SUMMARY
Antiviral immunity in Drosophila involves RNA interference and poorly characterized inducible responses. Here, we showed that two components of the IMD pathway, the kinase dIKKβ and the transcription factor Relish, were required to control infection by two picorna-like viruses. We identified a set of genes induced by viral infection and regulated by IKKβ and Relish, which included an ortholog of STING. We showed that dSTING participated in the control of infection by picorna-like viruses, acting upstream of dIKKβ to regulate expression of Nazo, an antiviral factor. Our data reveal an antiviral function for STING in an animal model devoid of interferons, and suggest an evolutionarily ancient role for this molecule in antiviral immunity.
INTRODUCTION
Viral infections represent a major burden for all organisms. Because of their intimate association with host cells, from which they hijack the molecular machineries to replicate, these obligate intracellular pathogens offer few targets for sensing and neutralization and thus pose important challenges to the immune system of the host. Furthermore, high mutation rates promote the rapid evolution of viruses, and adaptation to antiviral mechanisms. This results in a permanent arms race between host and viruses, which favors the diversification of host-defense mechanisms. Investigating virus-host interactions in a broad range of animals can therefore reveal innovative strategies of antiviral immunity (Marques and Imler, 2016).
The fruit fly Drosophila melanogaster has been a useful model to decipher host-pathogen interactions, revealing unexpected conservation between innate immunity pathways in mammals and insects. Indeed, bacterial and fungal infections in Drosophila are controlled by the Immune deficiency (IMD) and Toll pathways, which share several similarities with the Tumor Necrosis Factor receptor and interleukin-1-Toll like receptor pathways in mammals (reviewed in Hoffmann, 2003; Hoffmann et al., 1999; Lemaitre and Hoffmann, 2007). In the case of viral infections, insects largely rely on RNA interference (RNAi), whereas mammalian antiviral innate immunity is predominantly orchestrated by the strong and rapid induction of cytokines of the interferon family (reviewed in Ding, 2010; Paro et al., 2015; Schneider et al., 2014; tenOever, 2016). Yet, analysis of the transcriptome of virus-infected flies revealed deregulated expression of large sets of genes (Kemp et al., 2013; Merkling et al., 2015b; Xu et al., 2012). Some of these changes in gene expression might reflect responses to stress or altered physiology (Chtarbanova et al., 2014; Merkling et al., 2015b). Interestingly, the IMD (Avadhanula et al., 2009; Costa et al., 2009; Huang et al., 2013), Toll (Zambon et al., 2005) but also Jak-STAT (Dostert et al., 2005; Merkling et al., 2015a) pathways have been proposed to play a role in antiviral immunity in Drosophila. However, the mechanism of activation of these pathways in the context of viral infection and the function of the induced genes remain poorly characterized (Lamiable and Imler, 2014).
We have recently characterized the function of the gene diedel (die), which is upregulated after infection by several viruses in Drosophila (Lamiable et al., 2016). Die encodes a 12kDa immunomodulatory cytokine that down-regulates the IMD pathway, preventing its sustained activation and potentially detrimental consequences for the fly (Lamiable et al., 2016). Significantly, several insect DNA viruses belonging to different families (Ascoviridae, Baculovoridae and Entomopoxviridae) express genes homologous to die (Zaghloul et al., 2017). The identification of a suppressor of the IMD pathway within viral genomes provides useful indications of the restrictive pressures mounted by this pathway against viruses and prompted us to analyze its contribution in antiviral immunity (Marques and Imler, 2016). We report here that two components of the IMD pathway, the kinase dIKKβ and the NF-ĸB transcription factor Relish, but not the pathway as a whole, were critical in the control of infections by picorna-like viruses in Drosophila.
RESULTS
dIKKβ and Relish, but not dIKKγ participate in the control of DCV infection
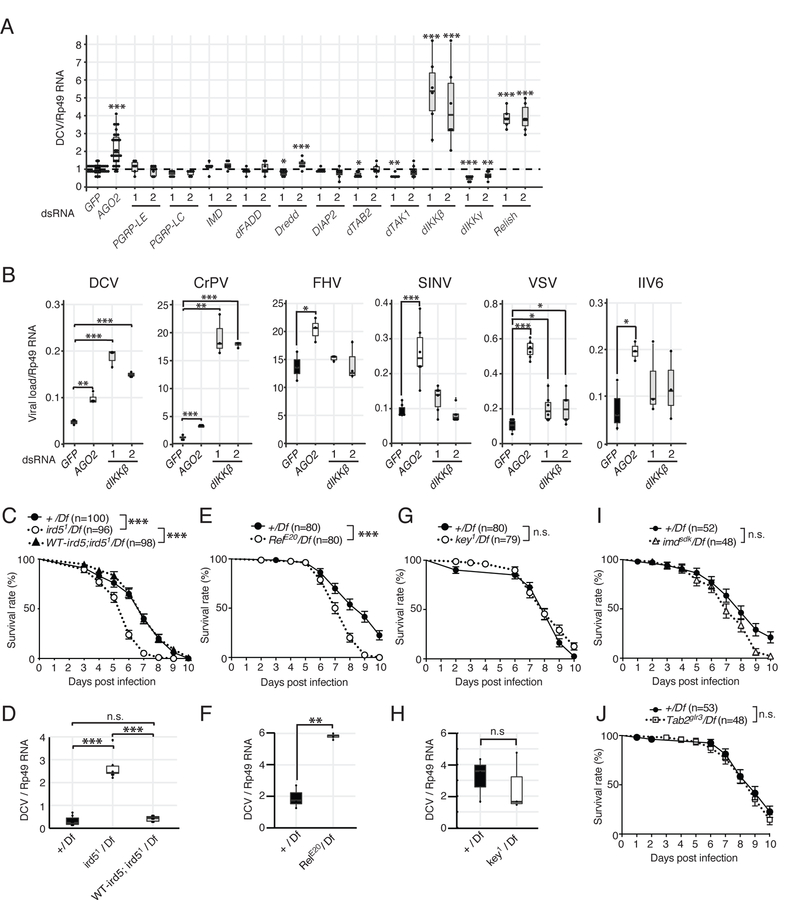
To address the involvement of the IMD pathway in antiviral immunity, we used macrophage-like S2 cells in which we individually knocked down by RNAi canonical components of the pathway, from the receptors PGRP-LE and -LC to the NF-ĸB transcription factor Relish. The cells were then challenged with Drosophila C Virus (DCV), a natural fly pathogen. To rule out possible off-target effects, each gene was knocked down by two independent dsRNAs, targeting a different region of the gene. Silencing of all genes of the pathway, with the exception of PGRP-LE, which is not expressed in this cell line, resulted in a significant decrease of the expression of the antimicrobial peptide Cecropin A1 upon stimulation with heat-killed Escherichia coli (Figure S1A). By contrast, accumulation of DCV RNA was not affected