Abstract
Erythropoietin (EPO), a cytokine molecule, is best-known for its role in erythropoiesis. Preclinical studies have demonstrated that EPO has robust neuroprotective effects that appear to be independent of erythropoiesis. It is also being clinically tested for the treatment of neuropsychiatric illnesses due to its behavioral actions. A major limitation of EPO is that long-term administration results in excessive red blood cell production and increased blood viscosity. A chemical modification of EPO, carbamoylated erythropoietin (CEPO), reproduces the behavioral response of EPO in animal models but does not stimulate erythropoiesis. The molecular mechanisms involved in the behavioral effects of CEPO are not known. To obtain molecular insight we examined CEPO induced gene expression in neuronal cells. PC- 12 cells were treated with CEPO followed by genome-wide microarray analysis. We investigated the functional significance of the gene profile by unbiased bioinformatics analysis. The Ingenuity pathway analysis (IPA) software was employed. The results revealed activation of functions such as neuronal number and long-term potentiation. Regulated signaling cascades included categories such as neurotrophin, CREB, NGF and synaptic long-term potentiation signaling. Some of the regulated genes from these pathways are CAMKII, EGR1, FOS, GRIN1, KIF1B, NOTCH1. We also comparatively examined EPO and CEPO-induced gene expression for a subset of genes in the rat dentate gyrus. The CEPO gene profile shows the induction of genes and signaling cascades that have roles in neurogenesis and memory formation, mechanisms that can produce antidepressant and cognitive function enhancing activity.
Keywords: Gene regulation, Neurotrophic signaling, CREB signaling, PC-12, Antidepressant, Cognition
1. Introduction
Erythropoietin (EPO), a 30.4 kDa glycoprotein, is widely prescribed to treat anemia. Progress in understanding EPO’s biological actions over the past two decades has shown that it has robust neurotrophic and neuroprotective actions on the brain (Brines and Cerami, 2005). Clinical studies have attempted to harness the neurotrophic properties of EPO to treat schizophrenia and depression. Results from rigorous clinical trials in treatment resistant depression are promising, and indicate that EPO could be developed as an antidepressant and cognition enhancing agent (Miskowiak et al., 2014). Interestingly, neuroimaging and volumetric analyses indicate that the behavioral effects could be due to EPO’s actions on the hippocampus (Miskowiak et al., 2015), potentially providing direction for mechanistic understanding and hypothesis testing.
Despite the encouraging results from neuropsychiatry studies and the overall safety of EPO, it should be noted that EPO is a potent inducer of red blood cell production and could produce adverse hematological consequences in non-anemic individuals. Carbamoylated EPO (CEPO), a chemically engineered in vitro post- translational modification of EPO, has emerged as an attractive trophic molecule because it is devoid of erythropoietic activity. Yet, it retains the neurotrophic effects in the central nervous system (Leist et al., 2004). The precise mechanism involved in CEPO’s cellular actions are not well understood. It has been proposed that CEPO is non-erythropoietic, as it signals via an EPO receptor (EPOR)- beta common receptor (βcR\CD131) heteromer rather than the classical EPOR-EPOR dimer bound by EPO (Leist et al., 2004). We therefore hypothesized that the downstream signal transduction pathways and gene profile induced by CEPO would differ from EPO due to lack of erythropoietic activity, but exhibit overlap due to their shared neurotrophic activity. We used the neuronal phenotype PC12 cell line in a genome wide microarray analysis of gene expression to gain insight into CEPO’s mechanism of action in neurons. This cell line has been used to examine multiple aspects of EPO signaling (Um et al., 2007; Wu et al., 2007) and gene profiling analysis (Renzi et al., 2002). We subjected the CEPO gene profile to bioinformatics analysis to examine the functional implications of CEPO regulated genes and to shed light on mechanisms involved in its behavioral effects. We also conducted independent secondary validation of array data using quantitative PCR analysis.
2. Materials and Methods
2.1. Carbamoylation of EPO
Erythropoietin was purchased from Prospec Bio (Israel) and carbamoylated in 1 mg aliquots as per (Mun and Golper, 2000) with mild modifications. Briefly, EPO was deprotonated in a high pH (pH = 8.9) borate buffer and then exposed to potassium cyanate for 16h at 36°C. CEPO was exhaustively dialyzed for 6h against PBS. CEPO concentration was determined using the Qubit protein assay (ThermoFisher). CEPO purity was verified by silver staining after electrophoretic gel analysis.
2.2. Cell Culture
Rat pheochromocytoma cells (PC-12 cells) were obtained from American Type Culture Collection (ATCC). The cells were grown in suspension in RPMI-1640 (ATCC) with 10% heat inactivated horse serum, 5% fetal bovine serum (Gibco) at 37°C and 5% CO2. To differentiate the cells into neuronal cells PC-12 cells were plated in collagen coated dishes (Corning) and were grown in RPMI-1640 with NGF (100ng/ml, Alomone Labs) and 1% Horse Inactivated serum (Gibco). The cells were grown for 10 days and the medium was changed every 2 days. Neuronal morphology and robust neurite outgrowth was confirmed by microscopy. NGF was removed overnight before the day of experiment. PC-12 cells were treated with CEPO 100ng/ml for 3hrs. Vehicle-treated (PBS) cells were used as control.
