SUMMARY
Programmed cell death (PCD) plays critical roles in plant immunity but must be regulated to prevent excessive damage. The E3 ubiquitin ligase SPL11 negatively regulates PCD and immunity in plants. We show that SPL11 cell-death suppressor 2 (SDS2), an S-domain receptor-like kinase, positively regulates PCD and immunity in rice by engaging and regulating SPL11 and related kinases controlling defense responses. An sds2 mutant shows reduced immune responses and enhanced susceptibility to the blast fungus Magnaporthe oryzae. Conversely, SDS2 over-expression induces constitutive PCD accompanied by elevated immune responses and enhanced resistance to M. oryzae. SDS2 interacts with and phosphorylates SPL11, which in turn ubiquitinates SDS2, leading to its degradation. In addition, SDS2 interacts with related receptor-like cytoplasmic kinases, OsRLCK118/176, that positively regulate immunity by phosphorylating the NADPH oxidase OsRbohB to stimulate ROS production. Thus, a plasma membrane-resident protein complex consisting of SDS2, SPL11, and OsRLCK118/176 controls PCD and immunity in rice.
Graphical Abstract
In Brief
Plant cell death and immunity must be strictly controlled. Fan et al. show that the monocot-specific RLK SDS2 phosphorylates the E3 ligase SPL11 to positively regulate cell death and immunity. SDS2 also interacts with the receptor-like cytoplasmic kinases RLCK118 and RLCK176 to regulate immunity via the NADPH oxidase OsRbohB.
INTRODUCTION
Plants rely largely on innate immunity to defend against pathogen invasion. To achieve this, plants have evolved in a multitier immune system to recognize non-self or modified-self molecules using plasma membrane-associated pattern recognition receptors (PRRs). These PRRs can perceive pathogen-associated molecular pattern (PAMPs), host-derived damage-associated molecular patterns (DAMPs), or other molecules to initiate P/DAMP-triggered immunity (PTI). The recognition between PRRs and PAMPs/DAMPs leads to a series of immune responses, including Ca2+ influx, reactive oxygen species (ROS) production, activation of mitogen-activated protein kinases (MAPKs), ethylene production, callose deposition, and pathogenesis-related (PR) gene expression (Jones et al., 2016; Zipfel, 2014). These responses result in transcriptional reprograming in the nucleus to establish PTI.
In Arabidopsis, FLS2, EFR, LORE, CERK1/LYM1/LYM3, CERK1/LYK5, and RLP23 are the PRRs of PAMPs flagellin, elongation factor Tu (EF-Tu), lipopolysaccharides (LPS), peptidoglycan (PGN, chitin, and Nep1-like protein 20 (nlp20)), respectively (Albert et al., 2015; Couto and Zipfel, 2016; Ranf et al., 2015). FLS2 and EFR are leucine-rich repeat receptor-like kinases (LRR-RLKs), and the others are lysin-motif (LysM) RLKs, except LORE, which is an S-domain-1-type RLK. These PRRs mediate pattern recognition with different binding partners such as receptor-like proteins (RLPs) and LysM-RLKs to form PRR complexes (Couto and Zipfel, 2016). PRR perception induces immune signaling that branches into ROS production and MAPK cascades via different receptor-like cytoplasmic kinases (RLCKs). For example, RLCK AtBIK1/AtPBL1 directly transmit AtFLS2/AtEFR signals to AtRbohD to produce ROS, while AtPBL27 links CERK1 to MAPKKK5, which activates the MAPK cascade to induce PR gene expression (Kadota et al., 2014; Li et al., 2014; Yamada et al., 2016). For attenuation of PTI, FLS2 is ubiquitinated by the U-box E3 ligases PUB12/13 for degradation (Lu et al., 2011). Besides the detection of dangerous non-self, PRRs can also recognize host-derived DAMPs to trigger PTI (Böhm et al., 2014).
In rice, the PTI recognition and signaling pathway are much less understood. Upon chitin binding, the rice LysM-RLP chitin elicitor-binding protein (OsCEBiP) forms a homodimer, then followed by heterodimerization with OsCERK1, which creates a signaling-active sandwich-type receptor system (Hayafune et al., 2014; Shimizu et al., 2010). The LysM-containing RLPs LYP4 and LYP6, the dual-specificity receptors for both chitin and PGN, associate with OsCERK1 in a ligand-dependent manner (Ao et al., 2014; Liu et al., 2012). OsRacGEF1 and OsRac1 are the key components of OsCEBiP1/OsCERK1-mediated signaling pathway (Akamatsu et al., 2013). OsRLCK176 and OsRLCK185 are signal transducers of both OsCEBiP1/OsCERK1 and LYP4/6/OsCERK1 complexes.
Study of lesion mimic mutants and their suppressors has greatly increased our understanding of the molecular basis of PCD and immunity in plants (Bruggeman et al., 2015; You et al., 2016). SPL11, encoding a plant U-box (PUB) type E3 ubiquitin ligase, negatively regulates PCD and innate immunity, and the rice spl11 mutant is lesion mimic (Zeng et al., 2004). SPL11 interacts with the Rho GTPase-activating protein (RhoGAP) SPL11-INTERACTING PROTEIN 6 (SPIN6), which negatively regulates the Rho GTPase OsRac1, a central regulator of rice PTI and ETI signaling (Kawano and Shimamoto, 2013; Liu et al., 2015). Many RLKs are substrates of PUB proteins, and their interactions have been well studied. For example, the PUB ARC1 and the RLK SRK are involved in brassica self-incompatibility. PUB12/13, together with FLS2 or LYK5, regulate Arabidopsis PTI responses. OsPUB15 and Pid2 module both rice PTI and ETI. The Medicago PUB1 regulates root nodulation and symbiosis of Rhizobial and Arbuscular Mycorrhizal together with LYK3 and DMI2 (Trujillo, 2018; Wang et al., 2015). In addition, PUB proteins are reported to form conserved modules with S-domain-1-type RLKs in Arabidopsis (Samuel et al., 2008).
We previously isolated three spl11 cell death suppressors (sds1–3) (Shirsekar et al., 2014). Here, we report map-based cloning and characterization of SDS2 that encodes an S-domain RLK. Mutation of SDS2 leads to reduced resistance to the blast fungus Magnaporthe oryzae. Conversely, SDS2 over-expression enhances resistance to M. oryzae and induces defense gene expression. We also find that SDS2 interacts with and phosphorylates SPL11, which in turn ubiquitinates SDS2. SDS2 positively regulates rice immunity and interacts with RLCKs OsRLCK118 and OsRLCK176, which are also positive regulators of rice immunity. Furthermore, OsRLCK118 interacts with and phosphorylates the NADPH oxidase OsRbohB to induce ROS burst during pathogen infection. Taken together, our results show that SDS2 is a monocot-specific RLK that plays a positive role in the regulation of PCD and immunity by complexing with the E3 ligase SPL11 and OsRLCK118/176 in rice.
RESULTS
SDS2 Encodes an SD-1 Type RLK
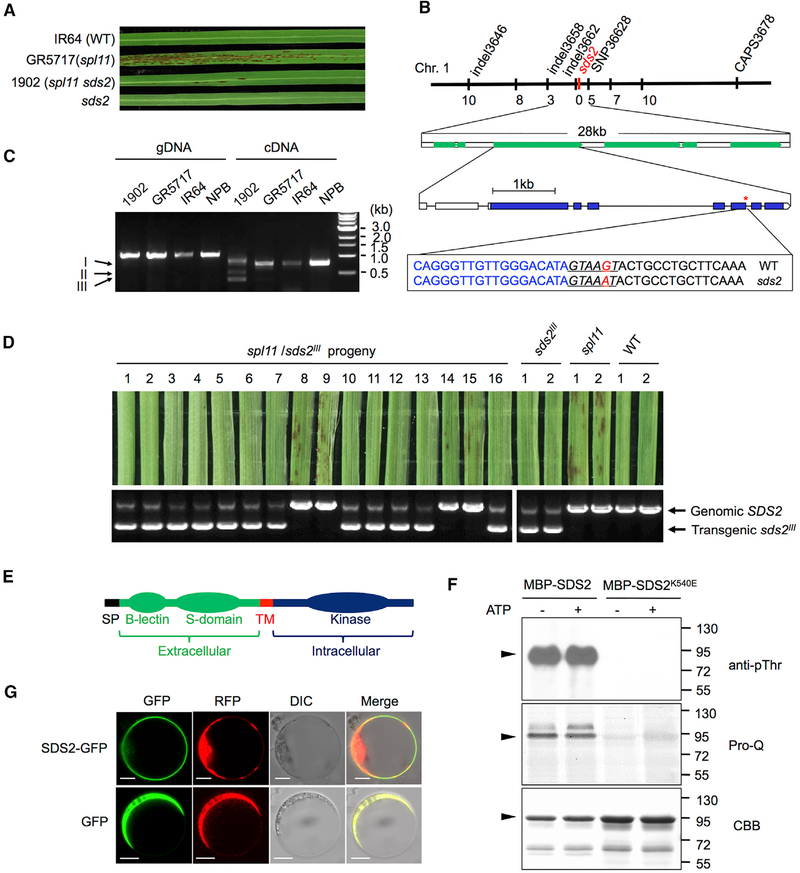
Compared to the spl11 mutant line GR5717, its suppressor line 1902 (spl11 sds2) had significantly fewer cell death lesions (Figure 1A). The sds2 mutation in 1902 was previously determined to be located between the markers In34_72 and In34_58 on chromosome 1 (Shirsekar et al., 2014). We fine-mapped sds2 to a 28 kb interval flanked by markers Indel3658 and SNP36628. Six protein-coding genes were annotated in this region (Table S1), with only one (LOC_Os01g57480) harboring a G-to-A substitution in sds2 in comparison with the wild-type (WT) (Figure 1B). This mutation occurs in a conserved splicing donor sequence (GTAAGT) of the 7th intron, resulting in three alternatively spliced transcripts, bands I, II, and III (Figures 1C, lane 5), which were derived from the retention of the 7th intron, removal of 8th exon, and alternative donor site-mediated splicing (the 6th exon), respectively (Figure S1A). We did not detect the WT full-length transcripts of LOC_Os01g57480 in the sds2 mutant.