in most conditions, indicating that the IMD pathway does not restrict replication of this virus in these cells (Figure 1A). Strikingly however, silencing of two genes, ird5 encoding the ortholog of IKKβ (dIKKβ) and Relish encoding the p105-like NF-ĸB transcription factor activated by the IMD pathway, resulted in significant increases of viral RNA (Figure 1A). Of note, silencing of the gene kenny (key), which encodes the ortholog of NEMO (also known as IKKγ) (dIKKγ), the regulatory subunit of the IĸB kinase, resulted in the opposite phenotype, namely a significant decrease of DCV replication. Monitoring of the accumulation of the viral coat protein and of the infectious titer confirmed that dIKKβ restricted DCV replication (Figures S1B–D). We next investigated the impact of dIKKβ on the replication of other viruses. Silencing dIKKβ resulted in a strong increase of viral RNA when cells were infected with Cricket Paralysis Virus (CrPV), which belongs to the same family as DCV (Dicistroviridae). A small but significant increase of viral RNA was also observed in the case of the RNA virus VSV (Vesicular Stomatitis Virus) when expression of dIKKβ was knocked down (Figure 1B). However, silencing of dIKKβ did not affect replication of the three other viruses tested, namely the RNA viruses Flock house virus (FHV) and Sindbis Virus (SINV), and the DNA virus Invertebrate iridescent virus 6 (IIV6) (Figure 1B).
Figure 1. dIKKβ and Relish, but not the regulatory subunit dIKKγ, participate in resistance to DCV infection.
(A) S2 cells were treated with dsRNAs for each major component of the IMD pathway, infected with DCV and its replication was monitored by RT-qPCR. Two different regions of each gene designated as (1) and (2) were targeted by dsRNAs to detect possible off-target effects. GFP and AGO2 were used as negative or positive controls, respectively.
(B) S2 cells were treated with control or dIKKβ targetting dsRNAs, infected with the indicated viruses, and viral load was monitored by RT-qPCR 16h (DCV, CrPV, FHV) or 48h (VSV, SINV, IIV6) later.
(C and D) Control (+/Df), dIKKβ null mutant (ird51/Df) and genomic rescue (WT-ird5;ird51/Df) flies were infected with DCV and survival rates (C) and viral RNA loads (D) were monitored.
(E–J) Similar analyses were performed on null mutant flies for Relish (RelE20/Df) (E and F), dIKKγ (key1/Df) (G and H), imd (imdsdk/Df) (I) and TAB2 (Tab2glr3/Df) (J). (A and B) Data are representative from 3–5 independent experiments, each containing at least 3 biological replicates, except for IIV6 and SINV (n=2). (C-J) Mean of at least three independent experiments is shown. For survival curves, the numbers of flies is indicated and log-rank (Mantel Cox) test was used. (D,F and H) Mean of 3 independent experiments, each involving at least two biological replicates. Boxplots represent the median (horizontal line) and 1st/3rd quartiles, with whiskers extending to points within 1.5 times the interquartile range. Student t-test, p*<0.05. p**<0.01. p***<0.001. n.s. indicates statistically non significant. See also Figure S1.
We confirmed these results in vivo with mutant flies. Flies hemizygote for the ird51 null mutation (ird51/Df(3R)Exel7328) succumbed more rapidly to DCV injection than controls and contained increased amount of viral RNA at 3 days post-infection (dpi). Importantly, this phenotype was rescued by a transgene containing a genomic copy of dIKKβ (Figures 1C and 1D). The resistance of Relish mutant flies to DCV was also impaired (Figures 1E and 1F). By contrast, resistance to DCV infection was not affected in flies mutant for three other established members of the IMD pathway, dIKKγ, imd and Tab2 (Figures 1G–1J).
Taken together, these results reveal that two components of the IMD pathway namely dIKKβ and Relish, restrict infection by two picorna-like viruses in S2 cells and in flies, in addition to their well-characterized role in antibacterial immunity.
Identification of virus-induced genes differentially regulated by dIKKβ and dIKKγ
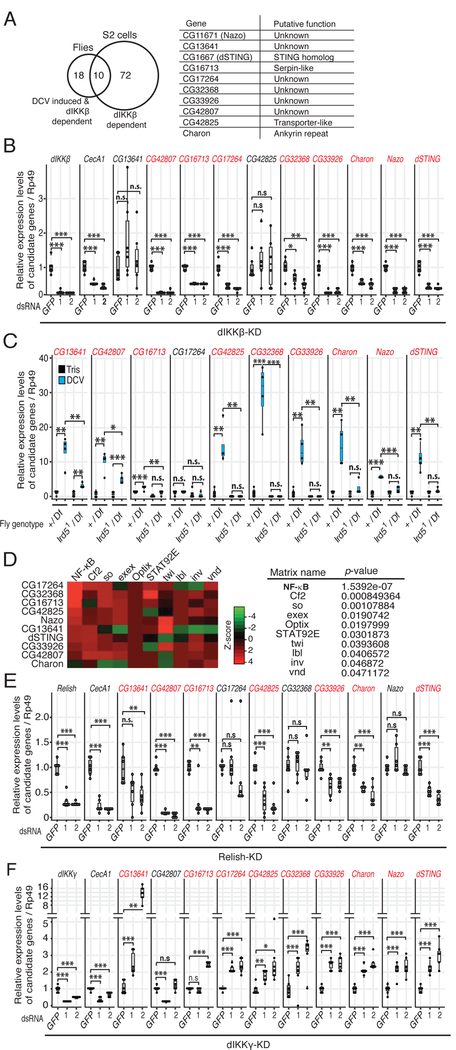
We next performed transcriptomic analysis to identify genes regulated by dIKKb in the context of viral infection in flies and S2 cells (Data deposited to GSE99043 and GSE99044). Using the in vivo infection model, we identified 28 genes induced by DCV in a dIKKβ-dependent manner (Table S1). In S2 cells, only few genes were up-regulated by DCV and CrPV infection, with a strong heat shock response signature, as previously reported (Merkling et al., 2015b). Upregulation of these genes was not affected by dIKKβ silencing. This led us to consider that dIKKβ may regulate constitutive expression of antiviral genes in S2 cells. Indeed, 82 constitutively expressed genes were significantly down-regulated in S2 cells when expression of the kinase was knocked-down (Table S2). Of note, ten of these genes are induced in a dIKKβ-dependent manner in DCV infected flies (Figure 2A). RT-qPCR analysis confirmed that these ten genes are regulated by dIKKβ in S2 cells or in DCV-infected flies (Figures 2B and 2C).