2.3. Animals
Adult male Sprague-Dawley rats (n=6 per group, mass 220 – 240 gm; Envigo) and pair-housed according to treatment group (Vehicle, EPO and CEPO) for the duration of the experiments. Rats were maintained on a standard 12-hr light-dark cycle with free access to food and water. All procedures were carried out in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approval by the USD Institutional Animal Care and Use Committee. Every effort was made to minimize the number of animals used. Rats received single daily i.p. injections of either vehicle (PBS), EPO or CEPO (30 μg/kg) for 4 consecutive days. Five hours after the last dose animals were decapitated according to American Veterinary Medical Association guidelines and the brains were frozen on dry ice.
2.4. Laser microdissection
Cryocut hippocampal sections (coronal, 16 μm) were collected on PEN- Membrane slides (Leica) and processed for laser microdissection. Sections were fixed in histochoice fixative (Sigma) for 3 min followed by brief rinses in PBS, 70% ethanol and milliQ water. Sections were stained with cresyl violet (2 min), followed by dehydration in 95% and 100% ethanol. The dentate gyrus was delineated using the free draw tool on a Wacom Cintiq high resolution monitor and microdissected using the Leica LMD 7000 system. Microdissected tissue was collected directly into lysis solution and processed for RNA isolation using the RNAqueous micro kit (ThermoFisher). The RNA samples were then used for quantitative-PCR analysis.
2.5. RNA extraction and Microarray analysis
RNA was isolated using the RNAqueous kit (ThermoFisher) and quantitated using the NanoDrop spectrophotometer (ThermoFisher). RNA quality was tested using the Nano-assay on the Bio-analyzer (Agilent). RNA integrity numbers were ≥ 9 (see supplementary material). A dual color (Cy3 vs Cy5) microarray experiment (N=4) was performed using the Agilent rat whole genome expression array (Agilent rat (v3) GE, 4×44K-028282). Data was processed using Agilent feature extraction software and LOWESS dye normalization. GeneSpring software (Agilent) was then used to perform summarization, transformation, normalization, fold change calculation and regulation (up vs down).
2.6. Ingenuity Pathway Analysis (IPA)
Functional analysis of microarray data was conducted using IPA software. The dataset containing the gene identifiers and their corresponding average expression fold change values were uploaded as an excel spreadsheet to the IPA server. A fold change cutoff was set at 1.3 (both up and down regulated) to identify molecules which were differentially regulated. The network containing the genes from the dataset are either algorithmically generated or the genes from the dataset were assigned to biologically relevant networks and canonical pathways based on the information present in Ingenuity Pathway Knowledge Base. IPA uses two classes of statistical treatments for its functional analysis. 1) For all types of functional analysis IPA calculates p-values based on the association between molecules from the dataset and the pathways generated. The p-value is calculated using right-tailed Fisher’s Exact test and p-value ≤ 0.05 is considered as significant. 2) IPA also calculates z-scores for some of the functional analysis like canonical pathway analysis, diseases and functional analysis, and upstream regulator analysis based on correlation between what is known in IPA knowledge base and the data. The z-score predicts whether a pathway is activated or inhibited. A z-score of ≥ 2 means a pathway is activated and a score of ≤ −2 means a pathway is inhibited.
2.7. Quantitative PCR
Based on the results of IPA analysis and data from previous array analysis of EPO-induced gene expression in the hippocampus (Girgenti et al., 2009), we chose representative target genes for secondary validation. We focused our attention on genes with established neurobiological value, essentially genes that have been functionally validated in the literature by behavioral experiments. Quantitative PCR analysis was used for independent secondary validation as previously described (Sathyanesan et al., 2017). Briefly, reverse transcribed cDNA was amplified employing SYBR Green chemistry (Qiagen) and gene-specific primers in the Mastercycler realplex Real-time PCR machine (Eppendorf). Specificity of product was determined by melt curve analysis. Data were normalized using the housekeeping genes cyclophilin, p-actin and GAPDH.
2.8. Statistical Analysis
Statistically significant differential expressing genes in our dataset was determined by Significance Analysis of Microarray (SAM) in rstudio. Two class unpaired test was performed and false discovery rate (FDR) less than 1.5% was selected as acceptable. Quantitative PCR based gene regulation was calculated using the ΔΔCt method. Gene expression comparisons were considered statistically significant at p < 0.05. The results were replicated in an independent set. Data are presented as mean +/− s.e.m.
The quantitative-PCR data for in-vitro studies were statistically analyzed using the t-test and in-vivo studies were analyzed using ANOVA statistical analysis followed by Holm-Sidak tests using SigmaStat 4.0 software (Systat).
3. Results
3.1. Nervous system functions regulated by CEPO
Employing a cutoff of ±1.3 for the fold change values 2868 genes showed differential expression. There were 1535 upregulated genes and 1333 that were downregulated. In order to understand the functional significance of the genes expressed in our microarray study we used the diseases and functions tool under the IPA core analysis module. IPA generated list of top 10 up and downregulated genes are shown in Table 1. We then examined the nervous system related functions that were enriched in our dataset. The prominent functions that were activated included quantity of neurons or neuronal number (p ≤1.09E-10 and z-score = 2.84), long-term potentiation (p ≤ 0.00000296 and z-score = 2.506), and neurotransmission (p ≤ 6.79E-10 and z- score = 2.017) as shown in Table 2. Genes represented in the functional categories of neuronal number (Fig. 1A) and long-term potentiation (Fig. 1B) are shown along with cellular location of protein expression.
Table 1.