Figure 1. SDS2 Encodes an SD-1 Type RLK.
(A) Lesion phenotypes of sds2, spl11 sds2, spl11 (GR5717), and IR64 on 3rd leaves.
(B) The genetic and molecular maps of the SDS2 locus on chromosome 1. The markers and numbers of recombinants are labeled up and down of the chromosome, respectively. Annotated genes in the region are indicated by green boxes. The mutation is a G-to-A transition that locates to the 7th intron of LOC_OS01g57480. Lines and boxes indicate introns and exons, respectively. Blue boxes indicate coding region of the candidate gene. Scale bar represents 1 kb. The sequences highlight the location of the mutation. Blue and black letters indicate the sequences of exons and introns, respectively. Red letter highlights the G/A mutation. Underlined italic letters indicate conserved splicing donor sites of introns.
(C) Amplification of the candidate gene from sds2, GR5717, IR64 and Nipponbare (NPB) genomic DNA (gDNA), and cDNA templates.
(D) Co-segregation of lesion suppression with the sds2III transgene in the progeny.
(E) Schema of the SDS2 structure. SP, signal peptide; TM, transmembrane.
(F) Auto-phosphorylation analysis of SDS2. – and + indicate absence or presence of ATP in the reaction. Pro-Q staining and CBB staining were done with the same gel. Molecular weights are labeled on the right. Black triangles indicate MBP-SDS2 protein.
(G) Subcellular localization of SDS2. RFP-fused Rac1 was used as a plasma membrane marker. GFP, RFP, differential interference contrast (DIC), and merge channels are labeled on the top of the picture. Scale bars represent 10 mm. See also Figure S1 and Table S1.
To confirm that LOC_Os01g57480 is the candidate of SDS2, we carried out a co-segregation analysis. We detected LOC_Os01g57480 transcripts with 32 plants consisting of both lesion-positive (L+) and lesion-negative (L−) individuals (11 L+ and 21 L−). All of the L+ individuals showed only the WT band, while the L− individuals showed mutant-specific bands (Figure S1B). Since the sds2 mutation is dominant, we cannot introduce the WT gene to complement the sds2 mutant phenotype. Instead, we introduced a mutated SDS2, band III splicing isoform of SDS2 (named sds2III, missing 1,623–2,009 nucleotides), to suppress lesion formation in the spl11 background. The missing region is located in 541–669 aa, the central part of the kinase domain, which may result in a compromised kinase. We generated transgenic plants that over-expressed sds2III in the TP309spl11 background. We obtained two TP309spl11/sds2III lines that showed no or few lesions (Figure S1C, 5th and 6th leaf from the left). qRT-PCR analysis showed that both lines accumulated high levels of sds2III transcripts (Figure S1D). Phenotypic analysis of the F3 progenies indicated that lesion formation was negatively correlated with the sds2III transgene (Figure 1D). The ratio of the L+ to L− plants was 1:3 (n = 16, χ2 = 0, p = 1), suggesting a single T-DNA insertion in transgenic plants. Together, these results demonstrate that LOC_Os01g57480 is the SDS2 gene.
SDS2 encodes an S-domain family (SD-1a type) RLK consisting of an extracellular B-lectin domain, an S-domain, a single-transmembrane domain, and an intracellular kinase domain (Figure 1E). Interestingly, SDS2 appears to be present only in monocots, but not in dicots (Xing et al., 2013). To examine whether SDS2 is a functional kinase, we conducted an in vitro kinase assay using the intracellular domain of SDS2 fused with maltose-binding protein (MBP). The kinase-inactive mutant, SDS2K540E, which bears a K-to-E substitution in the conserved ATP-binding site (K540E, leading to loss of kinase activity), was included as a control. Immuno-blotting with a phosphothrenine-specific antibody (anti-pThr) and phospho-protein-specific dye staining (Pro-Q Diamond Dye) showed that phosphorylation signals were detected for MBP-SDS2, but not MBP-SDS2K540E (Figure 1F), suggesting that SDS2 is an active kinase. Notably, strong phosphorylation signals were detected for MBP-SDS2, but not MBP-SDS2K540E, in the absence of ATP, which is likely attributed to phosphorylation in E. coli. In addition, we examined the SDS2 subcellular localization by expressing SDS2 fused with green fluorescent protein (GFP) in rice protoplasts. Red fluorescent protein (RFP) plasmids were co-transfected to indicate cytoplasm. Signals from SDS2-GFP only localized to the plasma membrane (Figure 1G).
SDS2 Is a Positive Regulator of Rice Innate Immunity
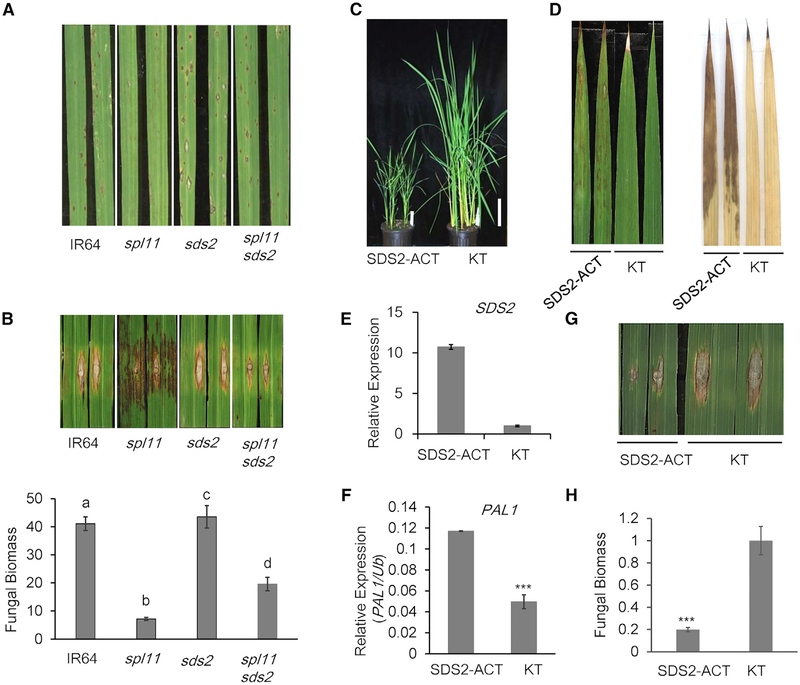
To investigate the function of SDS2, we obtained the sds2 mutant in the non-spl11 background from a cross between IR64 and the suppressor line 1902 (spl11 sds2). The sds2 plants did not show any cell-death lesions on leaves (Figure 1A). We spray and punch inoculated the sds2 plants with M. oryzae isolate PO6–6. Compared to WT IR64 plants, spl11 plants showed enhanced resistance to PO6–6, while sds2 plants showed reduced resistance with larger lesions and elevated fungal biomass (Figures 2A and 2B). To investigate how SDS2 regulates rice immunity, we measured ROS generation in response to PAMP treatments. As shown in Figures S2A and S2B, chitin or flg22-triggered ROS burst were reduced in sds2 plants compared to those in WT plants, suggesting that SDS2 positively regulates PTI responses upstream of ROS burst in rice.
Figure 2. SDS2 Positively Regulates Rice Immunity.
(A and B) sds2, IR64 (WT), spl11 (GR5717), and spl11/sds2 (1902) plants were inoculated with blast isolate PO6–6 by spray method (A) and punch method (B), respectively. Spray- and punch-inoculated plants were recorded 5 and 9 days post-inoculation (dpi), respectively. Quantification of fungal biomass of punch inoculation is shown in (B), lower panel. Values are means ± SD, n = 3 (technical repeats). Data were analyzed by one-way ANOVA followed by Tukey test. Different letters indicate significant differences at p < 0.05. Both inoculations have been replicated three times with similar results.
(C) Growth suppression of SDS2-ACT plants (2-month-old plants).
(D) SDS2-ACT plants show cell death on leaves (left panel) and accumulate higher level of ROS as stained by DAB (right panel). Leaves are from 2-month-old plants.
(E) SDS2 transcript levels in the SDS2-ACT and wild type (Kitaake or KT) plants.
(F) qRT-PCR analysis of PR gene (PAL1) expression in SDS2-ACT plant. Values are means ± SD, n = 3 (technical repeats), p = 1.5e-05 < 0.001.
(G) Enhanced resistance of SDS2-ACT plants to M. oryzae upon punch inoculation with blast isolate RO1–1 at 9 dpi. The inoculation has been replicated three times with similar results.
(H) Fungal biomass of punch inoculated leaves in (G). Values are means ± SD, n = 3, p = 4.0e-04. Asterisks represent significant difference determined by Student’s t test (*** p < 0.001) for all diagrams. See also Figure S2.
To confirm SDS2 function in rice immunity, we generated SDS2 over-expression transgenic lines. SDS2-GFP or the kinase-inactive mutant variant SDS2K540E-GFP driven by the maize ubiquitin promoter was introduced into Nipponbare (NPB). We obtained 5 and 16 independent lines for the SDS2-GFP and SDS2K540E-GFP constructs, respectively. All of the SDS2-GFP plants showed severe growth retardation, while all of the SDS2K540E-GFP plants were indistinguishable from WT plants (Figure S2C). The transgene transcript accumulation was about 15–100 times greater than endogenous SDS2 gene (Figure S2D). Considering SDS2K540E exhibited undetectable auto-phosphoryaltion activity (Figure 1F), it is likely that the kinase activity of SDS2 is required to trigger growth retardation phenotypes in transgenic plants. In addition, the defense-associated genes, including phenylalanine ammonia lyase 1 (PAL1) and pathogenesis-related 5 (PR5), were elevated in SDS2-GFP plants, but not in the SDS2K540E-GFP or WT plants (Figures S2E and S2F).