Figure 2. A subset of virus-induced genes are regulated positively by dIKKβ and Relish, and negatively by dIKKγ.
(A) The Venn diagram presents the genes positively regulated by dIKKβ in DCV infected flies and in S2 cells, as determined by DNA microarray analyses. The table shows the list of the 10 common genes and their putative function. Note that these genes are extracted from biological duplicates with the criteria of S.D.≦20% and FDR<0.05.
(B and C) Validation of the microarray data for the 10 genes by RT-qPCR analysis in dIKKβ knock- down (KD) cells (B) and in dIKKβ null mutant flies (ird51/Df) (C). GFP dsRNA and +/Df flies were used as controls.
(D) Pscan analysis of the proximal −500 bp promoter of the 10 genes, showing the top 10 putative transcription factors.
(E and F) Expression of the 10 dIKKβ-dependent genes in Relish-KD and dIKKγ-KD cells. CecA1 expression was used as a control. Data are from 2 (B, E and F) or 4 (C) independent experiments, each containing 6 (B, E and F) or 4 (C) biological replicates. A color code highlights the dIKKβ, dIKKγ and Relish dependent genes in panels (B,C,E,F). Statistics are the same as in Figure 1. See also Figure S2.
Promoter analysis of the ten genes revealed a strong enrichment for consensus NF-kB binding sites (Figure 2D). Indeed, RT-qPCR analysis confirmed that expression of at least 7 out of the 10 genes depends on Relish (Figure 2E). Knock-down of dIKKγ resulted in the opposite phenotype, namely increased gene expression for at least 8 of the 10 genes (Figure 2F). Using luciferase reporter plasmids, we confirmed that the activity of the promoter of the gene CG1667 (dSTING) decreased when dIKKβ or Relish were silenced, and increased when dIKKγ was silenced (Figures S2A and S2B). The activity of the promoter decreased gradually when its sequences were truncated from −900 to −150bp, but this did not affect the regulation by the three components of the IMD pathway. However, this regulation was lost when the promoter was truncated to −100bp. Mutation of the consensus NF-ĸB-binding motif (−23 to −13 nt) resulted in a significant decrease of promoter activity and a complete loss of regulation by dIKKβ, Relish and dIKKγ (Figure S2C).
In summary, these data indicate that dIKKβ and Relish regulate a subset of virus-induced genes independently of dIKKγ. Furthermore, the opposite effects of dIKKβ and dIKKγ silencing on viral RNA accumulation (Figure 1A) and on regulation of gene expression (Figures 2B and 2F) suggest that the 10 virus-induced genes we identified are associated with the control of viral replication.
Identification of antiviral factors regulated by dIKKβ
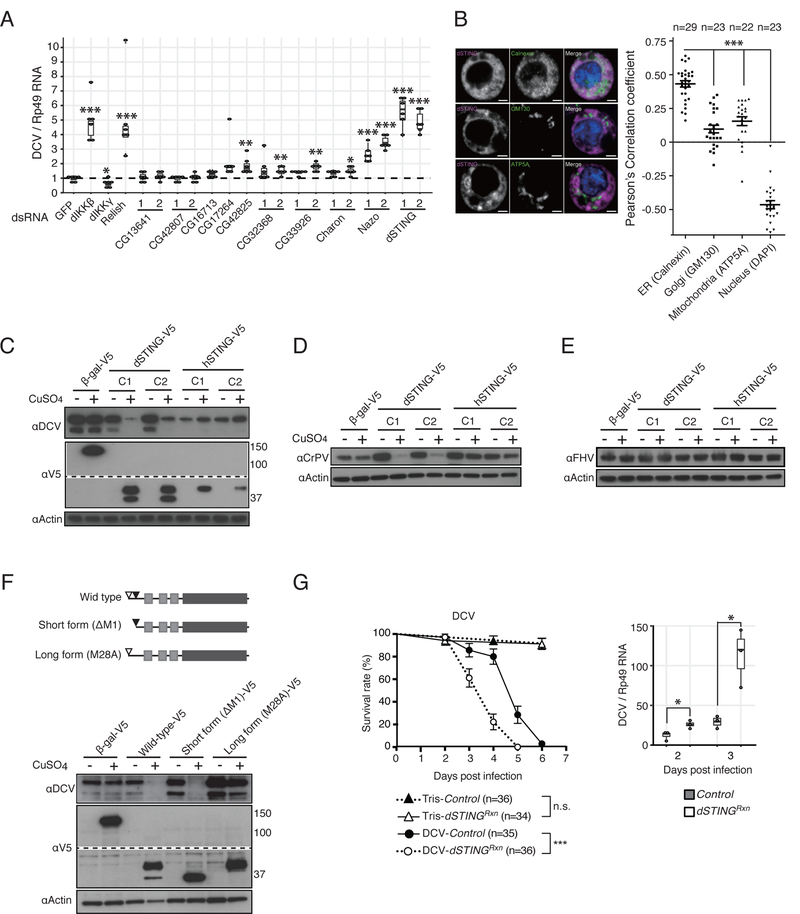
We individually knocked-down expression of the 10 dIKKβ-regulated genes in S2 cells and monitored DCV replication by RT-qPCR. We observed a striking increase in viral RNA accumulation when expression of the gene CG11671 or CG1667 was silenced. In addition, silencing of four other genes resulted in significant increase of DCV RNA with one of the two dsRNAs preparation tested (Figure 3A). CG1667 is an orthologue of the mammalian gene stimulator of interferon genes (STING) and we hereafter refer to it as dSTING. CG11671 is an uncharacterized Drosophila gene that we named Nazo, meaning enigma in Japanese. Viral titration assays confirmed that knock-down of both dSTING and Nazo resulted in increased production of DCV infectious viral particles (Figure S3A).