Representative top molecules identified by IPA analysis after CEPO treatment
| Gene Symbol | Fold Change |
Genebank Accn |
Description | Molecular Function |
|---|---|---|---|---|
|
Up-regulated genes |
||||
| Ddx3x | 14.599 | NM_ 0011082 46 | DEAD-box helicase 3, X-linked | Helicase |
| KAT2B | 12.401 | XM_ 0037506 17 | lysine acetyltransferase 2B | Transcription regulator |
| CIRBP | 10.999 | NM_031147 | cold inducible RNA binding protein | Translation regulator |
| CHRDL1 | 10.636 | NM_199502 | chordin like 1 | plays a role in neuronal differentiation |
| POGZ | 10.337 | NM_ 0011076 93 | pogo transposable element derived w ith ZNF domain | transposase |
| ALY REF | 9.963 | NM_ 0011096 02 | Aly/REF export factor | Transcription regulator |
| Cts8 | 9.288 | NM_ 0011282 16 | cathepsin 8 | Peptidase |
| ELAC1 | 9.193 | NM_ 0011074 06 | elaC ribonuclease Z 1 | ribonuclease |
| PPP1R3A | 9.023 | NM_ 0011092 22 | protein phosphatase 1 regulatory subunit 3A | Phosphatase |
| PAQR7 | 8.931 | NM_ 0010340 81 | progestin and adipoQ receptor family member 7 | Steroid membrane receptor |
|
Down-regulated genes |
||||
| CHMP4B* | −6.315 | NM_0012764 56 |
charged multivesicular body protein 4B | protein homodimerization activity |
| NPM3 | −4.762 | XM_0010585 48 | nucleophosmin/nucleoplasmin 3 | chaperone protein |
| GABRA5 | −3.948 | NM_017295 | gamma-aminobutyric acid type A receptor alpha5 subunit | Ion Channel |
| Raet11 | −3.785 | NM_0010130 63 | retinoic acid early transcript 1L | Peptide antigen binding |
| KLK1* | −3.73 | NM_0010053 82 | kallikrein 1 | Peptidase |
| GJD2 | −3.517 | NM_019281 | gap junction protein delta 2 | Transporter |
| SNW1 | −3.497 | NM_0011092 79 | SNW domain containing 1 | Transcription regulator |
| KIF1C | −3.353 | NM_145877 | kinesin family member 1C | microtubule motor activity |
| GZMB | −3.294 | NM_138517 | granzyme B | Peptidase |
| Olr1124 | −3.127 | NM 0010004 26 | olfactory receptor 1124 | G-protein coupled receptor |
Table shows top 10 upregulated and downregulated molecules with their gene symbol, fold change value, gene bank accession number, description and molecular function.
Table 2.
Functions related to Nervous system enriched in the dataset.
| Functions Annotation | p-Value | Activation z- score | Predicted Activation State | # Molecules |
|---|---|---|---|---|
| Neuronal Number | 1.09E-10 | 2.84 | Increased | 117 |
| long-term potentiation | 0.0000029B | 2.506 | Increased | 66 |
| Neurotransmission | 6.79E-10 | 2.017 | Increased | 108 |
The p-values and z scores were generated by IPA for categories neuronal number, long term potentiation and neurotransmission after core analysis. The significance and z scores are calculated by the IPA software as described in the methods section. The z ≥2 predicts increase in activation of that function. Differentially expressed genes in the dataset for each function is represented as number of molecules.
Figure 1. Most relevant neuronal functions and their interaction with the differentially expressed genes as predicted by IPA.
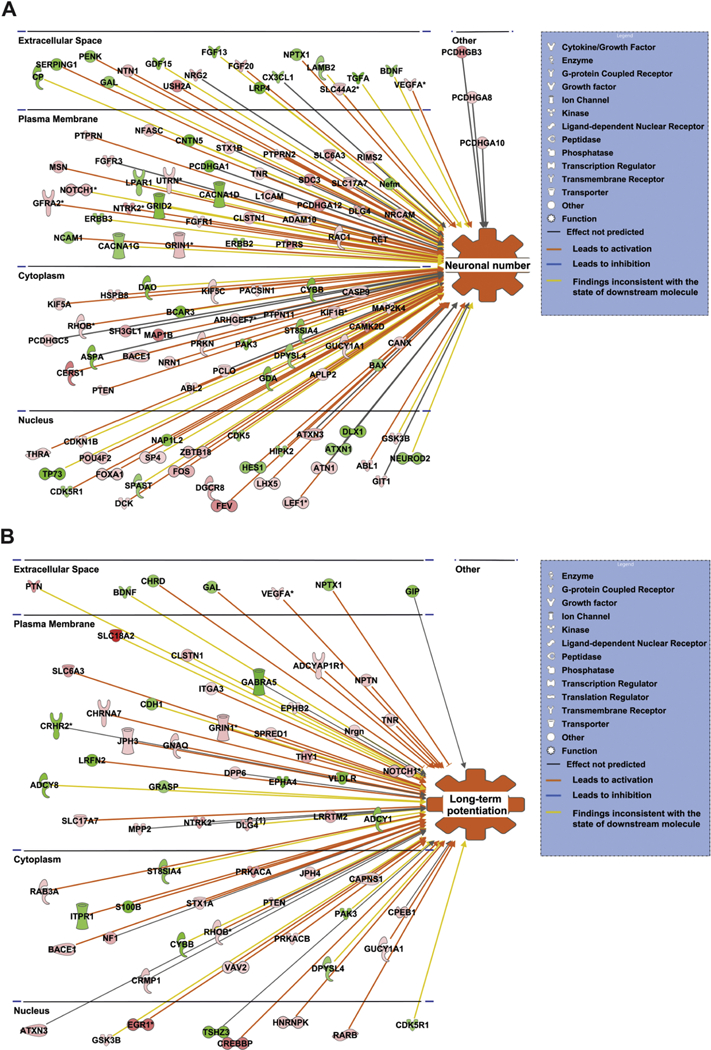
(A) Shows the genes associated with neuronal number. (B) Shows the genes associated with long term potentiation. Red and green nodes represent the differentially expressed genes. A red node denotes an up-regulated gene, and a green node denotes a down-regulated gene. The intensity of the node color indicates the degree of up or down regulation. Different structures of the nodes represent different functional class of the gene product. Orange line indicates that it leads to activation, blue line indicates that it leads to inhibition, grey line indicates that the effect is not predicted, and yellow line indicates that the findings are inconsistent with the state of the downstream molecule.
