this review, analytic validity of EndoPredict was evaluated in only one study [15]. The test showed high reproducibility with a success rate of >95%.
clinical validity/utility
There was a clear heterogeneity in the patient population in which these genomic tests were investigated. This includes variability in tumor stage (i.e. lymph node positive and negative), treatment modalities (systemically untreated, tamoxifen- or chemotherapy-treated), and ER expression (i.e. positive and negative). The source of information was mainly retrospective studies. In a few incidences, the genomic test was carried out on retrospectively collected samples of patients who were enrolled in a prospective randomized trial. As expected, this was feasible only for genomic tests that can be carried out on FFPE such as Oncotype Dx™, EndoPredict, and BCI (supplementary Table S2, available at Annals of Oncology online). Of note, sample size and power calculation were not addressed in any of the evaluated studies.
A total of 36 multivariate models were evaluated across the six genomic tests. In 34 models (94%), the genomic test was significantly associated (P < 0.05) with prognosis. Of note, several models did not adjust for relevant clinicopathologic features like progesterone receptor (PgR). Only a few models adjusted for the proliferation marker Ki67. Table 3 summarizes the different factors that were adjusted for in the different models. Table 4 summarizes the methods of assessing incremental predictive ability for each genomic test.
Table 3.
Evaluable multivariate models
| Oncotype Dx™ | MammaPrint® | GGI | Breast Cancer Index | PAM50 (ROR-S) | EndoPredict | |
|---|---|---|---|---|---|---|
| Number of patients | 4219 | 1465 | 1284 | 539 | 1496 | 1702 |
| Number of multivariate models | 13 | 13 | 4 | 2 | 2 | 2 |
| Adjustment factors, n (%) | ||||||
| Tumor size | 12 (92) | 10 (77) | 4 (100) | 2 (100) | 2 (100) | 2 (100) |
| Nodal status | 13 (100) | 13 (100) | 3 (75) | 2 (100) | 2 (100) | 2 (100) |
| Histologic grade | 10 (77) | 10 (77) | 4 (100) | 2 (100) | 2 (100) | 2 (100) |
| Age | 10 (77) | 9 (70) | 3 (75) | 1 (50) | 2 (100) | 2 (100) |
| ER | 13 (100) | 10 (77) | 4 (100) | 2 (100) | 2 (100) | 2 (100) |
| PgR | 2 (15) | 2 (15) | 3 (75) | 1 (50) | 0 | 2 (100) |
| HER2 | 3 (23) | 3 (23) | 1 (25) | 1 (50) | 1 (50) | 2 (100) |
| Ki67 | 1 (7.5) | 0 | 1 (25) | 0 | 0 | 2 (100) |
| Treatment | 13 (100) | 9 (70) | 2 (50) | 2 (100) | 2 (100) | 2 (100) |
| AOL | 2 (15) | 4 (30) | 0 | 2 (100) | 0 | 2 (100) |
GGI, Genomic Grade Index; ER, estrogen receptor; PgR, progesterone receptor; AOL, Adjuvant Online.
Table 4.
Incremental predictive ability for each genomic test
| Oncotype Dx™ | MammaPrint® | GGI | Breast Cancer Index | PAM50 (ROR-S) | EndoPredict | |
|---|---|---|---|---|---|---|
| Adequate documentations of multivariable regressions | ||||||
| Number of multivariate models | 13 | 13 | 4 | 2 | 2 | 2 |
| Genomic test is significant (P < 0.05) | 12 | 12 | 4 | 2 | 2 | 2 |
| Added value demonstrated using the likelihood ratio test (P < 0.05) | 5 | 2a | 0 | 0 | 1 | 2 |
| Adequate documentation of AUC in ROC analysis or c-index | ||||||
| Presented the AUC values (± additional predictor) | 1 | 2 | 0 | 1 | 2 | 2 |
| Presented P-value for comparison | 0 | 0 | 0 | 0 | 1b | 2 |
| Reclassification analysis versus standard risk categories (AOL, NPI, St Gallen) | 2 | 6 | 0 | 0 | 0 | 0 |
aCarried out in a logistic regression model.
bNot significant.
GGI, Genomic Grade Index; AUC, area under the curve; ROC, receiving operating characteristics; c-index, concordance index; AOL, Adjuvant! Online; NPI, Nottingham Prognostic Index.
There was inconsistency in the end point that was investigated across the different models. End points evaluating distant relapse (i.e. distant metastasis free survival, distant relapse free survival, and time to distant relapse) were the most commonly investigated, up to 26 multivariate models (72%).