Figure 3. dSTING, a dIKKb-regulated gene, is necessary and sufficient to restrict replication of Dicistroviridae.
(A) Expression of the 10 dIKKβ-regulated genes was silenced using dsRNA targeting one or two regions of the genes (labelled 1 and 2). Cells were infected with DCV and viral replication was monitored. A representative from two experiments is shown, each containing 6 biological replicates.
(B) Intracellular localization of V5-tagged version of dSTING using the indicated markers for co-localization.
(C–E) Stable cell lines for inducible expression of dSTING or hSTING were established, infected with DCV (C), CrPV (D) or FHV (E) and analyzed by immunoblot using the indicated antibodies. A b-galactosidase (b-gal) expressing cell line was used as control. Results for two independent clones (labelled C1 and C2) are shown.
(F) Stable cell lines expressing the two isoforms of dSTING (DM1 and M28A) were established, infected with DCV and the cell lysates were analyzed by immunoblot using the indicated antibodies.
(G) dSTING null mutant (dSTINGRxn) and control flies were challenged with DCV and survival rate and viral RNA load were monitored at the indicated time points. Data representative of two independent experiments each involving 3 biological replicates are shown. Statistics are the same as in Figure 1. See also Figure S3 and S4A–C.
We next generated stable Drosophila cell lines expressing tagged versions of dSTING or, as a control, human STING (hSTING). Both proteins exhibit a vesicular pattern in S2 cells, and co-localize with the endoplasmic reticulum (ER) marker calnexin (Figure 3B and Figure S3B). Overexpression of dSTING, but not hSTING, resulted in a strong inhibition of DCV infection revealing that dSTING is both necessary and sufficient to control DCV (Figure 3C). In agreement with the virus-specific effect of dIKKβ (Figure 1B), overexpression of dSTING also resulted in a strong decrease of CrPV replication but did not affect FHV (Figures 3D and 3E). We noted that, while hSTING was expressed as a ~42 kDa protein, two bands of ~37 kDa and ~42 kDa were reproducibly observed for dSTING, which has an expected molecular weight of 40 kDa (Figure 3C and Figure S3C,D). Edman degradation analysis revealed that the long form starts at the predicted Methionine residue, while the short form starts at a downstream Methionine corresponding to position 28 in the full-length protein (Figure S3H). Accordingly, mutating the methionine at position 1 or 28 resulted in expression of the short or long isoform, respectively (Figure 3G). Western blot analysis with an antiserum raised against recombinant dSTING confirmed the existence of the long and short isoforms in S2 cells (Figure S3C). In addition, we identified four putative N-glycosylation sites in dSTING and found by LC-MS/MS analysis that at least one of them is glycosylated in the long form but not in the short form (Figures S3D–F, Table S3).
STING proteins are transmembrane (TM) proteins with their active domain corresponding to the C-terminal half of the protein facing the cytosol (Chen et al., 2016). Of note, both isoforms of dSTING have three predicted TM domains instead of four in mammalian STING. This odd number of TM domains raises the question of whether the C-terminal region faces the lumen of the ER or the cytosol. To clarify this point, we expressed dSTING and hSTING with N- or C-terminally fused AVI tag in S2 cells expressing the bacterial BirA enzyme in the cytosol. As expected, we observed that hSTING was predominantly biotinylated when the tag was inserted at its C-terminus, even though some labeling was also observed for the N-terminal tag (Figure S3G). The short form of dSTING, but not the long form, was also biotinylated when the tag was inserted at the C-terminus. We conclude that the long and short isoforms of dSTING have different membrane topologies, consistent with their different glycosylation patterns (Figure S3H). Finally, we tested the function of the short and long isoforms of dSTING and observed that only the short form efficiently represses viral replication (Figure 3F and Figure S3I).
We created a Drosophila mutant fly line containing a deletion of the gene encoding dSTING (Roxanne allele) by mobilizing a transposable element (Figure S4A). dSTINGRxn mutant flies were more susceptible to DCV infection than control flies in the same genetic background in which the transposable element was precisely excised and contained an increased viral RNA load at 2 and 3 dpi (Figure 3G). As observed for dIKKβ mutants, dSTINGRxn mutant flies also showed high susceptibility to CrPV, but not to FHV and SINV infections (Figure S4B). Increased viral titers were also observed when the dSTINGRxn allele was tested in hemizygous flies, using the Df(2R)BSC133, which covers dSTING. Furthermore, transheterozygote flies containing the deletion allele Orc635, which deletes the whole Orc6 gene and the 3’ end of dSTING (Balasov et al., 2009), and the dSTINGRxn allele succumbed more rapidly than control containing the Orc635 allele in front of the chromosome with the precise excision of the EP(2)06491 element, when infected with DCV (data not shown). dSTINGRxn mutant flies resisted like controls infection by the bacteria Escherichia coli and Micrococcus luteus or by the fungus Beauveria bassiana. In addition, the mutation did not impair the induction of antimicrobial peptides by these infections (Figure S4C). We conclude that dSTING participates in an antiviral response, but is largely dispensable for antibacterial or antifungal immunity in Drosophila.