3.2. Neuronal signaling pathways regulated by CEPO
We then investigated the nervous system signaling pathways that were induced in our dataset. We utilized the canonical pathways tool in IPA’s core analysis module to generate canonical signaling pathways. Figure 2 shows that there are thirteen canonical signaling pathways related to nervous system signaling that are significantly upregulated and activated. The signaling pathways include gonadotropin-releasing hormone (GNRH) signaling (-log p-value=8.39 and z score=3.592), nerve growth factor (NGF) signaling (-log p-value=7.62 and z score=3.667), cAMP response element- binding protein (CREB) signaling in neurons (-log p-value=6.62 and z score=3.683), neurotrophin/tyrosine receptor kinase(TRK) signaling (-log p-value=6.49 and z score= 3.266), neuropathic pain signaling in dorsal horn neurons (-log p-value =6.16 and z score=3.656), glial cell-derived neurotrophic factor (GDNF) family ligand-receptor interactions (-log p-value =6.02 and z score=3.838), ErbB4 signaling (-log p-value=4.79 and z-score=2.985), ephrin receptor signaling (-log p-value=4.61 and z-score=4.426), cholecystokinin/gastrin-mediated signaling (-log p-value=4.55 and z-score=3.272), dopamine-DARPP32 feedback in cAMP signaling (-log p-value=3.5 and z- score=2.475), synaptic long-term potentiation signaling (-log p-value =2.75 and z- score=2.746), ciliary neurotrophic factor (CNTF) signaling (-log p-value=2.27 and z- score=2.84), and melatonin signaling (-log p-value=1.73 and z-score=2.84).
Figure 2. The canonical pathways associated with nervous system signaling that are significantly upregulated by CEPO.
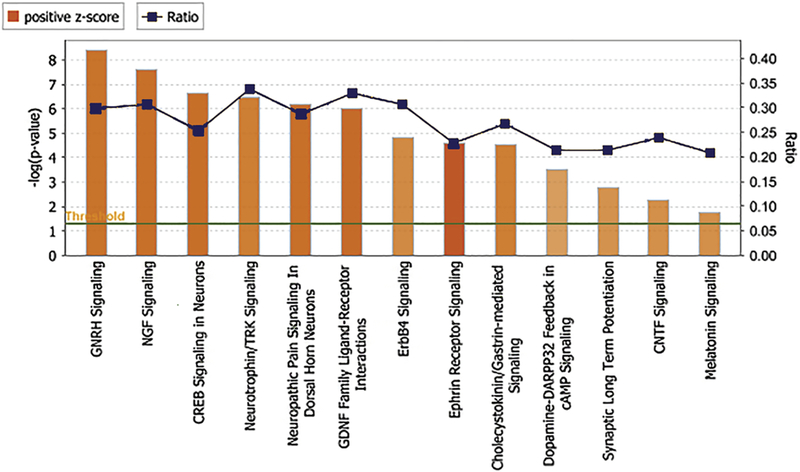
The canonical pathways are represented on the x axis. The y axis represents the significance scores as -log p-value. The threshold line in dark green indicates the significance (p<0.05) cutoff The height of the bar shows the level of significance. The ratio number line in dark blue represents number of molecules present in the dataset divided by the total number of molecules in the pathway.
Among the above signaling cascades we were interested in the pathways related to neuronal number and long-term potentiation. The pathways associated with neuronal number that are being activated include CREB signaling in neurons, neurotrophin/TRK signaling (Fig. 3A) and NGF signaling. The pathway involved in long-term potentiation function that is being activated include synaptic long-term potentiation signaling (Fig. 3B).
Figure 3. Canonical pathways involved in nervous system development and function.