The severe dwarfisms in SDS2-GFP plants made it unfeasible for pathogen inoculation and ROS assays. Fortunately, we obtained an SDS2 activation tagging (SDS2-ACT) line that possesses a T-DNA insertion at 0.4 kb upstream of the transcription start site of SDS2. The T-DNA insertion contains a quadruple CaMV 35S enhancer which leads to the over-expression of SDS2 (Figure S2G). SDS2-ACT plants showed clear but much reduced growth retardation compared with the SDS2-GFP transgenic plants (Figure 2C). In addition, SDS2-ACT plants showed spontaneous cell death and ROS accumulation without infections (Figure 2D). The SDS2 transcripts in SDS2-ACT plants accumulated moderately with about 11 times higher than those in WT plants (Figure 2E). Similar to SDS2-GFP transgenic plants, SDS2-ACT plants also had elevated expression of PAL1 (Figure 2F). In response to PAMP treatments, SDS2-ACT plants produced more ROS than WT plants (Figure S2H). SDS2-ACT plants showed enhanced resistance to M. oryzae with reduced disease lesion sizes and fungal biomass compared to WT plants (Figures 2G, 2H, and S2I). Taken together, the data suggest that SDS2 is a positive regulator in PTI signaling, cell death control, and immunity to fungal infection in rice.
SDS2 Interacts with SPL11 in a Kinase Activity-Dependent Manner
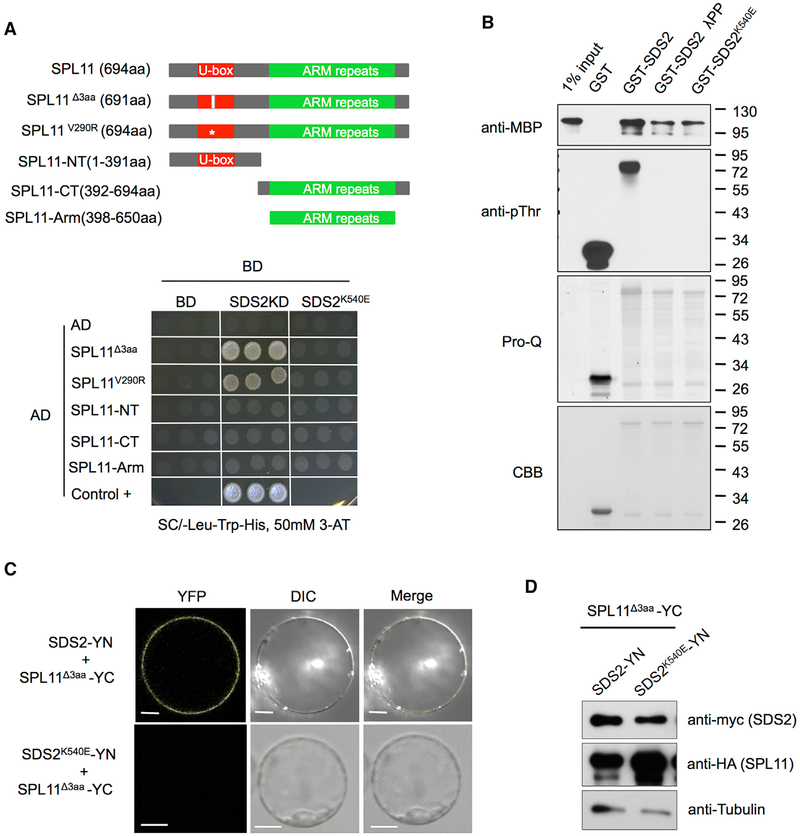
A yeast two-hybrid (Y2H) assay was conducted to determine whether SDS2 and SPL11 interact with each other. Full-length E3 inactive mutants SPL11Δ3aa (314–316 aa deletion in the U-box domain) and SPL11V290R (V290R substitution in the U-box domain) of SPL11 were used in the Y2H assay to detect the SDS2-SPL11 interaction (Zeng et al., 2004). The N-terminus (SPL11-NT, 1–391 aa), C-terminus (SPL11-CT, 392–694 aa), and ARM repeats (SPL11-Arm, 398–650 aa) of SPL11 were used in the same assay to determine which part of SPL11 interacts with SDS2 (intracellular domain only) in yeast. At the same time, SDS2 kinase-inactive mutant SDS2K540E is used to investigate whether SDS2 kinase activity is involved in potential interaction. The results showed that WT SDS2 interacted with both SPL11 mutants (SPL11Δ3aa and SPL11V290R), but not with truncated SPL11 variants (SPL11-NT and SPL11-Arm), while SDS2K540E didn’t interact with any forms of SPL11 (Figure 3A). Another Y2H vector system (pGBK-T7 and pGAD-T7) is also applied to confirm SDS2-SPL11 interaction. We took one homologue of SPL11, OsPUB12 (LOC_Os06g01304), as specificity control. The result is similar to the above. Interaction only occurs between WT SDS2 and SPL11 mutants, but not between all the other combinations (Figure S3A). Detection of SDS2 and SPL11 proteins in the yeast by western blotting shown that all the SDS2 and SPL11 isoforms are expressed in yeast with the WT SPL11 accumulating to a lower level (Figure S3B).
Figure 3. SDS2 Interacts with SPL11 in a Kinase Activity-Dependent Manner.
(A and B) Effect of SDS2 kinase activity on the SDS2-SPL11 interaction detected by yeast two-hybrid (A) and by pull-down assay (B).
(A) E3 inactive mutant SPL11Δ3aa (C314 P315 T316 deletion in the U-box domain) and SPL11V290R (V290R substitution in the U-box domain) and truncated SPL11 were fused with the activation domain (AD) of GAL4. SPL11-NT, -CT, and -Arm indicate N terminus (1–391 aa), C terminus (392–694 aa), and ARM repeats (398–650 aa) of SPL11, respectively. Schema, not in scale, is shown in upper panel. The white bar and star in U-box domain of SPL11 mutants indicate 3 aa deletion and V290R substitution, respectively. Intracellular domain (435–828 aa) of SDS2 (KD) and SDS2K540E (kinase-inactive mutant) were fused with the binding domain (BD) of GAL4. Shown is the growth of co-transformants on selection media (SC/-Leu-Trp-His with 50 mM 3-AT) for 5 days. Positive control (Control +) is the intact GAL4 transcription factor transformant.
(B) Lambda protein phosphatase (lPP)-treated and -untreated GST-SDS2 and GST-SDS2K540E were used as the baits. MBP-SPL11 was used as a prey. Prey was detected by immunoblotting (anti-MBP). CBB staining shows loading amount of the bait. Immunoblotting (anti-pThr) and Pro-Q staining were applied to detect the phosphorylation level of the bait proteins. Pro-Q staining and CBB staining were performed with the same gel. Molecular weights are labeled on the right.
(C) BiFC detection of the SDS2-SPL11 interaction in vivo. SDS2 (or SDS2K540E) and SPL11Δ3aa were fused to the N and C termini of YFP (YN and YC), respectively. Co-transfected protoplasts were observed by a confocal microscope 48 hr after transfection. YFP, DIC, and merged channels were labeled on the top of the pictures. Scale bars represent 10 mm.
(D) Detection of protein levels for SDS2 and SPL11 in protoplasts in BiFC assay shown in (C). See also Figures S3 and S4.
In addition, an in vitro pull-down assay was applied to confirm the findings above. SDS2 intracellular domain fused with the GST tag (GST-SDS2) was used as the bait, and full-length SPL11 fused with the MBP tag was used as the prey. Glutathione Sepharose beads with GST-SDS2 could effectively pull down MBP-SPL11 proteins. The beads with GST-SDS2K540E or GST-SDS2 pre-treated with Lambda Protein Phosphatase (λPP) were much less effective than that of SDS2 (Figure 3B). The phosphorylation levels of baits were determined by immune-blot with anti-pThr and Pro-Q Diamond dye staining. Apparently, the interaction between SDS2 and SPL11 depends on the SDS2 kinase activity (auto-phosphorylation).
Next we used bimolecular fluorescence complementation (BiFC) to further confirm that SDS2 and SPL11 interact with each other in vivo. SDS2 and SDS2K540E was fused to the N-terminus of yellow fluorescence protein (YFP) (SDS2-YN), and mutated SPL11 was fused to the C terminus of YFP (SPL11Δ3aa-YC). The BiFC assay showed that YFP signal was evident on the plasma membrane of the protoplasts transfected with SDS2-YN and SPL11Δ3aa-YC plasmids, but not with SDS2K540E-YN and SPL11Δ3aa-YC plasmids (Figure 3C). Protein levels of SDS2 and SPL11 were detected by western blot, showing that both of them are expressed in the protoplasts (Figure 3D). These results indicate that SDS2 interacts with SPL11 both in vitro and in vivo.
To determine the auto-phosphorylation sites and their role in the SDS2-SPL11 interaction, purified GST-SDS2 proteins were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Interestingly, with 68% protein coverage, only one phosphorylation site on threnine 667 residue (T667) in the activation-loop of SDS2 was detected (Figures S3C and S3D). Importantly, unlike the WT SDS2, SDS2T667A was no longer interacted with SPL11Δ3aa in yeast (Figure S3E), suggesting that T667 is an important auto-phosphorylation residue required for the SDS2-SPL11 interaction.
We also tested the interaction between the intracellular domains of SDS2 and several known rice PRR components, i.e., OsCERK1 (LOC_Os08g42580), OsSERK1 (LOC_Os08g07760), OsSERK2 (LOC_Os04g38480), and OsFLS2 (LOC_Os04g52780) by Y2H assays. The assays showed that SDS2 did not interact any of those proteins (Figure S3F). In Arabidopsis, PUB12/13 interact with FLS2 and LYK5 (Liao et al., 2017; Lu et al., 2011). Unlike in Arabidopsis, we found that SPL11 did not interact with OsCERK1, OsSERK1, OsSERK2, and OsFLS2 in yeast (Figure S3G). Co-immunoprecipitation (coIP) assay also showed that SPL11 interacts with SDS2, but not OsCERK1, with or without chitin treatment (Figure S3H). These results show that SDS2 might not be associated with the PRRs of chitin and flg22 in rice but possibly that is a PRR of unknown DAMPs or effectors from rice pathogens. It is also possible that SDS2 is a general component in the SPL11-mediated cell death and defense pathway.