dSTING acts upstream of dIKKβ and Relish in an antiviral pathway
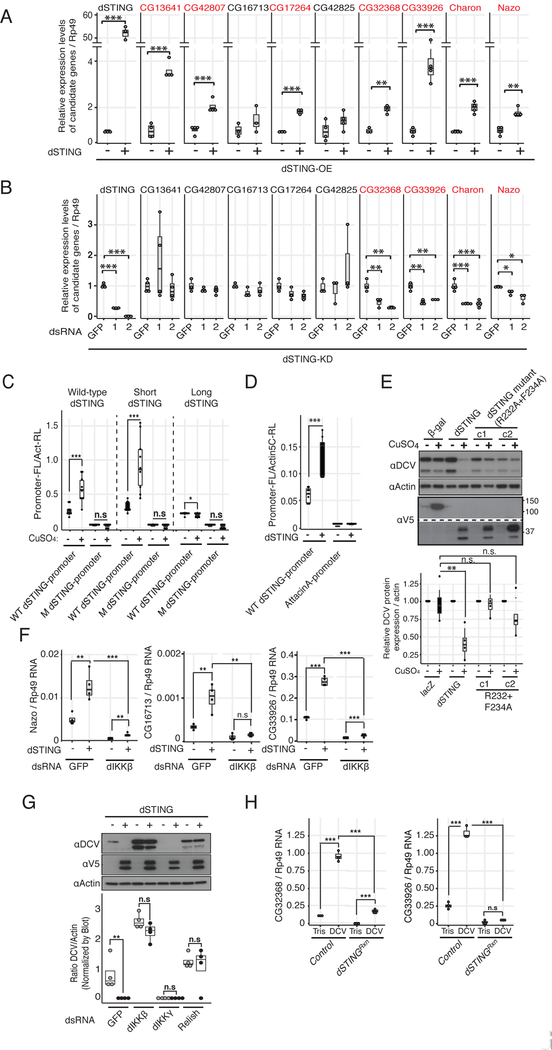
We next investigated whether dSTING is involved in the regulation of dIKKβ-dependent genes. Overexpression of dSTING in stably transfected cells was sufficient to significantly up-regulate 7 out of 9 genes regulated by dIKKβ in addition to dSTING (Figure 4A). Furthermore, silencing of dSTING resulted in significant down-regulation of four of these genes in unstimulated S2 cells (Figure 4B). These experiments suggest that dSTING and dIKKβ regulate a similar subset of genes. Importantly, dSTING overexpression activated the dSTING promoter in a luciferase reporter assay and this was abrogated when the putative NF-ĸB binding site was mutated (Figure 4C). Similar induction was observed when the short isoform of dSTING was expressed. By contrast, the long isoform was inactive (Figure 4C). Of note, dSTING overexpression did not induce the promoter of the Attacin-A gene, a target of the canonical IMD pathway (Figure 4D). Interestingly, mutating the residues R232 and F234, corresponding in hSTING to R238 and Y240, which are involved in cyclic dinucleotide (CDN) binding (Chen et al., 2016), abrogated the antiviral activity of dSTING (Figure 4E).
Figure 4. dSTING acts upstream of dIKKb and Relish in an antiviral pathway.
(A) Expression of dIKKβ-regulated genes was monitored before and after dSTING overexpression (dSTING-OE).
(B) Same as in (A), but in cells in which expression of dSTING was knocked-down by RNAi (dSTING-KD). Data are representative of at least 3 experiments except for CG32368 (n=2), each containing 3 or 4 biological replicates. A color code highlights dSTING dependent genes.
(C) Reporter plasmids expressing Firefly luciferase (FL) under the control of the dSTING proximal promoter wild-type (WT) or mutated (M) for the consensus NF-kB binding site were transfected in cell lines stably expressing the two isoforms (wild-type) or the short or long isoforms separately under the control of a methallothionein promoter. A co-transfected Actin5C-Renilla (Act-RL) vector was used to normalize for transfection efficiency. Luciferase activity was monitored 48h post-induction. The average of 3 experiments is shown.
(D) The indicated Firefly luciferase reporters were transfected in cell lines expressing dSTING, and their activity was monitored 48h post-induction. A co-transfected Actin5C-Renilla (Act-RL) vector was used as normalization. The average of 3 experiments is shown.
(E) Stable cell lines (c1 and c2) expressing a dSTING mutant with changes of two amino acids in the putative cGAMP binding site (R232A +F234A) were established and infected with DCV. The cell lysates prepared 24h post-infection were analysed by Western blot. The detected bands were quantified relative to actin signal (lower panel, n = 4 biological replicates).
(F and G) Epistasis analysis of dSTING and dIKKβ. dSTING overexpressing cells were treated with the indicated dsRNAs and expression of the dIKKβ-regulated genes Nazo, CG16713 and CG33926 was monitored. Data are representative of at least two experiments, each involving 4 biological replicates (F). dSTING-V5 expressing cells were treated with the indicated dsRNAs, infected with DCV and viral replication was monitored. aActin was used as loading control. The normalized ratios of DCV/Actin band intensities, from three independent experiments, were compared with two-way ANOVA, followed by multiple comparisons between induced and non-induced cells (G).
(H) dSTINGRxn and control flies were injected with Tris or DCV and expression of CG32368 and CG33926 was monitored. Data are representative of 3 experiments, each containing 3 biological replicates. Statistics are the same as in Figure 1. See also Figure S5.
We next conducted an epistasis analysis and observed by RT-qPCR that expression of Nazo, CG16713 and CG33926 in dSTING overexpressing cells was significantly reduced when dIKKβ was silenced (Figure 4F). In agreement with this finding, inhibition of virus replication by dSTING overexpression was much reduced or blocked in dIKKβ and Relish silenced cells (Figure 4G). In mammals, STING activates IKKβ and TBK1, which in turn activate NF-ĸB and IRF3, respectively. Whereas the mechanism of activation of IKKβ by STING is poorly characterized, TBK1 phosphorylates STING and this phosphorylation is critical for the subsequent activation of IRF3 (reviewed in Chen et al., 2016). dSTING lacks the C-terminal tail containing the phosphorylated amino-acids in mammalian STING (Liu et al., 2015) (Figure S5A), and we failed to detect phosphorylated peptides in dSTING. In addition, silencing of dIKKε, the drosophila orthologue of TBK1, had no impact on DCV replication (Figure S5B). We finally confirmed in vivo that the induction by DCV of the dIKKb and Relish regulated genes, CG32368 and CG33926, was completely blocked in dSTINGRxn mutant flies (Figure 4H). Taken together, these results indicate that dSTING functions in an antiviral pathway acting upstream of dIKKβ and Relish. Induction of the IKKb and STING-dependent genes CG32268, CG33926 and CG13641 was not dependent on AGO2, revealing that this pathway can be activated independently of RNAi in antiviral immunity (Figure S5C–D).
Nazo is a antiviral effector regulated by dSTING and IKKβ.