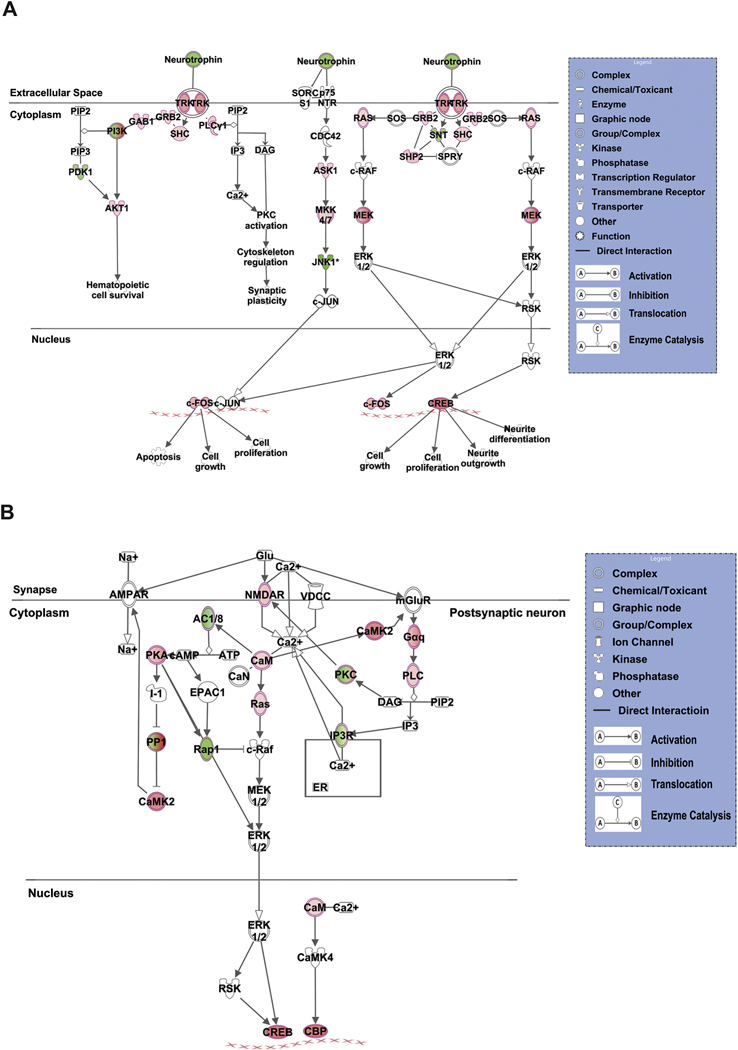
(A) Neurotrophin/TRK Signaling. (B) Synaptic long-term potentiation signaling. Red and green nodes represent the differentially expressed genes. A red node denotes an up-regulated gene, and a green node denotes a down-regulated gene. The intensity of the node color indicates the degree of up or down regulation. White colored nodes represent the genes that were not differentially expressed in our experiment but are the part of the signaling cascade as proposed by IPA. Different structures of the nodes represent different functional class of the gene product. Edges represent the type of relationship between the nodes. Filled arrow head indicates activation, bar-headed line indicates inhibition, hollow arrow head indicates translocation and perpendicular arrowheads indicate enzyme catalysis reaction.
3.3. Induction of CREB binding protein (CREBBP or CBP) and CBP-mediated functions
As signaling cascades converge to activate specific transcription factors that then regulate entire programs of gene expression, we sought to identify the transcription factors that were being activated by CEPO. To this end we employed the upstream analysis tool in IPA. This analysis revealed that several transcription factors were regulated expressed in our dataset. The list of transcription factors includes CTNNB1, KLF5, CBP, RELA, ID2, FOS, PAX4, ETV5, HNRNPK, EPAS1, LEF1, REST, CCND1, MAFB, ZEB2, POU3F3, PPARGC1B, SP4, IRF1, SPDEF, GMNN, WT1 and TP73. The upstream analysis also indicated that CBP transcription factor (p≤ 0.00018, z- score=2.739) was being activated by CEPO. Given the importance of CBP in neuroscience investigations we mined the data for genes whose expression is regulated by CBP (Fig 4). Some of the important genes that were regulated by CBP includes CAMKII, EGR1, FOS, GRIN1, KIF1B, NOTCH1, NTN1 and RARB.
Figure 4. Subcellular localization of differentially expressed genes regulated by CBP.
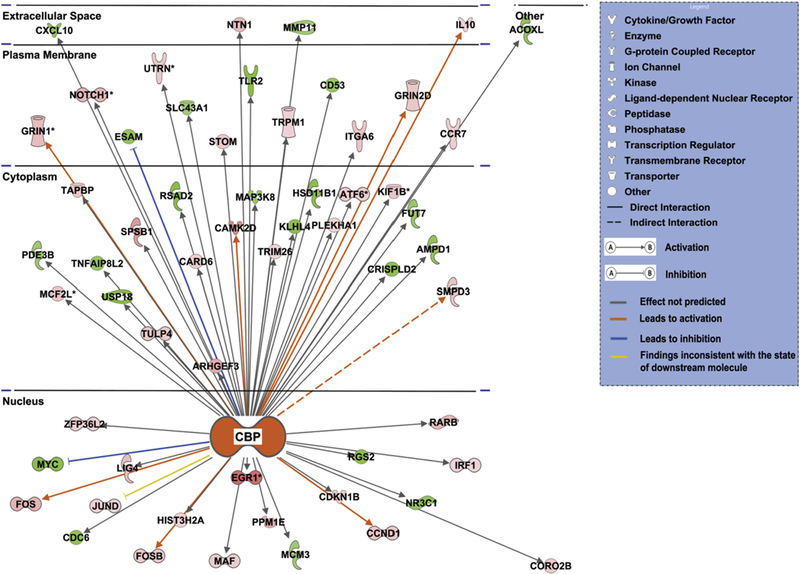
Red and green nodes represent the differentially expressed genes. A red node denotes an up-regulated gene, and a green node denotes a down-regulated gene. The intensity of the node color indicates the degree of up or down regulation. Different structures of the nodes represent different functional class of the gene product. Edges represent the type of relationship between the nodes. Filled arrow head indicates activation and bar-headed line indicates inhibition. Orange line indicates that it leads to activation, blue line indicates that it leads to inhibition, grey line indicates that the effect is not predicted, and yellow line indicates that the findings are inconsistent with the state of the downstream molecule.
In order to obtain insight into the biological functions regulated by CBP we employed the regulator effects tool in IPA. This tool can algorithmically assign biological functions (based on the large body of curated information from the IPA knowledge base) to group genes present in the dataset which are regulated by few important molecules also present in the dataset. These key molecules are identified as regulator molecules and the biological functions they produce are termed regulator effects. In our study CBP is the regulator molecule and by interacting with CAMKII, CCND1, CDKN1B, EGR1, FOS, GRIN1, ITGA6, KIF1B, LIG4, MAF, NOTCH1, NR3C1, NTN1, RARB, and UTRN it can activate functions such as long-term potentiation of brain, neuronal number, and inhibit function like perinatal death (Fig. 5).