SDS2 Is Ubiquitinated and Degraded by SPL11 but Phosphorylates the Latter
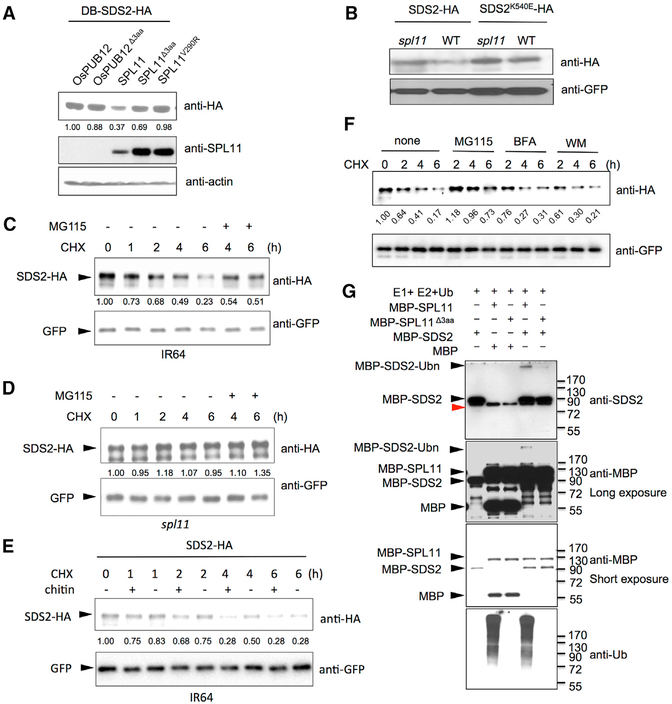
SDS2 interacts with the E3 ligase inactive mutant variants of SPL11, but not with the WT SPL11 in yeast (Figures 3A and S3A). In addition, transgenic plants over-expressing SDS2 showed lesion mimic phenotypes similar to that of spl11 mutant plants (Figure 2D). These observation prompted us to test whether SPL11 ubiquitinates SDS2, leading to its subsequent degradation. To test this hypothesis, we performed a degradation assay in yeast. SDS2-HA plasmids were co-transformed with SPL11, OsPUB12, and their mutant variants (SPL11Δ3aa, SPL11V290R, and OsPUB12Δ3aa). Immunoblotting analysis showed that SDS2-HA accumulated to a much lower level with the WT SPL11 than with the mutated SPL11s, OsPUB12, or OsPUB12Δ3aa (Figure 4A). WT SPL11 also accumulates to a much lower level than that of mutants, which is often seen for E3 (Figure 4A, middle panel). We also detected time-course degradation of SDS2-HA in yeast. The result showed that SDS2-HA was degraded faster at the present of WT SPL11 than that of the mutant SPL11Δ3aa (Figure S4A). In addition, we carried out a degradation assay in rice protoplasts. WT and spl11 rice protoplasts were transfected with SDS2-HA or SDS2K540E-HA. SDS2-HA accumulated to a lower level in WT rice protoplasts than in spl11 protoplasts (Figure 4B). However, SDS2K540E-HA accumulated at a similar level in both WT and spl11 protoplasts. The data suggest that SDS2-HA, but not SDS2K540E-HA, is degraded by SPL11. Furthermore, we conducted a time-course degradation assay in rice protoplasts. The rice protoplasts were treated with the protein synthesis inhibitor cycloheximide (CHX) to block SDS2-HA synthesis 24 hr after the transfection, and the SDS2-HA protein levels were determined by immunoblotting at different time points. 26S Proteasome inhibitor MG115 was added to determine whether SDS2 is degraded via the ubiquitin-proteasome system. The assay showed that the SDS2-HA level declined faster in WT rice protoplasts than in spl11 protoplasts (Figures 4C and 4D), suggesting that SDS2-HA is degraded by the endogenous SPL11 in WT rice protoplasts. Together, these results demonstrate that SDS2 is degraded by SPL11 in a kinase activity-dependent manner via the 26S protea-some system. We also compared SDS2-HA degradation in WT protoplasts between chitin treated and the control. The results showed that chitin treatment enhanced SDS2-HA degradation (Figure 4E). To investigate whether SDS2 is degraded via the vacuolar degradation pathway like FLS2 (Robatzek et al., 2006), we treated rice protoplasts with endocytosis inhibitors Brefeldin A (BFA) and Wortmannin (WM) and detected protein levels at different time points after treatments. The assay showed that both inhibitors did not affect SDS2-HA degradation at the tested conditions (Figure 4F). It will be interesting to investigate whether SDS2 undergoes endocytic and/or autophagic degradation, or not, with additional inhibitors.
Figure 4. SDS2 Is Degraded by SPL11 and Phosphorylates the Latter.
(A) SDS2 degradation by SPL11 in yeast. SDS2-HA was co-transformed with SPL11, OsPUB12, and their mutants (SPL11Δ3aa, SPL11V290R, and OsPUB12 Δ3aa) into yeast. SDS2-HA protein level was determined by immunoblotting. Relative band intensity of each lane is labeled below the image determined by the ImageJ program.
(B) SDS2 degradation by SPL11 in rice protoplasts. Plasmids of SDS2-HA or SDS2K540E -HA was transfected into wild-type or spl11 rice protoplasts. GFP plasmids were co-transfected as a control. SDS2-HA and SDS2K540E -HA protein levels were determined by immunoblotting.
(C and D) Time course degradation of SDS2-HA in wild-type IR64 (C) and spl11 (D) rice protoplasts. Co-transfected rice protoplasts were treated with (50 μg/mL) cycloheximide (CHX) to block protein synthesis, and SDS2-HA levels were monitored by immunoblotting. 26S proteasome inhibitor MG115 (100 μM) is added to determine whether SDS2-HA is degraded via 26S Proteasome pathway after 4 and 6 hr treatment. Bands intensities determined by Image Lab software (Bio-Rad) are labeled below the bands.
(E) SDS2-HA degradation is enhanced by PAMP treatment. Transfected IR64 protoplasts were treated with 50 μg/mL CHX, 5 mg/mL chitin (+), or no chitin (−). Protoplasts are harvested at different time points after treatments and applied to western blot.
(F) Endocytosis inhibitors do not affect SDS2 degradation at the tested condition. After transfection, rice protoplasts were treated with CHX (50 mg/mL) alone or together with MG115 (100 μM), Brefeldin A (BFA, 2.0 μg/mL), or Wortmannin (WM, 0.5 μM), respectively. Protoplasts were sampled at 2, 4, and 6 hr after the treatment. Band intensities were determined by ImageJ program.
(G) In vitro ubiquitination assay of SDS2 by SPL11. MBP was used as a control substrate to show specificity. Mutant SPL11Δ3aa was used as a negative control. − and + indicate absent or present of proteins labeled on the left. Ubiquitination of MBP-SDS2 was detected by anti-SDS2 and anti-MBP antibodies. Anti-Ub antibody is applied to determine SPL11 E3 activity. Black triangles indicate corresponding protein bands labeled on the left. MBP-SDS2-Ubn denotes ubiquitinated SDS2 band.
Next, we determined whether SPL11 could directly ubiquitinate SDS2 by an in vitro ubiquitination assay. SPL11Δ3aa was used as a negative control. A specific high-molecular-weight band was detected in the lane of MBP-SPL11 with MBP-SDS2 (Figure 4G, top panel, lane 4), but not in the lane of the mutated SPL11Δ3aa (Figure 4G, top panel, lane 5). These results demonstrate that SPL11 ubiquitinates SDS2 in vitro. To confirm this finding, we carried out an in vitro ubiquitination assay with a mutant ubiquitin in which all Lys residues are substituted to Arg and only one ubiquitin molecule can be added on to a substrate. The result showed that a specific 8 kD band was detected in the reaction with WT MBP-SPL11, but not with the mutated SPL11Δ3aa, confirming the ubiquitination of SDS2 by SPL11 (Figure S4B). The weak ubiquitination of SDS2 by SPL11 may be due to E2/E3 interaction specificity.
To determine whether SDS2 phosphorylates SPL11, we carried out an in vitro phosphorylation assay using MBP-SPL11 as the substrate and MBP-SDS2 as the kinase. As shown in Figure S4B, MBP-SDS2, but not MBP-SDS2K540E, phosphorylated MBP-SPL11 in vitro. Compared to the strong phosphorylation of Arabidopsis BAK1 (cytoplasmic domain) to PUB13, the phosphorylation of MBP-SDS2 to MBP-SPL11 was rather weak but clearly detected (Figure S4C).
SDS2 Interacts with and Phosphorylates OsRLCK118 and OsRLCK176
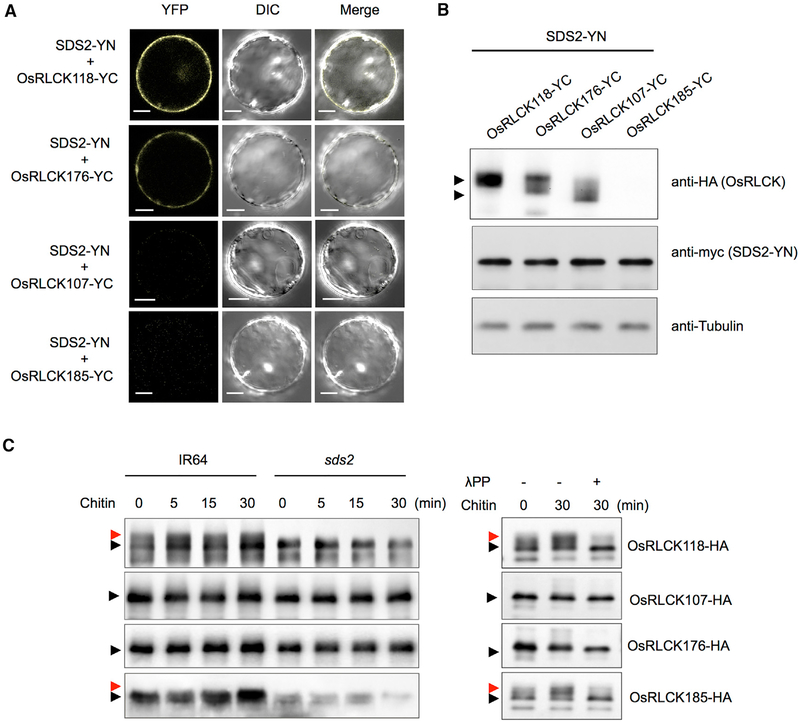
SDS2 belongs to the SRK (S-locus receptor-like kinase) family that controls pistil self-incompatibility in Crucifers (Takayama and Isogai, 2005). Because SRK interacts with MLPK (Modifier locus protein kinase), which is a RLCK in the VIIa subgroup, we hypothesized that SDS2 may interact with related RLCKs in rice. We used the MLPK sequence as a query to BLAST search the rice genome. The first three genes with highest similarity are OsRLCK176 (LOC_Os05g02020), OsRLCK107 (LOC_ Os03g16740), and OsRLCK118 (LOC_Os03g60710); all of them belong to the same clade (VIIa) as MLPK in Brassica and as BIK1 in Arabidopsis. We performed a BiFC assay to detect interaction between them and SDS2. We also selected OsRLCK185 (LOC_Os05g30870) that is involved in rice immunity in the same assay (Wang etal., 2017). The results showed that SDS2 interacted with both OsRLCK118 and OsRLCK176, but not OsRLCK107 and OsRLCK185 in vivo (Figures 5A and 5B). We also conducted a pull-down assay to show in vitro interaction between SDS2 and OsRLCK118/176. Indeed, SDS2 interacted with both of them (Figure S5A). We next determined whether OsRLCK118/176 are true RLCKs. Subcellular localization analysis showed that they are localized at the plasma membrane (Figure S5B). An in vitro kinase assay showed that WT OsRLCK118, but not kinase-inactive mutant OsRLCK118K116E, exhibited kinase activity (Figure S5C). OsRLCK176 did not show detectable kinase activity in the in vitro assay (Figure S5D). Together, these results demonstrate that SDS2 interacts with OsRLCK118/176.