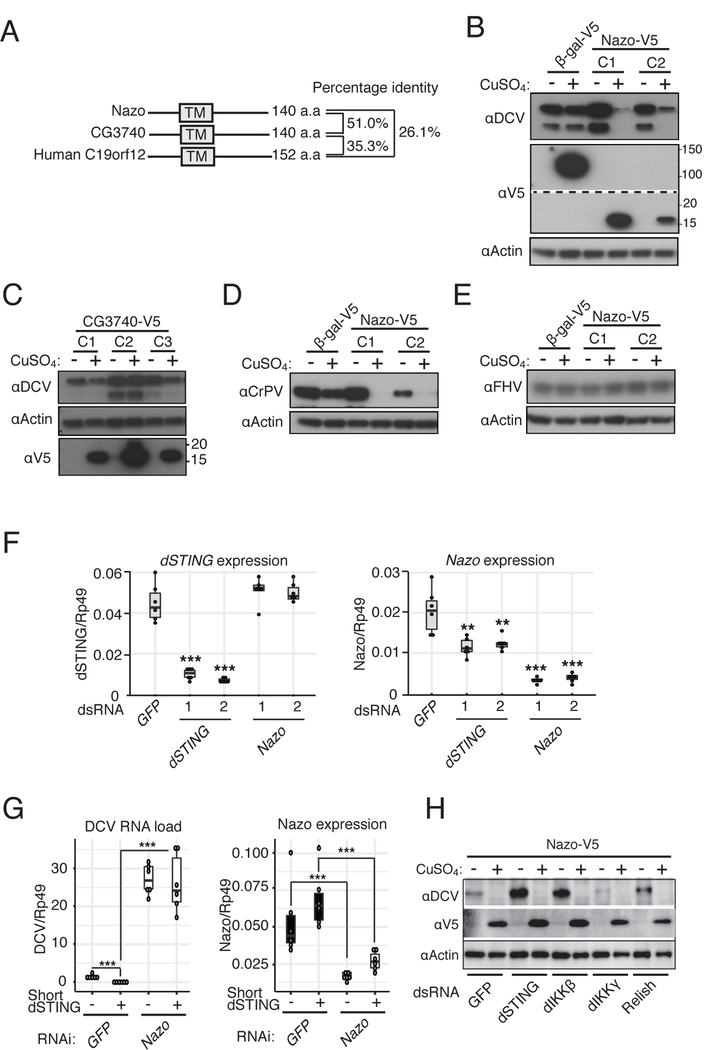
Nazo was the gene with the second highest increase of DCV replication when it was silenced (Figure 3A). Nazo encodes a 140 amino acid protein containing a predicted transmembrane domain. It is orthologous to the poorly characterized human protein C19orf12 (Figure 5A). A duplication occurred in the Drosophila lineage, resulting in two paralogues, Nazo and CG3740.
Figure 5. Nazo encodes a dSTING-regulated antiviral effector.
(A) Schematic representation and sequence identity between Nazo, CG3740 and human C19orf12.
(B – E) Stable cell lines expressing Nazo or CG3740 under the control of the CuSO4 inducible promoter were established and infected with DCV (B and C), CrPV (D) or FHV (E). The cell lysates prepared 24h post-infection were analyzed by immunoblot using the indicated antibodies. Data are representative of at least two independent experiments.
(F) Expression of both dSTING and Nazo was monitored by RT-qPCR in cells silenced for either dSTING or Nazo. Data are representative of 2 independent experiments, each containing 6 biological replicates.
(G) The impact of short dSTING overexpression on DCV replication was monitored in cells treated with a control dsRNA (GFP) or silenced for Nazo. The qRT-PCR results shown are the average of two independent experiments (three biological replicates per experiment).
(H) Nazo-V5 overexpressing cells were treated with the indicated dsRNAs, infected with DCV and viral replication was monitored by immunoblot. Data are a representative from at least two independent experiments. Statistics are the same as in Figure 1. See also Figure S4D,E.
We constructed stable cell lines expressing either Nazo or its paralog to test their function. Strikingly, Nazo overexpression was sufficient to strongly repress DCV or CrPV replication, although FHV replication was not affected (Figures 5B, 5D and 5E). By contrast, CG3740 overexpression had no effect on viral replication (Figure 5C). We generated Nazo mutant flies and challenged them with DCV and CrPV (Figure S4D,E). We only observed a mild increase of viral replication when flies were infected with DCV. Because Nazo is one of 28 genes induced by DCV in an IKKβ-dependent manner in vivo (Figure 2A), we hypothesize that functional redundancy accounts for this difference with S2 cells.
Silencing of dSTING resulted in decreased Nazo expression, in agreement with a signaling function of dSTING. However, dSTING expression was not affected by the silencing of Nazo, suggesting that Nazo does not carry a regulatory function in this new dSTING/dIKKβ pathway (Figure 5F). In addition, dSTING overexpression no longer repressed DCV replication when Nazo was silenced (Figure 5G). Finally, the strong antiviral activity associated with Nazo overexpression was not modified when dSTING, dIKKβ or Relish were silenced (Figure 5H), indicating that Nazo acts either downstream of or independently from these genes. Based on these results, we propose that Nazo is a novel antiviral factor specifically targeting picorna-like viruses upon activation of dSTING-dIKKβ.
DISCUSSION
Previous reports proposed a role for the IMD pathway in antiviral immunity in Drosophila (Avadhanula et al., 2009; Costa et al., 2009; Lamiable et al., 2016). Our results point to a critical role for the kinase dIKKβ and Relish in the control of infection by two members of the Dicistroviridae, DCV and CrPV. These two viruses express potent suppressors of RNAi (van Rij et al., 2006; Wang et al., 2006) and we propose that this inducible response represents a second layer of defense. Although induction of the STING-IKKβ-Relish pathway does not depend on AGO2, we cannot at this stage rule out that this pathway is completely independent from antiviral RNAi. For example, some of the induced genes may encode regulatory components of the siRNA pathway. The other known components of the IMD pathway, by contrast, are not involved. In particular, the regulatory subunit of the IKK kinase, dIKKγ, even seems to favor replication of these two viruses. Indeed, we observed an increased expression of a subset of virus-induced genes, and a corresponding decrease in replication of DCV, when dIKKγ was silenced. The recent discovery that in flies dIKKγ serves as an autophagy receptor and mediates the turnover of the IKK complex (Tusco et al., 2017) could explain how the inhibition of dIKKγ results in increased levels of dIKKβ, leading to decreased viral replication. An alternative explanation is that, as described in mammals, dIKKγ functions as a specificity scaffold to recruit IĸB proteins to IKKβ and restrict phosphorylation of alternative substrates (Schrofelbauer et al., 2012).