Figure 5. CBP regulation of different neuronal functions through its interaction with differentially expressed genes.
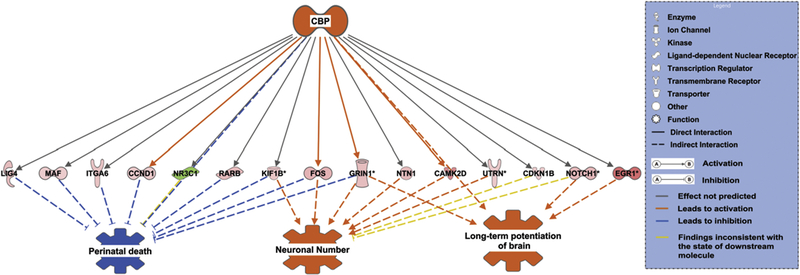
Red and green nodes represent the differentially expressed genes. A red node denotes an up-regulated gene, and a green node denotes a down-regulated gene. The intensity of the node color indicates the degree of up or down regulation. Different structures of the nodes represent different functional class of the gene product. Edges represent the type of relationship between the nodes. Filled arrow head indicates activation and bar-headed line indicates inhibition. Orange line indicates that it leads to activation, blue line indicates that it leads to inhibition, grey line indicates that the effect is not predicted and yellow line that the findings are inconsistent with the state of the downstream molecule.
3.4. Independent validation of microarray data
Secondary validation was performed using quantitative PCR. We chose a list of 10 genes from our IPA analysis of microarray data, including genes with established neurobiological value. Rather than choosing primers to overlap the probe sequences featured on the array we sought to conduct biological validation of the results by using independently designed primer sets. Results showed that the genes FOS, EGR1, ARC, VGF, BDNF, CD131, and CBP (P< 0.05) mRNA expression increased significantly as compared to control. However, for the downregulated genes CAMKIIA, HSD11 and TLR2, only CAMKIIA reached statistical significance (Fig. 6a).
Figure 6. Independent secondary validation of microarray data and Comparative analysis of EPO and CEPO-induced gene regulation in the hippocampus.
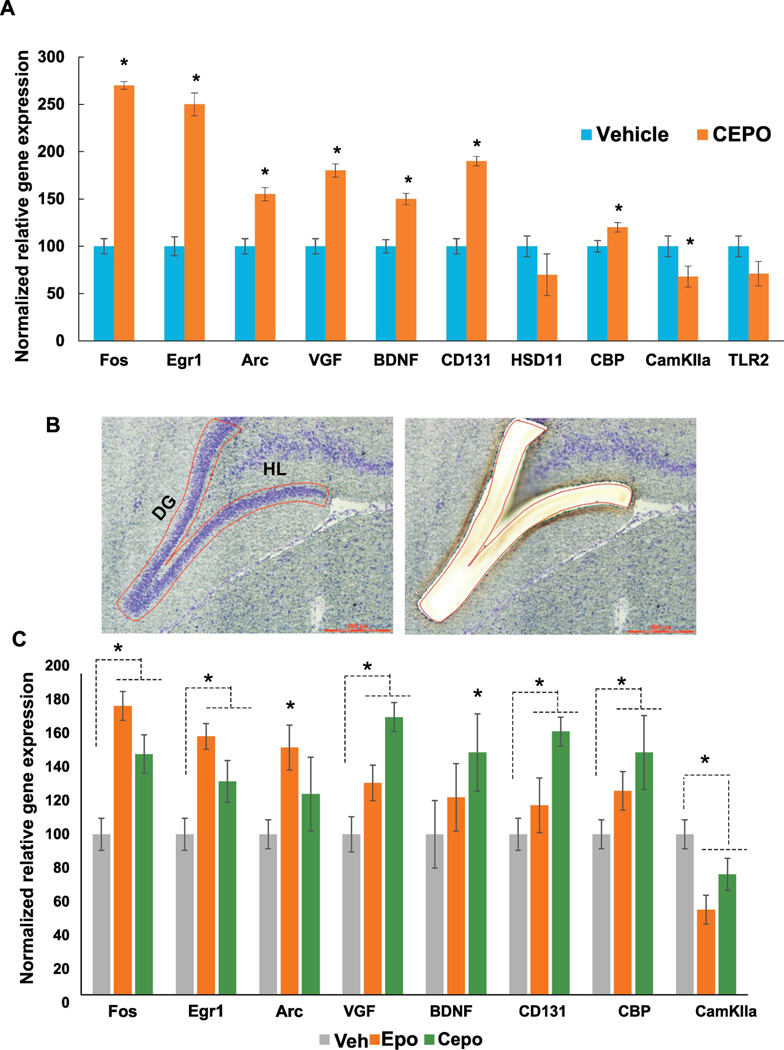
A. Quantitative PCR analysis was performed using gene specific primers. Gene regulation is expressed as percentage of vehicle levels (N=6). Error bars are +/− SEM; *p < 0.05 vs Vehicle, t-test. B. Laser microdissection of the dentate gyrus showing before (left) and after (right) images of the same section. C. Quantitative PCR analysis of EPO and CEPO-induced gene regulation in the dentate gyrus. Gene regulation is expressed as percentage of vehicle levels. Error bars are +/− SEM; *p < 0.05 ANOVA. Egr1- early growth response 1, Arc - activity regulated cytoskeleton-associated protein, VGF- non-acronymic, BDNF- brain derived neurotrophic factor, CD131 - beta common receptor, HSD11- l1β hydroxysteroid dehydrogenase, CBP- CREB binding protein, CamKIIa- calcium/calmodulin dependent protein kinase type 2 alpha, TLR2- toll-like receptor 2. DG - dentate gyrus, HL- hilus.