Figure 5. SDS2 Interacts with and Phosphorylates OsRLCK118.
(A) BiFC analysis of the SDS2-OsRLCK118 or OsRLCK176 interactions in rice protoplasts. SDS2 and OsRLCKs were fused to the N and C termini of YFP, respectively. Protoplasts were observed with a confocal microscope 48 hr after transfection. YFP, DIC, and merged channels were labeled on the top of the pictures. Scale bars represent 10 μm.
(B) Detection of protein levels in BiFC assays in (A). Anti-HA and anti-myc were used to detect OsRLCKs-YC and SDS2-YN, respectively. Anti-tubulin was used to determine protein loading amount. Arrows indicate OsRLCK118, OsRLCK176, and OsRLCK107. OsRLCK185 was too low to detect.
(C) Detection of OsRLCKs in vivo phosphorylation by SDS2. Plasmids of OsRLCKs with HA tag are transfected into wild-type (IR64) and sds2 mutant protoplasts, separately. Transfected protoplasts are treated with 5 μg/mL chitin for 0, 5, 15, and 30 min. Phosphorylation of OsRLCK is determined by band shift. Black and red arrows indicate unphosphorylated and phosphorylated band, respectively (left panel). The phosphorylation is confirmed by treatment of lambda protein phosphatase (λPP) (right panel). − and + indicate with or without λPP treatment. See also Figure S5.
We conducted an in vivo phosphorylation assay to determine whether the four RLCKs above can be phosphorylated by SDS2. We transfected RLCK-HA plasmids into both WT (IR64) and sds2 protoplasts and treated the protoplasts with chitin 16 hr after transfection. Samples are collected at different time points and applied to western blotting. The results showed that OsRLCK118-HA, but not the other 3 OsRLCKs, is specifically phosphorylated by SDS2 (Figure 5C, left panel). The phosphorylation was confirmed by the treatment with λPP (Figure 5C, right panel). Notably, phosphorylation of OsRLCK118-HA is constitutive under the tested condition, which could be due to overex-pression of OsRLCK118 or constitutive activation of SDS2 in the protoplasts. Unexpectedly, SDS2-ACT plant did not show any phosphorylation on OsRLCK118-HA, possibly because of strong auto-immunity (cell death) in the SDS2-ACT plant (Figure S5G). In addition, an in vitro kinase assay indicated that SDS2 and OsRLCK118 trans-phosphorylated each other (Figure S5E, lane 2; Figure S5F, lanes 2 and 3). However, no phosphorylation can be detected between SDS2 and OsRLCK176 by immunoblotting or Pro-Q staining, even though they interacted with each other (Figures S5E and S5F). Together, these results suggest that SDS2 complexes with and trans-phosphorylates OsRLCK118.
OsRLCK118 and OsRLCK176 Are Positive Regulators of Rice Immunity, and SDS2 Signaling Is Dependent on OsRLCK118
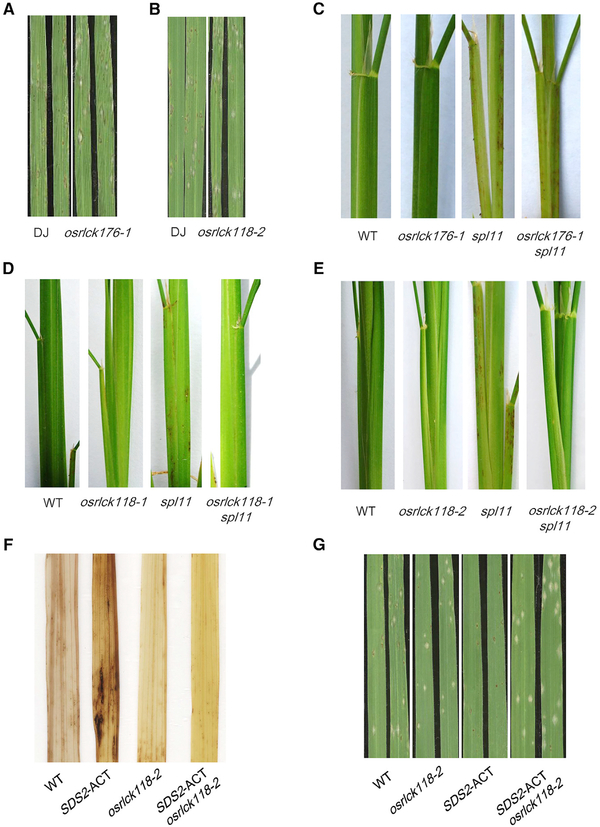
To analyze the function of OsRLCK118/176, we obtained T-DNA insertion mutants for the two genes in which the transcriptions were abolished (Figures S6A, S6B, and S6F). We then detected ROS generation with these two mutants to investigate OsRLCK118/176 functions in PTI responses. The assay showed that the osrlck118/176 mutants generated less ROS upon the chitin and flg22 treatments (Figures S6C and S6G), suggesting that OsRLCK118/176 positively regulate PTI responses. In addition, osrlck118/176 mutant plants showed enhanced susceptibility to M. oryzae isolate RO1–1 with larger lesions (Figures 6A, 6B, S6D, and S6H) and greater fungal biomass (Figures S6E and S6I). These results demonstrate that, similar to SDS2, both OsRLCK118 and OsRLCK176 positively regulate rice PTI.
Figure 6. OsRLCK118 and OsRLCK176 Are Positive Regulators of Rice Immunity, and SDS2 Signaling Is Dependent on OsRLCK118.
(A and B) Spray inoculation of osrlck176 and osrlck118 mutants with M. oryzae isolate RO1–1 at 6 dpi.
(C) osrlck176 is not able to suppress spl11-mediated lesion formation.
(D and E) osrlck118–1 and osrlck118–2 suppresses spl11-mediated cell death (7-week-old plants).
(F) osrlck118–2 suppresses SDS2 over-expression-induced cell death on leaves of 2-month-old plants detected by DAB staining.
(G) osrlck118–2 suppresses SDS2 over-expression-mediated blast resistance in SDS2-ACT/osrlck118–2 plants upon spray inoculation at 6 dpi. See also Figure S6.
To investigate the genetic relationship between SPL11 and OsRLCK118/176, we made crosses between osrlck118/176 and spl11 plants. Interestingly, double mutants of osrlck118–1 spl11 and osrlck118–2 spl11 from the F2 populations had much fewer lesions than spl11 plants (Figures 6D and 6E), suggesting that osrlck118 suppresses spl11-mediated cell death and OsRLCK118 is downstream of SPL11. In contrast, osrlck176 spl11 double mutant plants had amounts of lesions similar to those of the spl11 mutant, suggesting that osrlck176 may not suppress spl11-mediated cell death (Figure 6C). To determine whether OsRLCK118 can also suppress SDS2 over-expression-mediated cell death and resistance to blast, we generated a double mutant of osrlck118–2 SDS2-ACT. Lesions were less abundant in osrlck118–2 SDS2-ACT plants than in SDS2-ACT plants (Figure 6F). Meanwhile, osrlck118–2 SDS2-ACT plants are more susceptible to blast than SDS2-ACT plants are (Figure 6G). These results demonstrate that OsRLCK118 suppresses SPL11-mediated and SDS2 over-expression-mediated cell death and functions downstream of the SDS2 signaling pathway.