Intriguingly, our data indicate that Relish participates in both antibacterial and antiviral immunity regulated by dIKKβ. Of note, activation of Relish is complex and involves several regulatory steps, which are still incompletely understood (Erturk-Hasdemir et al., 2009; Kim et al., 2014; Kleino and Silverman, 2014). Clearly, additional studies are required to understand the regulation of cleavage, phosphorylation, nuclear translocation and transcriptional activity of Relish in the context of viral (dIKKγ-independent pathway), but also bacterial (dIKKγ-dependent pathway) infections. In the meantime, we can hypothesize that the antibacterial versus antiviral role of dIKKβ involves cooperation of Relish with other transcription factors. For example, nuclear factors such as Akirin, which bridge NF-ĸB proteins to chromatin remodeling (Bonnay et al., 2014; Tartey et al., 2014), may participate in the selectivity of Relish activity, although our preliminary results indicate that Akirin is required both in the context of bacterial and viral infections. NF-ĸB transcription factors bind DNA as homo- or hetero-dimers, which regulate distinct yet overlapping programs (Zhang et al., 2017). Of note, one of the ten genes that we identified as regulated by DCV and dIKKβ in cells and in flies is Charon, also known as pickle (Ji et al., 2016; Morris et al., 2016). This gene encodes an IĸB protein, which acts as a selective inhibitor of Relish homodimers, without affecting the heterodimers formed by Relish with the two other NF-ĸB proteins in flies, namely Dorsal and DIF (Morris et al., 2016). It will therefore be interesting to investigate the role of Charon in the regulation of virus-induced genes, and its possible dependency on dIKKβ. Finally, induction of an antiviral program of gene expression may involve cooperation between Relish and a member of another family of transcription factors, as occurs between NF-ĸB and IRF3 on the IFNβ promoter in mammals (Panne et al., 2007). The fact that the genes that we identified are not strongly induced by bacterial infections and that sequences other than the consensus NF-ĸB binding site control expression of dSTING support this latter hypothesis.
STING has emerged in recent years as a key component of a pathway driving antiviral immunity in mammals through induction of type I interferons in response to sensing of cytosolic DNA (Chen et al., 2016; Roers et al., 2016). One receptor for cytosolic DNA is the enzyme cGAS, which synthesizes upon activation a second messenger, the cyclic dinucleotide (CDN) composed of guanosine monophosphate (GMP) and adenosine monophosphate (AMP) connected by one non canonical 2’−5’ phosphodiester bond and one canonical 3’−5’ phosphodiester bond (2’3’cGAMP) (Sun et al., 2013). cGAMP binds to the endoplasmic reticulum resident protein STING, triggering interaction with the kinase TBK1 and signaling (Chen et al., 2016). TBK1 phosphorylates residues of the C-terminal tail (CTT) of STING, which allows for the recruitment of IRF3 and its subsequent phosphorylation by TBK1 (Liu et al., 2015). Of note, bacteria also synthesize CDNs such as c-di-AMP and c-di-GMP, which can be sensed by STING (Burdette et al., 2011; Woodward et al., 2010).
The presence of STING in invertebrates, which do not have interferon genes, had been noted, raising the question of the ancestral function of this gene (Kranzusch et al., 2015; Margolis et al., 2017; Wu et al., 2014). Indeed, the CTT of STING, which mediates activation of IRF3 and induction of interferons (none of which are encoded by the Drosophila genome) is a feature acquired in vertebrates (Margolis et al., 2017). In addition, cGAS-like molecules in invertebrates, including Drosophila, lack the zinc ribbon domain required for DNA binding (Margolis et al., 2017; Wu et al., 2014). This indicates that the main function currently known of the cGAS-STING axis, namely sensing of cytosolic DNA and downstream signaling, has been acquired in vertebrate lineages. Therefore, the ancient origin of STING indicates that this molecule must have other functions in invertebrates. Our data reveal that dSTING is associated with the control of infection by two RNA viruses of the Dicistroviridae family in Drosophila, suggesting that dSTING has long been associated with antiviral immunity. Control of DCV and CrPV by dSTING does not involve the drosophila orthologue of TBK1, and we failed to detect interaction between dSTING and IKKβ, suggesting that additional components of the pathway remain to be identified. Interestingly, an ancient function of STING in resistance to RNA viruses may still be operating in mammals (Aguirre et al., 2012; Chen et al., 2011; Franz et al., 2018; Ishikawa and Barber, 2008; Schoggins et al., 2014).