3.5. Dentate gyrus gene regulation
A comparative analysis of EPO and CEPO-induced gene regulation in the dentate gyrus region using laser microdissection (Fig. 6b) revealed substantial overlap between them (Fig. 6c) and also with CEPO-treated PC12 cells (Fig. 6a). With the exception of Arc, genes that were significantly regulated in PC12 cells were also regulated by CEPO in the dentate gyrus. Arc, Fos and Egr1 were more strongly induced by EPO than CEPO. CD131 was significantly induced by CEPO but not by EPO (Fig 6c).
4. Discussion
We conducted a transcriptome analysis of non-erythropoietic, carbamoylated erythropoietin induced gene regulation in neuronal cells and investigated the functional significance of the gene profile by bioinformatics analysis employing the Ingenuity Pathway Analysis suite. Our dataset revealed the prominent enrichment of functional categories pertaining to neuronal number, long term potentiation and neurotransmission.
EPO has been previously been shown to play an essential role in developmental neurogenesis (Tsai et al., 2006), and is known to be involved in cell proliferation in multiple cell types (Bahlmann et al., 2003; Chamorro et al., 2013). It also elevates adult neurogenesis via a rather unique mechanism, increasing neuronal and oligodendrocyte number by 20% in the hippocampus but without substantially impacting cell proliferation (Hassouna et al., 2016). The EPO-induced increase in hippocampal volume noted in the clinic could be in part driven by this cellular mechanism (Miskowiak et al., 2015). As CEPO regulates numerous genes that play key roles in progenitor cell proliferation or differentiation, including CBP, Fos, KIF1B, BDNF and NTN1 to name a few, it is reasonable to expect that it would elicit similar cellular effects in the hippocampus.
EPO has shown robust cognitive improvement in schizophrenia (Ehrenreich et al., 2007) and treatment resistant depression (Miskowiak et al., 2014). Rodent studies designed to understand these cognitive enhancing effects have demonstrated that EPO enhances long term potentiation (LTP) in the CA1 (Adamcio et al., 2008) and the cognitive performance improvements persist for at least 3 months after the last dose (El-Kordi et al., 2009). Interestingly, it was recently shown that CEPO produces notable cognitive effects in the social defeat model, preferentially regulating neurotrophic gene expression in the dorsal hippocampus (Sathyanesan et al., 2018). Given the relationship between hippocampal LTP and cognition it is interesting to note that CEPO regulated over 66 genes with known roles in modulating LTP, such as DLG4, EGR1, GRIN1, NOTCH1, VEGFA and Arc. The elevation in Arc is of particular interest due to its known roles in regulating synaptic plasticity (Shepherd and Bear, 2011), cognitive function (Zhang et al., 2015) and LTP (Messaoudi et al., 2007).
IPA also revealed the increase in neurotrophin signaling pathways by CEPO. Apart from independently exhibiting trophic actions CEPO appears to also regulate other neurotrophic factors that act via their respective receptors. The CEPO-induced increase in VGF and BDNF is significant as these genes are also induced in the hippocampus by exercise (Hunsberger et al., 2007) electroconvulsive seizure (Newton et al., 2003) and EPO (Girgenti et al., 2009). Both genes were also induced in the rat dentate gyrus by CEPO in the current study. They are downregulated with stress exposure and independently produce antidepressant-like effects when infused into the hippocampus (Rasmusson et al., 2002; Shirayama et al., 2002; Thakker-Varia et al., 2007). Among the several nervous system signaling cascades activated by CEPO, the regulation of CREB signaling is pertinent to antidepressant activity as the cAMP pathway and CREB phosphorylation has been heavily investigated in psychiatric neuroscience research as a key transcription factor and convergence hub for different classes of antidepressants activation (Blendy, 2006). CREB activation increases neuronal proliferation in the dentate gyrus (Nakagawa et al., 2002), which could be a cellular mechanism that mediates its behavioral effects.
CBP, a transcriptional coactivator, was prominently indicated by IPA and its regulation was confirmed by secondary validation in PC12 cells and in the hippocampus. Previous work in PC12 cells has demonstrated the induction of CBP after CEPO treatment and its role in increasing neurite length and spine density (Choi et al., 2014). CBP is a diverse, multifunctional molecule with roles in several cellular processes. Apart from possessing intrinsic acetyltransferase activity it also associates with several transcription factors and transcriptional activators and functions as a crucial regulatory hub for CREB-dependent gene expression (Vo and Goodman, 2001). CBP has been shown to be essential for hippocampal neurogenesis and the associated improvement in cognitive performance (Lopez-Atalaya et al., 2011; Gouveia et al., 2016). The induction of CREB and CBP mediated gene expression suggests that this pathway and downstream genes could be important for CEPO’s behavioral effects, including both antidepressant and cognitive enhancing activity. Elucidating how CEPO activates CREB and CBP will be an interesting and worthwhile endeavor as it is not indicated by the canonical JAK-STAT pathway utilized by EPO. It is however not surprising that CEPO activates the CREB pathway as it is well known that neurotrophic factors (BDNF) can activate CREB by multiple cAMP-independent pathways, the calcium/calmodulin dependent kinase IV (CaMKIV) pathway and the Ras pathway (Finkbeiner et al., 1997). Perhaps the first line of investigation would be to interrogate the role of the erythropoietin receptor and the intracellular adaptor molecules that associate with it upon CEPO binding.