OsRLCK118 Interacts with and Phosphorylates the NADPH Oxidase OsRbohB
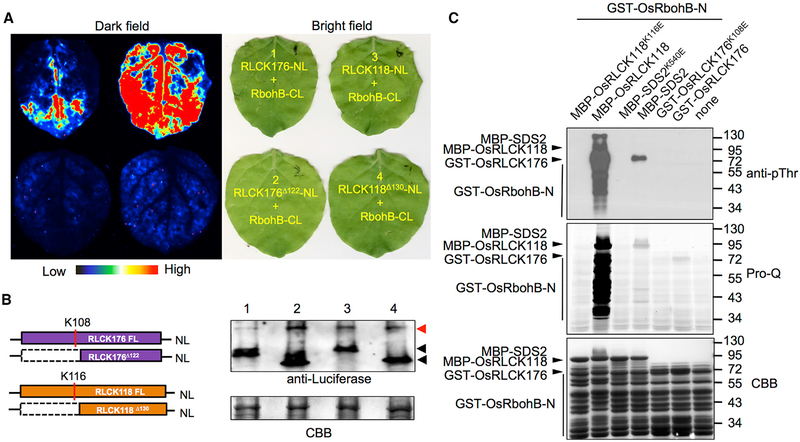
OsRbohB (LOC_Os01g25820) is the major NADPH oxidase responsible for ROS production on the plasma membrane in rice (Nagano et al., 2016; Wong et al., 2007). OsRbohB is also the orthologue of Arabidopsis RbohD, which is regulated by BIK1, which belongs to the same clade as OsRLCK118/176 (Shiu et al., 2004). We hypothesized that OsRLCK118/176 regulate OsRbohB activity. To test this hypothesis, we carried out GST pull-down and luciferase complementation imaging (LCI) assays to detect the interactions between OsRLCK118/176 and OsRbohB. The assays showed that OsRLCK118/176 interacted with OsRbohB in vitro and in vivo (Figures 7A and S7A). To further confirm the regulatory role of OsRLCK118/176, we conducted an in vitro phosphorylation assay. GST-OsRbohB-N (N-terminus 1–355 aa, the regulatory motif of OsRbohB) was co-incubated with or without OsRLCK118/176 and was subjected to phospho-protein visualization by immunoblotting and Pro-Q staining. The assays showed that GST-OsRbohB-N was strongly phosphorylated by OsRLCK118, but not by SDS2 or OsRLCK176 (Figure 7B), suggesting that OsRbohB activity might be regulated by OsRLCK118. Mass spectrometry analysis was applied to identify phosphorylation sites on GST-OsRbohB-N. The result showed that, with a coverage of 80.56%, 10 phosphor-Ser/Thr sites have been identified (Figure S7B), of which only one site is found to be conserved between rice OsRbohB (Ser32) and Arabidopsis AtRbohD (Ser39). This result suggests regulatory variation on ROS production between rice and Arabidopsis.
Figure 7. OsRLCK118 Interacts with and Phosphorylates the NADPH Oxidase OsRbohB.
(A) Luciferase complementation imaging (LCI) analysis of the OsRLCK118/176-OsRbohB interactions in N. benthamiana. OsRLCK118/176 and OsRbohB were fused to the N terminus (NL) and C terminus (CL) of luciferase, respectively. Co-infiltrated leaves were treated with 1 mM luciferin and applied to fluoresce imaging 3 days after infiltration. Light spectrum bar indicates signal intensity.
(B) Protein levels of OsRLCK118/176 and OsRbohB in LCI analysis are determined by western blot. CBB staining is used to show loading amount. Red and black arrows indicate OsRbohB-CL and OsRLCK118/OsRLCK176-NL, respectively.
(C) Phosphorylation of OsRbohB by OsRLCK118 in vitro. GST-OsRbohB-N was used as the substrate to detect phosphorylation by OsRLCK118/176 and SDS2. GST-OsRbohB-N phosphorylation was visualized by immunoblotting (anti-pThr) and Pro-Q staining. CBB staining shows loading amounts. “None” (lane 7) indicates no kinase was added. See also Figure S7.
DISCUSSION
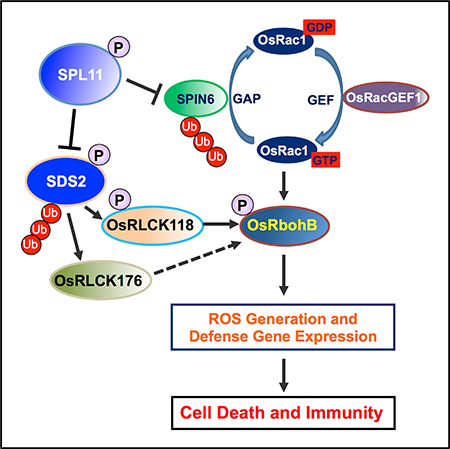
Via a cell death suppressor screen, we demonstrated that SDS2 is a monocot-specific SD-1-type RLK that functions as a positive regulator of cell death and innate immunity. A series of genetic and biochemical analysis further revealed that a plasma-membrane-associated complex consisting of SDS2, the E3 ubiquitin ligase SPL11, and RLCKs OsRLCK118/176 regulates plant cell death, immunity to fungal infections, and PTI signaling in rice. The architecture of the SDS2-mediated signaling bears several similarities with the Arabidopsis PRR signaling. First, activation of SDS2 induces PTI responses and enhanced resistance, which is similar to PRR activations in Arabidopsis by PAMP treatments (Figures 2 and S2). Second, constitutive activation of SDS2 in SDS2-GFP transgenic plants leads to rice growth suppression, which is reminiscent of Arabidopsis growth suppression caused by continuous treatment with PAMPs (Figures 2 and S2) (Gómez-Gómez and Boller, 2000; Liu et al., 2013; Zipfel et al., 2006). Third, SDS2 is negatively regulated by SPL11, which mirrors the regulation of AtFLS2 by PUB12/13 (Figure 4) (Lu et al., 2011). SDS2 interacts with and phosphorylates RLCK members OsRLCK118/176, which are downstream of SDS2 and upstream of the NADPH oxidase OsRbohB, similar with Arabidopsis PRRs (Figure 5) (Couto and Zipfel, 2016). These similarities suggest that SDS2 may function as a receptor or co-receptor to sense PAMPs and/or DAMPs in rice. However, our Y2H assays showed that SDS2 does not interact with the PRRs and partners of chitin and flg22 in rice, ruling out the possibility that it is a co-receptor (Figure S3F). Notably, plants evolve lineage-specific PRRs, suggesting recognition differences exist among plants. For example, EFR and LORE are Brassicaceae-specific PRRs recognizing bacterial EF-Tu and LPS, respectively, while CORE is a Solanaceae-specific PRR recognizing bacterial cold-shock protein (Ranf et al., 2015; Saur et al., 2016; Wang et al., 2016). Thus, SDS2 represents a RLK of PTI in monocots.
In response to pathogen infection, rice immunity is initiated by PRR complexes such as the chitin and PGN receptor, OsCERK1. OsCERK1 activates immunity via OsRacGEF1 and OsRLCK185 (Couto and Zipfel, 2016). OsRacGEF1 positively regulates the activity of the key immune molecule OsRac1 for ROS production and defense gene activation (Akamatsu et al., 2013). OsRLCK185 phosphorylates MAPKKKε and MAPKKK18 to initiate rice immunity (Wang et al., 2017; Yamada et al., 2017). In our previous study, we reported that SPL11 interacts with SPIN6, which negatively regulates the activity of OsRac1 (Liu et al., 2015). In this study, we found that SDS2 interacts with and phosphorylates SPL11, which in turn ubiquitinates SDS2, leading to its degradation. These findings suggest several important areas of future investigation on the mode of action and regulation of SDS2 in rice immunity. First, the ligand of SDS2 should be identified. Second, the co-receptor of SDS2, if any, for defense signaling after ligand recognition is not known. Third, the regulation of SDS2 by both SPL11-mediated ubiquitination and OsRLCK118-mediated phosphorylation should be further investigated. Finally, the relationship between SDS2 and other rice PRRs should be determined.
In rice, both OsRLCK176 and OsRLCK185 interact with OsCERK1 and positively regulate responses to PGN and chitin (Ao et al., 2014; Yamaguchi et al., 2013). In this study, we demonstrate that the SDS2 signaling pathway depends on OsRLCK118. OsRLCK118 functions immediately downstream of SDS2 and upstream of OsRbohB to transduce SDS2 signaling and to activate ROS burst. OsRLCK118 probably doesn’t link other PRRs to OsRbohB, because phosphorylation of OsRLCK118 depends on SDS2 (Figure 5C). For PAMP-induced ROS production, OsRLCK118 functions equivalent to AtBIK1 in Arabidopsis that interacts AtRbohD upon PRR activation (Kadota et al., 2014; Li et al., 2014). In fact, it remains challenging to identify the AtBIK1 orthologue in rice based on phylogenetic analysis, because multiple paralogues are present in both genomes (Shiu et al., 2004). OsRLCK176 may have a minor role but similar to that of OsRLCK118, because ROS production upon PAMP induction is also reduced in osrlck176 plants (Figure S6G). Alternatively, OsRLCK176 may have a specific role in other aspects of immune responses, because it has been shown to work downstream of chitin and PGN sensing and BR signaling (Ao et al., 2014; Zhou et al., 2016). Further investigation of the roles of OsRLCK118 and OsRLCK176 in the immune signaling, as well as their relationship with OsRbohB in ROS generation and defense gene activation, will be of great interest for future explorations.