Oncotype Dx™ was developed in patients with ER-positive and node-negative disease treated with tamoxifen [16]. Slightly more than 4000 patients were investigated in 13 multivariate models, some of which were fitted using the same patient series. All but one showed a significant association between Oncotype Dx™ and BC outcome. Of note, this was the only study that included patients with ER-negative disease, which constituted up to 31% of the 149 included patients [17]. Two studies investigated the ability of Oncotype Dx™ in determining patients who would benefit from chemotherapy [18, 19]. Paik et al. [18] reported that patients with node-negative ER-positive BC with high Oncotype Dx™ score benefit more of CMF chemotherapy compared with tamoxifen (P-value for interaction = 0.038). This study included 651 patients constituting nearly 28% of those who were previously treated in the context of the NSABP-B20 randomized trial. Importantly, the tamoxifen arm of this study was already used as a training set in the development of Oncotype Dx™ [16], which increases the probability of over-fitting. This re-use of development data might partly explain the treatment interaction seen with Oncotype Dx™ [20, 21]. Albain et al. [19] addressed a similar question but in patients with node-positive BC who were randomly assigned to receive CAF chemotherapy followed by tamoxifen or tamoxifen alone in the SWOG S8814 phase III trial. In this retrospective study, 40% of the original samples (n = 367) were available to address the predictive ability of Oncotype Dx™. The results showed that high Oncotype Dx™ scores (118 patients; 54 events) were associated with higher benefit from CAF particularly during the first 5 years (P-value for interaction = 0.029) following BC diagnosis.
MammaPrint® was evaluated in 13 multivariate models, which included a total of 1465 patients. Similar to the observation made with Oncotype Dx™, several models were based on patients who were included in prior studies as well. All but one model [22] showed a significant association between MammaPrint® and BC outcome. The Dutch group had first shown that the MammaPrint® is associated with distant metastasis-free survival independent of clinical variables in a population that included both ER-positive and ER-negative patients [23, 24]. These findings were subsequently confirmed by the TRANSBIG consortium in a study that included 307 independent patients [13]. In the latter, patients with a discordant gene signature and AOL risk classification, the sensitivity of AOL was found to be very poor compared with MammaPrint®. Pooled analysis of patients with tumors <2 cm has further shown that MammaPrint® can identify a low-risk group among this group of patients independent of nodal status, histologic grade, treatment, ER and HER2 status [25].
EndoPredict was evaluated in only two models conducted by the same group of investigators, but included large number of patients (n = 1702) [15]. The two models were homogenous, and were carried out on patients who were included in two randomized phase III trials (ABCSG-6 and ABCSG-8). All patients had ER-positive and HER2-negative disease and were treated with adjuvant hormonal therapy. Unlike any of the other tests, the models were adjusted for all possible confounding covariates including ki67 and PgR. EndoPredict showed a significant added value to classic prognostic factors as well as AOL, which were demonstrated using a likelihood ratio test and c-index.
PAM50 (ROR-S) was evaluated also in two models, including slightly >1400 patients [26, 27]. The first study included node-negative systemically untreated patients, while the second included both node-positive and node-negative patients treated with tamoxifen. While PAM50 (ROR-S) was associated with the risk of relapse in the multivariate model, it failed to show an added value to AOL, using c-index in node-positive or node-negative disease.
The GGI and BCI were evaluated in 1284 and 539 patients, respectively. For GGI, the patient population was heterogeneous within and across the studies in terms of treatment, disease stage, and ER status. HER2 status was not reported in at least three out of the four models [28, 29]. BCI was evaluated in two studies, which were restricted to patients with node-negative and ER-positive BC [30, 31]. The two genomic tests were associated with BC outcome in all multivariate models, but no data were published on their added value, using the likelihood ratio test or c-index.
IMPAKT 2012 Working Group Statement
analytical validity
The majority of the working group members found the available evidence on the analytical validity of Oncotype Dx™ and MammaPrint® to be convincing. The panel believed that further data are required for analytical validity of the other genomic tests.
clinical validity
The majority of the working group members found the available evidence on the clinical validity of Oncotype Dx™ and MammaPrint® to be convincing in patients with ER-positive BC. The panel concluded that both tests could add small but significant additional prognostic information to the currently available prognostic tools. While the other tests showed a clear association with prognosis, the working group concluded that further data are required to confirm their clinical validity. This includes testing on larger number of patients, independent groups, in addition to providing more robust data on their analytic validity before considering them as clinically valid.
clinical utility
The working group found none of the genomic tests demonstrated robust evidence of clinical utility. The panel concluded that it was not clear from the current evidence that modifying treatment decisions based on the results of a given genomic test would result in improved clinical outcome. Hence, the group did not endorse withholding chemotherapy in patients with ER-positive BC solely on the basis of being low risk by the genomic test.
recommendations/guidelines of using genomic tests by other groups
Over the past 5 years, other groups have made similar efforts to evaluate the medical usefulness of genomic tests in BC [9, 32–34]. Consensus reached by each group was not always concordant with the other groups owing to the different evaluation methodology and the date on which the evaluation was carried out. Among the six genomic tests evaluated here, only Oncotype Dx™ and MammaPrint® were previously evaluated. Table 5 summarizes the consensus and recommendations by other groups.