Among the important questions raised by our findings that need to be addressed now are (i) the signaling downstream of dSTING, in particular how it activates dIKKβ and Relish; (ii) the mechanism by which dSTING gets activated, since insect STINGs do not appear to bind CDNs (Kranzusch et al., 2015); and (iii) the mode of action of the antiviral molecules it regulates, such as the novel factor Nazo. The mutant dSTING fly line that we have established and the list of IKKβ- and STING-regulated genes that we have identified pave the way for the genetic and functional characterization of STING in Drosophila. This may possibly lead to discovery of conserved and still unknown functions for this molecule in mammals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms. A. Courtin for excellent technical assistance. P. Hamman and J. Chicher for dSTING proteomic approach. L. Coquet and T. Jouenne for Edman degradation sequencing. Microarray analyses were performed by the IGBMC Microarray and Sequencing platform, a member of the ‘France Génomique’ consortium (ANR-10-INBS-0009). This work was supported by the National Institute of Health (PO1 AI070167), the Agence Nationale de la Recherche (ANR-13-BSV3–0009 and ANR-17-CE15–0014), the Balzan Foundation (to J.A.H.), the Investissement d’Avenir Programs (ANR-10-LABX-0036 and ANR-11-EQPX-0022), the Sino-French Hoffmann Institute, CNRS and INSERM. N.M. was supported by a Marie Skłodowska Curie fellowship.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. (2012). DENV Inhibits Type I IFN Production in Infected Cells by Cleaving Human STING. PLoS pathogens 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avadhanula V, Weasner BP, Hardy GG, Kumar JP, and Hardy RW (2009). A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS pathogens 5, e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M, Huijbregts RP, and Chesnokov I (2009). Functional analysis of an Orc6 mutant in Drosophila. Proc Natl Acad Sci U S A 106, 10672–10677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnay F, Nguyen XH, Cohen-Berros E, Troxler L, Batsche E, Camonis J, Takeuchi O, Reichhart JM, and Matt N (2014). Akirin specifies NF-kappaB selectivity of Drosophila innate immune response via chromatin remodeling. EMBO J 33, 2349–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Sun H, You FP, Sun WX, Zhou X, Chen L, Yang J, Wang YT, Tang H, Guan YK, et al. (2011). Activation of STAT6 by STING Is Critical for Antiviral Innate Immunity. Cell 147, 436–446. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun L, and Chen ZJ (2016). Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17, 1142–1149. [DOI] [PubMed] [Google Scholar]
- Chtarbanova S, Lamiable O, Lee KZ, Galiana D, Troxler L, Meignin C, Hetru C, Hoffmann JA, Daeffler L, and Imler JL (2014). Drosophila C virus systemic infection leads to intestinal obstruction. J Virol 88, 14057–14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Jan E, Sarnow P, and Schneider D (2009). The Imd pathway is involved in antiviral immune responses in Drosophila. PloS one 4, e7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW (2010). RNA-based antiviral immunity. Nat Rev Immunol 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, and Imler JL (2005). The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6, 946–953. [DOI] [PubMed] [Google Scholar]
- Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stoven S, Meier P, and Silverman N (2009). Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A 106, 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz KM, Neidermyer WJ, Tan YJ, Whelan SPJ, and Kagan JC (2018). STING-dependent translation inhibition restricts RNA virus replication. Proc Natl Acad Sci U S A 115, E2058–E2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA (2003). The immune response of Drosophila. Nature 426, 33–38. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Kafatos FC, Janeway CA, and Ezekowitz RA (1999). Phylogenetic perspectives in innate immunity. Science 284, 1313–1318. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kingsolver MB, Avadhanula V, and Hardy RW (2013). An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J Virol 87, 4272–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, and Barber GN (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Thomas C, Tulin N, Lodhi N, Boamah E, Kolenko V, and Tulin AV (2016). Charon Mediates Immune Deficiency-Driven PARP-1-Dependent Immune Responses in Drosophila. Journal of immunology 197, 2382–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, et al. (2013). Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. Journal of immunology 190, 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Paik D, Rus F, and Silverman N (2014). The caspase-8 homolog Dredd cleaves Imd and Relish but is not inhibited by p35. J Biol Chem 289, 20092–20101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, and Silverman N (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol 42, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Wilson SC, Lee AS, Berger JM, Doudna JA, and Vance RE (2015). Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2’,3’ cGAMP Signaling. Mol Cell 59, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamiable O, and Imler JL (2014). Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol 20, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamiable O, Kellenberger C, Kemp C, Troxler L, Pelte N, Boutros M, Marques JT, Daeffler L, Hoffmann JA, Roussel A, and Imler JL (2016). Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc Natl Acad Sci U S A 113, 698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, and Hoffmann J (2007). The host defense of Drosophila melanogaster. Annu Rev Immunol 25, 697–743. [DOI] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, and Chen ZJ (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630. [DOI] [PubMed] [Google Scholar]
- Margolis SR, Wilson SC, and Vance RE (2017). Evolutionary Origins of cGAS-STING Signaling. Trends Immunol 38, 733–743. [DOI] [PubMed] [Google Scholar]
- Marques JT, and Imler JL (2016). The diversity of insect antiviral immunity: insights from viruses. Curr Opin Microbiol 32, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkling SH, Bronkhorst AW, Kramer JM, Overheul GJ, Schenck A, and Van Rij RP (2015a). The epigenetic regulator G9a mediates tolerance to RNA virus infection in Drosophila. PLoS pathogens 11, e1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkling SH, Overheul GJ, van Mierlo JT, Arends D, Gilissen C, and van Rij RP (2015b). The heat shock response restricts virus infection in Drosophila. Sci Rep 5, 12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris O, Liu X, Domingues C, Runchel C, Chai A, Basith S, Tenev T, Chen H, Choi S, Pennetta G, et al. (2016). Signal Integration by the IkappaB Protein Pickle Shapes Drosophila Innate Host Defense. Cell Host Microbe 20, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, and Harrison SC (2007). An atomic model of the interferon-beta enhanceosome. Cell 129, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro S, Imler JL, and Meignin C (2015). Sensing viral RNAs by Dicer/RIG-I like ATPases across species. Curr Opin Immunol 32, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Hiller B, and Hornung V (2016). Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 44, 739–754. [DOI] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, and Rice CM (2014). Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32, 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. (2014). Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Polley S, Behar M, Ghosh G, and Hoffmann A (2012). NEMO ensures signaling specificity of the pleiotropic IKKbeta by directing its kinase activity toward IkappaBalpha. Mol Cell 47, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LJ, Wu JX, Du FH, Chen X, and Chen ZJJ (2013). Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartey S, Matsushita K, Vandenbon A, Ori D, Imamura T, Mino T, Standley DM, Hoffmann JA, Reichhart JM, Akira S, and Takeuchi O (2014). Akirin2 is critical for inducing inflammatory genes by bridging IkappaB-zeta and the SWI/SNF complex. EMBO J 33, 2332–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR (2016). The Evolution of Antiviral Defense Systems. Cell Host Microbe 19, 142–149. [DOI] [PubMed] [Google Scholar]
- Tusco R, Jacomin AC, Jain A, Penman BS, Larsen KB, Johansen T, and Nezis IP (2017). Kenny mediates selective autophagic degradation of the IKK complex to control innate immune responses. Nat Commun 8, 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, and Andino R (2006). The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20, 2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, and Ding SW (2006). RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, and Portnoy DA (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu FH, Wang X, Wang L, Siedow JN, Zhang W, and Pei ZM (2014). Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res 42, 8243–8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Grant G, Sabin LR, Gordesky-Gold B, Yasunaga A, Tudor M, and Cherry S (2012). Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe 12, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul H, Hice R, Arensburger P, and Federici BA (2017). Transcriptome analysis of the Spodoptera frugiperda ascovirus in vivo provides insights into how its apoptosis inhibitors and caspase promote increased synthesis of viral vesicles and virion progeny. J Virol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Nandakumar M, Vakharia VN, and Wu LP (2005). The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A 102, 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Lenardo MJ, and Baltimore D (2017). 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell 168, 37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.