STAR★METHODS
KEY RESOURCES TABLE
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Phospho-Threnine (anti-pThr) | Cell Signaling Technology | Cat# 9831S |
| Rat anti-HA | Roche | Cat# 11867423001, RRID:AB_10094468 |
| Rabbit anti-myc | Cell Signaling Technology | Cat# 2278S, RRID:AB_10693332 |
| Mouse anti-MBP | New England Biolabs | Cat# E8032, RRID:AB_1559730 |
| Mouse anti-beta-Tubulin | Cell Signaling Technology | Cat# 86298, RRID:AB_2715541 |
| Rabbit anti-Luciferase | Sigma-Aldrich | Cat# L0159; RRID:AB_260379 |
| Rabbit anti-Ubiquitin | Enzo Life Sciences | Cat# BML-PW0930, RRID:AB_10998070 |
| Mouse anti-GFP | Sigma-Aldrich | Cat# 11814460001, RRID:AB_390913 |
| Rabbit anti-SDS2 (poly-clonal) | This paper | N/A |
| Mouse anti-SPL11 (mono-clonal) | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ubiquitin (human, recombinant) (untagged) | Enzo | Cat# BML-UW8795–0005 |
| Ubiquitin mutant (all K to R) (human, recombinant) (untagged) | Enzo | Cat# BML-UW0205–1000 |
| Wheat E1 | This paper | N/A |
| Arabidopsis E2 (UBC10) | This paper | N/A |
| Lambda Protein Phosphatase (λPP) | New England Biolabs | Cat# P0753 |
| Cellulase RS | Yakult Pharmaceutical | CAS: 9012-54-8 |
| Macerozyme R-10 | Yakult Pharmaceutical | CAS: 9032-75-1 |
| Chitin | Santa Cruz Biotechnology | Cat# SC-217876, CAS: 1398-61-4 |
| flagellin22 (flg22) | GenScript | Cat# RP 19986 |
| MG115 (26S Proteasome inhibitor) | AG Scientific | Cat# M-1155, CAS: 133407-86-0 |
| Cycloheximide (CHX) | Sigma-Aldrich | Cat# C7698, CAS: 66-81-9 |
| Pro-Q Diamond Dye | Invitrogen | Cat# P33301 |
| Luminol | Sigma-Aldrich | Cat# 123072, CAS: 521-31-3 |
| Luciferin | Goldbio | Cat# LUCK-1G, CAS: 15144-35-9 |
| 3,3-Diaminobenzidine (DAB) | Sigma-Aldrich | Cat# D8001, CAS: 91-95-2 |
| PEG4000 | Sigma-Aldrich | Cat# 81240, CAS: 25322-68-3 |
| Brefeldin A (BFA) | Cell Signaling Technology | Cat# 9972, CAS: 20350-15-6 |
| Wortmannin (WM) | Cell Signaling Technology | Cat# ,9951, CAS: 19545-26-7 |
| Experimental Models: Organisms/Strains | ||
| Rice: IR64 | This paper | N/A |
| Rice: Nipponbare (NPB) | This paper | N/A |
| Rice: spl11 (IR64 background) | This paper | N/A |
| Rice: sds2 (IR64 background) | This paper | N/A |
| Rice: sds2 spl11 (IR64 background) | This paper | N/A |
| Rice: TP309spl11 (spl11 in TP309 background) | This paper | N/A |
| Rice: TP309 | This paper | N/A |
| Rice: pUbi::sds2III/TP309spl11(transgenic line) | This paper | N/A |
| Rice: pUbi::SDS2-GFP/NPB | This paper | N/A |
| Rice: pUbi::SDS2K540E-GFP/NPB | This paper | N/A |
| Rice: SDS2-ACT (SDS2 activation tagging line, Kitaake background) | This paper | N/A |
| Rice: osrlck118–1 (Dongjin Background) | This paper | N/A |
| Rice: osrlck118–2 (Dongjin Background) | This paper | N/A |
| Rice: osrlck176–1 (Dongjin Background) | This paper | N/A |
| Rice: osrlck176–2 (Dongjin Background) | This paper | N/A |
| Rice: osrlck118–1 spl11 double mutant | This paper | N/A |
| Rice: osrlck118–2 spl11 double mutant | This paper | N/A |
| Rice: osrlck176–1 spl11 double mutant | This paper | N/A |
| Rice: osrlck118–2 SDS2-ACT double mutant | This paper | N/A |
| Magnaporthe oryzae: isolates RO1 −1 and PO6–6 | This paper | N/A |
| Oligonucleotides | ||
| Primers for SDS2 qRT-PCR, see Table S2 | This paper | N/A |
| Primers for SDS2 variants, see Table S2 | This paper | N/A |
| Primers for OsRLCK118 qRT-PCR, see Table S2 | This paper | N/A |
| Primers for OsRLCK118 qRT-PCR, see Table S2 | This paper | N/A |
| Primers for OsPAL1 qRT-PCR, see Table S2 | This paper | N/A |
| Primers for OsPR5 qRT-PCR, see Table S2 | This paper | N/A |
| Primers for rice blast fungal biomass assay, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| pCAMBIA-pUbi::SDS2-GFP | This paper | N/A |
| pCAMBIA-pUbi::SDS2K540E-GFP | This paper | N/A |
| pCAMBIA-pUbi::sds2lll | This paper | N/A |
| pYBA-35S::RFP | This paper | N/A |
| pYBA-35S::OsRLCK118-GFP | This paper | N/A |
| pYBA-35S::OsRLCK176-GFP | This paper | N/A |
| PSPYNE173-SDS2 (SDS2-YN) | This paper | N/A |
| pSPYNEI73-SDS2K540E (SDS2K540E-YN) | This paper | N/A |
| pSPYCE(M)-SPL11Δ3aa (SPL11Δ3aa-YC) | This paper | N/A |
| pSPYCE(M)-OsRLCK118 (OsRLCK118-YC) | This paper | N/A |
| pSPYCE(M)-OsRLCK176 (OsRLCK176-YC) | This paper | N/A |
| pSPYCE(M)-OsRLCK185 (OsRLCK185-YC) | This paper | N/A |
| pSPYCE(M)-OsRLCK107 (OsRLCK107-YC) | This paper | N/A |
| pYBA-35S::OsRLCK118-HA | This paper | N/A |
| pYBA-35S::OsRLCK176-HA | This paper | N/A |
| pYBA-35S::OsRLCK185-HA | This paper | N/A |
| pYBA-35S::OsRLCK107-HA | This paper | N/A |
| PYBA-35S:: SDS2-HA | This paper | N/A |
| pYBA-35S:: SDS2K540E-HA | This paper | N/A |
| PYBA-35S:: OsCERK1-HA | This paper | N/A |
| pDBIeu-SDS2KD | This paper | N/A |
| pDBIeu-SDS2K540E | This paper | N/A |
| pPC86-SPL11 | This paper | N/A |
| pPC86-SPL11Δ3aa | This paper | N/A |
| pPC86-SPL11V290R | This paper | N/A |
| pPC86-SPL11-NT | This paper | N/A |
| PPC86-SPL11-CT | This paper | N/A |
| pPC86-SPL11-Arm | This paper | N/A |
| pGBKT7-SDS2KD | This paper | N/A |
| pGBKT7-SDS2K540E | This paper | N/A |
| PGADT7-SPL11 | This paper | N/A |
| pGADT7-SPL11Δ3aa | This paper | N/A |
| pGADT7-SPL11V290R | This paper | N/A |
| pGADT7-OsPUB12 | This paper | N/A |
| pGADT7- OsPUB12Δ3aa | This paper | N/A |
| pMAL-c2-SDS2KD | This paper | N/A |
| pMAL-c2-SDS2K540E | This paper | N/A |
| pMAL-c2-SPL11 | This paper | N/A |
| pMAL-c2-SPL11Δ3aa | This paper | N/A |
| pMAL-c2-OsRbohB-N (N erminus, 355 aa) | This paper | N/A |
| pGEX-6p-SDS2KD | This paper | N/A |
| pGEX-6p-SDS2K540E | This paper | N/A |
| pGEX-6p-OsRLCK118 | This paper | N/A |
| pGEX-6p-OsRLCK118K116E | This paper | N/A |
| pGEX-6p-OsRLCK176 | This paper | N/A |
| pGEX-6p-OsRLCK176K108E | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | Ver 1.49a; RRID:SCR_003070 |
| Image Lab software | Bio-Rad | RRID:SCR_014210 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Guo-Liang Wang (wang.620@osu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rice (Oryza sativa)
Rice plants were cultured in a greenhouse for crosses and phenotype observations. Plants in growth chambers were grown for M. oryzae inoculations and ROS assays. Growth chambers were set as: 26°C at day and 20°C at night, 80% humidity, 12 h light and 12 h dark. Rice cultivars and mutants used in the study are: 1902, GR5717 (spl11), IR64, TP309 and TP309spl11 as described (Shirsekar et al., 2014; Zeng et al., 2004). The SDS2-ACT line (PFG_K-02938), OsRLCK176 mutant lines (PFG_K-03350, PFG_2A-60144 and PFG_2C-10348) and OsRLCK118 mutant lines (PFG_3D-01619 and PFG_4A-01155) were obtained from the POSTECH rice insertion mutant library. The methods of rice transformation for the generation of transgenic lines were described previously (Park et al., 2012).
The generation of TP309spl11/sds2III was done by transformation of TP309 with the sds2III construct. The derived transgenic plants, named TP309/sds2III, were crossed with TP309spl11. In the F2 generation, TP309spl11/sds2III segregants were obtained.
In the culture conditions mentioned above, spl11-mediated or SDS2-ACT mediated cell death lesions appeared on sheath and older leaves about 6 weeks after transplanting. The spontaneous cell death lesions showed brown color while blast disease lesions were diamond-shaped with gray color in the middle.
Blast Fungus (Magnaporthe oryzae)
The blast fungus isolates RO1–1 and PO6–6 were cultured at room temperature on oat meal medium [3% (w/v) oat and 1.5% (w/v) Agar]. The fungal cultures were grown in dark for 5 days and was then under light for sporulation for 4–5 days. The spores were scratched from the plates and suspended in water (with 0.03% v/v Tween-20). The suspension was filtered with Miracloth before inoculation. The concentrations for spray inoculation and punch inoculation were 1 × 105 and 5 × 105, respectively.
METHOD DETAILS
In Vitro Phosphorylation Assay
The in vitro phosphorylation assay was performed by following the method described previously with modifications (Lu et al., 2010). Briefly, 2–5μg affinity-purified proteins were incubated at 30°C for 3 h with gentle shaking in 30μl phosphorylation buffer (50mM Tris-HCl, pH7.5, 10mM MgCl2, 10mM MnCl2, 1mM DTT, 10μM ATP) with or without 5μCi of [32P]-γ-ATP. The reaction was stopped by adding the 4X SDS loading buffer. The phosphorylated proteins were visualized by autoradiography, Pro-Q staining or immunoblotting after being resolved in 10% SDS-PAGE gels. Pro-Q staining was based on the product instructions (Molecular Probes). Briefly, a SDS-PAGE gel was fixed with the fixation buffer (50% methanol, 10% acetic acid) twice, 30 min each, and then, the gel was washed with ultrapure water 3 times, 10 min each. The treated gel was stained with Pro-Q dye for 60–90 min. Then, the stained gel was de-stained with the destaining buffer (20% acetonitrile, 50mM sodium acetate, pH 4) for 3 times, 30 min each. At last, the gel was washed with ultrapure water twice, 5 min each. Pro-Q stained gels were scanned by a laser scanner Typhoon 9400 (Amersham Biosciences) with 532 nm excitations and 560 nm longpass filter.
M. oryzae Inoculation and Fungal Biomass
For spray inoculation, germinated seeds were sowed in small pots (20 seeds per pot) and the seedlings were grown in a growth chamber for two weeks until the 3.5-leaf stage. The plants were inoculated with M. oryzae spore suspension as described previously (Qu et al., 2006). The spray inoculation was scored at 5 days post inoculation (dpi). For punch inoculation, seedlings were cultured in big pots (6 plants per pot) for six weeks. Punch inoculation was done always on second leaf (count from top) by following the method described previously (Park et al., 2012). The phenotype was scored at 9 dpi. Fungal biomass was determined by quantitative PCR analysis of the M. oryzae transposable element Pot2.