Table 5.
Recommendations/guidelines from other groups including the 2012 IMPAKT Working Group
| Year | Signatures | Statement | |
|---|---|---|---|
| ASCO | 2007 | Oncotype Dx™ |
|
| MammaPrint® | |||
| Breast Cancer Gene Expression Ratio | |||
| Institut National du Cancer, France | 2009 | Oncotype Dx™ |
|
| MammaPrint® | |||
| uPA-PAI-1 | |||
| EGAPP | Oncotype Dx™ |
|
|
| MammaPrint® | |||
| Breast Cancer Gene Expression Ratio | |||
| St Gallen | 2011 | Oncotype Dx™ |
|
| MammaPrint® | |||
| IMPAKT Working Group | 2012 | Oncotype Dx™ |
|
| MammaPrint® | |||
| Genomic Grade Index | |||
| PAM50 (ROR-S) | |||
| Breast Cancer Index | |||
| EndoPredict |
ASCO, American Society of Clinical Oncology; ER+, estrogen receptor-positive; pN0, pathologic node negative; CMF, cyclophosphamide, methotrexate, 5-flourouracil; EGAPP, Evaluation of Genomic Applications in Practice and Prevention.
conclusions and future direction
Clinical variables, particularly nodal status and tumor size, remain highly clinically relevant even in the era of genomic testing [35] and are likely to remain important for evaluating risk of relapse for patients with BC. At present, adjuvant chemotherapy prescription is still based on a patient's absolute risk of relapse. Tools such as AOL have been validated to provide an accurate estimate of this risk [36, 37]. Given that genomic tests, particularly Oncotype Dx™ and Mammaprint®, were clearly associated with prognosis, it is probable that these tests can further aid treatment decisions by refining this risk estimate. This calls for adopting models that integrate important clinicopathologic factors along with genomic tests. Models incorporating such variables with Oncotype Dx™, EndoPredict, and PAM50 (ROR-S) have shown improvement in prognostic accuracy compared with the genomic test alone [15, 27, 38]. This encourages further validation of this concept, which could be a way to integrate these new markers into clinical practice.
To date, data on the predictive value of any of the genomic tests are scarce. However, it is important to note that none of these tests was developed to be a ‘predictive’ marker for chemotherapy benefit. A prospective randomized trial is regarded as the gold standard to robustly address the question of clinical utility or a survival benefit from altered chemotherapy decision making due to a genomic test. Two large phase III trials investigating Oncotype Dx™ (TAILORx) and MammaPrint® (MINDACT) in patients with lymph node-negative early BC have closed accrual. However, these trials were not designed, nor statistically powered, for evaluating the interaction between the magnitude of benefit of chemotherapy and the genomic test. Another study is currently investigating the clinical utility of Oncotype Dx™ (RxPONDER) in patients with node-positive disease, whereas the test was developed for node-negative patients. This study is powered to look into treatment interaction, and will include only patients with Oncotype DxTM scores ≤25. Whether similar trials should be carried out to demonstrate the clinical utility of the other genomic tests as well remains an open question. Less costly and time-consuming alternatives like retrospective genomic testing of prospectively collected samples in interventional phase III trials could be an option [39]. Few examples exist on this approach [40, 41]. However, a limitation of this strategy could reside in publication bias, as negative results are less likely to get published.
Another complementary method proposed by the panel is to establish a registry of follow-up data and the decision-making steps for patients who are subjected to genomic testing in daily practice. This could further elucidate the additional prognostic value of genomic testing to standard clinicopathologic variables in clinical practice.
funding
HAA is supported by a translational research grant from the European Society for Medical Oncology (ESMO). FZ is supported by a research grant from the Hellenic Society for Medical Oncology (HeSMO) [grant numbers are not available].
disclosure
MF is a consultant and received honoraria from Sividon Diagnostics GmbH. WFS is a cofounder, and has patents with royalty in Nuvera Biosciences. AT serves on the advisory board of Genomic Health and received research funding related to MammaPrint®. SL was involved in the development of GGI and MammaPrint®, but has neither financial conflicts nor patent interests to declare. The remaining authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
The authors would like to thank ESMO and the Breast International Group (BIG) headquarters for their help and support in coordinating the IMPAKT working group activities.