ROS Assay
The ROS detection method was described previously (Park et al., 2012). Briefly, rice leaf discs were cut and suspended in water overnight. To detect ROS production, one leaf disc was soaked in 100ml solution with luminol substrate, peroxidase and a PAMP. Real-time ROS production was recorded by Promage GloMax 20/20 single tube Luminometer. Three replicates were performed for each treatment.
Yeast Two-Hybrid Assay
The yeast two-hybrid screen used in the study was described previously with modifications (Park et al., 2012). Briefly, yeast (MaV203) competent cells were chemically induced with 100mM LiAc for 30min at 30°C. The competent cells were suspended with suspension solution (30% PEG3350, 100 mM LiAc). One to two micrograms of each plasmid DNA were mixed with 200μl competent cells. Salmon DNA (250ng/ml) was added and mixed well and then the mix was incubated for 30min at 30°C. Followed by heat shock of the competent cells at 42°C for 30min, the tube was inverted every 5 min during heat shock. After heat shock, the cells were transformed and were applied to the selection medium (SC/-Leu-Trp). Co-transformants were grown in 2–3 days at 30°C, and then cultured on selection medium (SC/-Leu-Trp- His, with 3-AT) to detect the interactions.
Protoplast Isolation and Transfection
Protoplasts were isolated by following the method developed by Zhang et al with modifications (Zhang et al., 2011). Etiolated, rice seedlings of 10–14 day-day old grown on ½ MS medium in the dark were used for protoplast isolation. The sheath and stem of seedlings were cut to 0.5mm strips and soaked in the cell wall digestion buffer (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.6 M Mannitol, 10mM MES, 10mM CaCl2, 0.1% BSA, pH5.7) for 6 h in the dark with gentle shaking (60–80 rpm). After digestion, strips were washed with W5 solution (154mM NaCl, 125mM CaCl2, 5mM KCl and 2mM MES, 0.5% Glucose, pH 5.7) and filtered with 40μm Nylon meshes. Protoplasts were collected by centrifuge at 1500 rpm for 3 min and washed with W5 for 3 times, then suspended in MMG solution (0.4 M mannitol, 15 mM MgCl2 and 4 mM MES, pH 5.7) to a concentration of 2 X 106 cells per minilitter.
Protoplast transfections were done using PEG-mediated method. Briefly, 5–15mg plasmids were mixed with 100ml protoplasts. Equal volume of PEG solution [40% (W/V) PEG 4000 (Sigma-Aldrich), 0.2 M Mannitol and 0.1 M CaCl2] were added and mixed by inverting tubes gently. The mixture was incubated at room temperature for 10 min in the dark. Then, twice volumes of W5 solution were added to that mixture and mix by inverting tubes. Protoplasts were collected by centrifuging and the pellet was suspended in W5 solution in the dark for 16 h.
In Vivo Degradation Assay
Wild-type (IR64) and spl11 protoplasts were transfected with equal amount of SDS2-HA and GFP plasmids at the same time. Twenty-four h after transfection, protoplasts were treated with 50mg/ml protein synthesis inhibitor Cycloheximide (CHX, Sigma-Aldrich). Equal volume of protoplasts suspension was taken to detect protein levels at different time points (0, 1, 2, 4 and 6h). 26S Proteasome inhibitor MG115 was applied at 100mM to suppress ubiqitination-mediated degradation. GFP protein was used as a control. Protoplast protein was extracted with extraction buffer (50mM Tris-HCl, pH 7.5, 0.5M Sucrose, 10mM EDTA, 5mM DTT and plant protease inhibitor cocktail). Protein levels were determined by immunoblotting.
In Vitro Ubiquitination Assay
In vitro ubiquitination of SDS2 by SPL11 was done by following the method described previously (Park et al., 2012). Briefly, the ubiquitination assay was carried out in a 30ml reaction system including 40ng of wheat E1, 100ng of Arabidopsis E2 (UBC10), 0.5μg of ubiquitin, 2μg MBP-SDS2 and 2μg MBP-SPL11 or MBP-SPL11Δ3aa. The ubiquitination assay was performed at 30°C in ubiquitination buffer (50mM Tris-HCl, 2mM ATP, 5mM MgCl2, 2mM DTT, 30mM creatine phosphate, and 50mg/ml creatine phosphokinase, pH 7.5). The reaction was stopped by adding 5X SDS loading buffer. Ubiquitinated protein was detected by immunoblotting.
SDS2 Antibodies Production
Three polymorphic region of SDS2 (indica allele) were selected to raise antibodies in rabbit. Two of them are located at extracellular domain and another is at intracellular domain. The peptides antigen were chemically synthesized and coupled with a single cysteine at N-termini. The sequences of peptides are: QKEGKEKSHPHHS, IPNARLPDNGKKLT and SAEMDAHDGSFSQN. The synthesis peptides and immunization of animals were carried by GenScript (Beijing, China).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters are reported in the figures and figure legends. Normally, the quantitative PCR analyses of genes transcripts and fungal biomass and ROS assays are calculated from 3 replicates and the values are presented as mean ± SD. Significance of differences were analyzed by two-tailed Student’s t test between two groups and by one-way ANOVA followed by Tukey test between multiple groups. Asterisks indicate the different statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Supplementary Material
Highlights.
The RLK SDS2 positively regulates plant cell death and immunity in rice
SDS2 phosphorylates E3 ligase SPL11, which in turn ubiquitinates SDS2 for degradation
SDS2 phosphorylates receptor-like cytoplasmic kinases RLCK118
RLCK118 interacts with and phosphorylates the NADPH oxidase OsRbohB
ACKNOWLEDGMENTS
We thank Dr. Anna Dobritsa from the Ohio State University for allowing us to use her confocal. This work is supported by grants the National Key Research and Development Program of China (#2016YFD0100600) and the USDA-NIFA Hatch Project (OHO00231) to the Ohio Agricultural Research and Development Center (OARDC), NIH (1R01GM097247), and the Robert A. Welch foundation (A-1795) to L.S. and by the Young Elite Scientist Sponsorship Program by China Association for Science and Technology (2015QNRC001) to Y.N. Rice mutants were obtained from Dr. G. An, Republic of Korea.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.chom.2018.03.003.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, and Shimamoto K (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Albert I, Bohm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, and Raaymakers TM (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1, 15140. [DOI] [PubMed] [Google Scholar]
- Ao Y, Li Z, Feng D, Xiong F, Liu J, Li JF, Wang M, Wang J, Liu B, and Wang HB (2014). OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, and Nürnberger T (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20, 47–54. [DOI] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, and Delarue M (2015). To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, and Zipfel C (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, and Boller T (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K, et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 111, E404–E413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Vance RE, and Dangl JL (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, and Zipfel C (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kawano Y, and Shimamoto K (2013). Early signaling network in rice PRR-mediated and R-mediated immunity. Curr. Opin. Plant Biol. 16, 496–504. [DOI] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Liao D, Cao Y, Sun X, Espinoza C, Nguyen CT, Liang Y, and Stacey G (2017). Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 214, 1646–1656. [DOI] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, et al. (2012). Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24, 3406–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, and Zhou JM (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Park CH, He F, Nagano M, Wang M, Bellizzi M, Zhang K, Zeng X, Liu W, Ning Y, et al. (2015). The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog. 11, e1004629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, and He P (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, and Shan L (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Ishikawa T, Fujiwara M, Fukao Y, Kawano Y, Kawai-Yamada M, and Shimamoto K (2016). Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell 28, 1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, and Wang G (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172, 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA, Sánchez-Carballo PM, Zähringer U, Hückelhoven R, Lee J, and Scheel D (2015). A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol 16, 426–433. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, and Boller T (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, and Goring DR (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147, 2084–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur IM, Kadota Y, Sklenar J, Holton NJ, Smakowska E, Belkhadir Y, Zipfel C, and Rathjen JP (2016). NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 113, 3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, and Shibuya N (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsekar GS, Vega-Sanchez ME, Bordeos A, Baraoidan M, Swisshelm A, Fan J, Park CH, Leung H, and Wang GL (2014). Identification and characterization of suppressor mutants of spl11- mediated cell death in rice. Mol. Plant Microbe Interact. 27, 528–536. [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Karlowski WM, Pan R, Tzeng Y-H, Mayer KFX, and Li W-H (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, and Isogai A (2005). Self-incompatibility in plants. Annu. Rev. Plant Biol. 56, 467–489. [DOI] [PubMed] [Google Scholar]
- Trujillo M (2018). News from the PUB: plant U-box type E3 ubiquitin ligases. J. Exp. Bot 69, 371–384. [DOI] [PubMed] [Google Scholar]
- Wang J, Qu B, Dou S, Li L, Yin D, Pang Z, Zhou Z, Tian M, Liu G, Xie Q, et al. (2015). The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 15, 49.25849162 [Google Scholar]
- Wang L, Albert M, Einig E, Furst U, Krust D, and Felix G (2016). The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2, 16185. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang G, Zhang C, Zhu P, Dai H, Yu N, He Z, Xu L, and Wang E (2017). OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol. Plant 10, 619–633. [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, and Shimamoto K (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19, 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Li M, and Liu P (2013). Evolution of S-domain receptor-like kinases in land plants and origination of S-locus receptor kinases in Brassicaceae. BMC Evol. Biol 13, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K, et al. (2016). The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35, 2468–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Yoshimura S, Terauchi A, and Kawasaki T (2017). Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol. 58, 993–1002. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, et al. (2013). A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13, 347–357. [DOI] [PubMed] [Google Scholar]
- You Q, Zhai K, Yang D, Yang W, Wu J, Liu J, Pan W, Wang J, Zhu X, Jian Y, et al. (2016). An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769. [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, and Wang GL (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang J, Peng C, Zhu X, Yin J, Li W, He M, Wang J, Chern M, Yuan C, et al. (2016). Four receptor-like cytoplasmic kinases regulate development and immunity in rice. Plant Cell Environ. 39, 1381–1392. [DOI] [PubMed] [Google Scholar]
- Zipfel C (2014). Plant pattern-recognition receptors. Trends Immunol. 35, 345–351. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, and Felix G (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.