references
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 5.Henry LR, Stojadinovic A, Swain SM, et al. The influence of a gene expression profile on breast cancer decisions. J Surg Oncol. 2009;99:319–323. doi: 10.1002/jso.21244. [DOI] [PubMed] [Google Scholar]
- 6.Lo SS, Mumby PB, Norton J, et al. Prospective multicenter study of the impact of the 21-gene recurrence score assay on medical oncologist and patient adjuvant breast cancer treatment selection. J Clin Oncol. 2010;28:1671–1676. doi: 10.1200/JCO.2008.20.2119. [DOI] [PubMed] [Google Scholar]
- 7.Linstone HA, Turoff M. The Delphi Method: Techniques and Applications. Reading, MA: Addison-Wesley; 1975. [Google Scholar]
- 8.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Recommendations from the EGAPP Working Group: can tumor gene expression profiling improve outcomes in patients with breast cancer? Genet Med. 2009;11:66–73. doi: 10.1097/GIM.0b013e3181928f56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habel LA, Shak S, Jacobs MK, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8:R25. doi: 10.1186/bcr1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronin M, Sangli C, Liu ML, et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 12.Drury S, Salter J, Baehner FL, et al. Feasibility of using tissue microarray cores of paraffin-embedded breast cancer tissue for measurement of gene expression: a proof-of-concept study. J Clin Pathol. 2010;63:513–517. doi: 10.1136/jcp.2010.075754. [DOI] [PubMed] [Google Scholar]
- 13.Buyse M, Loi S, van't Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 14.Ach RA, Floore A, Curry B, et al. Robust interlaboratory reproducibility of a gene expression signature measurement consistent with the needs of a new generation of diagnostic tools. BMC Genomics. 2007;8:148. doi: 10.1186/1471-2164-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filipits M, Rudas M, Jakesz R, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 16.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 17.Esteva FJ, Sahin AA, Cristofanilli M, et al. Prognostic role of a multigene reverse transcriptase-PCR assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. Clin Cancer Res. 2005;11:3315–3319. doi: 10.1158/1078-0432.CCR-04-1707. [DOI] [PubMed] [Google Scholar]
- 18.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 19.Albain KS, Barlow WE, Shak S, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michiels S, Koscielny S, Hill C. Interpretation of microarray data in cancer. Br J Cancer. 2007;96:1155–1158. doi: 10.1038/sj.bjc.6603673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioannidis JP, Allison DB, Ball CA, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 22.Mook S, Schmidt MK, Viale G, et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2009;116:295–302. doi: 10.1007/s10549-008-0130-2. [DOI] [PubMed] [Google Scholar]
- 23.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 24.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 25.Mook S, Knauer M, Bueno-de-Mesquita JM, et al. Metastatic potential of T1 breast cancer can be predicted by the 70-gene MammaPrint signature. Ann Surg Oncol. 2010;17:1406–1413. doi: 10.1245/s10434-009-0902-x. [DOI] [PubMed] [Google Scholar]
- 26.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 29.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 30.Jerevall PL, Ma XJ, Li H, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104:1762–1769. doi: 10.1038/bjc.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jankowitz RC, Cooper K, Erlander MG, et al. Prognostic utility of the breast cancer index and comparison to Adjuvant! Online in a clinical case series of early breast cancer. Breast Cancer Res. 2011;13:R98. doi: 10.1186/bcr3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 33.Institut National du Cancer; 2009. Rapport 2009 sur l'etat des connaissances relatives aux biomarqueurs tissulaires uPA-PAI-1, Oncotye DX et MammaPrint dans la prise en charge du cancer du sein. [Google Scholar]
- 34.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mook S, Schmidt MK, Rutgers EJ, et al. Calibration and discriminatory accuracy of prognosis calculation for breast cancer with the online Adjuvant! program: a hospital-based retrospective cohort study. Lancet Oncol. 2009;10:1070–1076. doi: 10.1016/S1470-2045(09)70254-2. [DOI] [PubMed] [Google Scholar]
- 37.Olivotto IA, Bajdik CD, Ravdin PM, et al. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23:2716–2725. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- 38.Tang G, Cuzick J, Costantino JP, et al. Risk of recurrence and chemotherapy benefit for patients with node-negative, estrogen receptor-positive breast cancer: recurrence score alone and integrated with pathologic and clinical factors. J Clin Oncol. 2011;29:4365–4372. doi: 10.1200/JCO.2011.35.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peeters M, Price TJ, Cervantes A, et al. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:4706–4713. doi: 10.1200/JCO.2009.27.6055. [DOI] [PubMed] [Google Scholar]
- 41.Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2010;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.