The elevation in CD131/βcR is intriguing as it is a receptor that is shared by the beta common family of cytokines, granulocyte-macrophage colony stimulating factor (GM-CSF), interleukin (IL)-3 and IL-5 (Broughton et al., 2015). The tissue protective actions of EPO and CEPO have been previously shown to act via an interaction between the pcR and EPO receptor (Brines et al., 2004). It is thought that CEPO is non- erythropoietic because it primarily signals via this βcR-EPOR heteroreceptor complex rather than the EPOR-EPOR dimer utilized by EPO (Leist et al., 2004). Hippocampal expression of βcR is low and is induced mainly in reactive microglia after seizures (Nadam et al., 2007). From our quantitative PCR analysis, we noted that basal expression levels of βcR were 3-fold lower than EPOR. It is therefore possible that the robust CEPO-induced increase in βcR is an effort to potentially facilitate stoichiometric pairing of both receptors. Of note from our bioinformatics analysis is the lack of gene categories pertaining to the hematopoietic cascade that is strongly activated by EPO. There could be multiple reasons for this. 1) CEPO does not activate this cascade, 2) activation of the hematopoietic cascade is not reflected in a PC-12 cells gene profile despite expression of EPOR. It would be useful to examine the precise role played by βcR in CEPO induced gene expression by genetically manipulating receptor expression levels. In the larger context it will be important to test the specific role played by βcR in the behavioral effects of CEPO. However, it is useful to note that there was substantial overlap in EPO and CEPO-induced gene regulation in the hippocampus despite differences in levels of regulation. Although it will be interesting to compare their behavioral actions in a battery of behavioral assays, the hematological effects of EPO (Coleman et al., 2006) are likely to limit its use for CNS disorders particularly when chronic administration is required.
5. Conclusion
Our gene profile sheds light on CEPO’s potential molecular mechanism of action. The induction of neurotrophin signaling, CREB signaling and LTP signaling related genes indicate that it activates multiple intracellular signal transduction pathways. The gene products of the target genes have been strongly implicated in both antidepressant activity and cognition, raising expectations that CEPO would produce these therapeutic effects in rodent models and humans. This strengthens the case for further development of CEPO as a candidate molecule for the treatment of neuropsychiatric disorders.
Highlights.
CEPO induces a neurotrophic gene profile and signaling cascades.
CEPO and EPO produce comparable gene regulation in the hippocampal dentate gyrus.
Genes regulated by CEPO have been implicated in antidepressant activity and cognitive function.
Acknowledgements
This work was supported by US Public Health Service [grant MH106640 (SSN)] and the use of facilities at the Sioux Falls VA Healthcare system. The project described was supported by the National Institute of General Medical Sciences, [1U54GM115458–01]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Financial Disclosures
The authors have no disclosures to report
Footnote
* Duplicate genes in the dataset.
Affirmation: We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Neeraj K. Tiwari, Division of Basic Biomedical Sciences, Sanford School of Medicine, University of South Dakota, Vermillion, SD 57069; Neeraj.Tiwari@coyotes.usd.edu
Monica Sathyanesan, Division of Basic Biomedical Sciences, Sanford School of Medicine, University of South Dakota, Vermillion, SD 57069; Sioux Falls VA Healthcare System, Sioux Falls, SD 57105, Monica.Sathyanesan@usd.edu.
William Schweinle, Physician Assistant Program, School of Health Sciences, University of South Dakota, Vermillion, SD 57069; William.Schweinle@usd.edu.
Samuel S Newton, Division of Basic Biomedical Sciences, Sanford School of Medicine, University of South Dakota, Vermillion, SD 57069; Sioux Falls VA Healthcare System, Sioux Falls, SD 57105; Samuel.Sathyanesan@usd.edu.
References
- 1.Adamcio B, Sargin D, Stradomska A, Medrihan L, Gertler C, Theis F, Zhang M, Muller M, Hassouna I, Hannke K, Sperling S, Radyushkin K, El-Kord i A, Schulze L, Ronnenberg A, Wolf F, Brose N, Rhee JS, Zhang W, Ehrenreich H (2008) Erythropoietin enhances hippocampal long-term potentiation and memory. BMC biology 6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahlmann FH, DeGroot K, Duckert T, Niemczyk E, Bahlmann E, Boehm SM, Haller H, Fliser D (2003) Endothelial progenitor cell proliferation and differentiation is regulated by erythropoietin. Kidney international 64:1648–1652. [DOI] [PubMed] [Google Scholar]
- 3.Blendy JA (2006) The role of CREB in depression and antidepressant treatment. Biol Psychiatry 59:1144–1150. [DOI] [PubMed] [Google Scholar]
- 4.Brines M, Cerami A (2005) Emerging biological roles for erythropoietin in the nervous system. Nat Rev Neurosci 6:484–494. [DOI] [PubMed] [Google Scholar]
- 5.Brines M, Grasso G, Fiordaliso F, Sfacteria A, Ghezzi P, Fratelli M, Latini R, Xie QW, Smart J, SuRick CJ, Pobre E, Diaz D, Gomez D, Hand C, Coleman T, Cerami A (2004) Erythropoietin mediates tissue protection through an erythropoietin and common beta-subunit heteroreceptor. Proc Natl Acad Sci U S A 101:14907–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broughton SE, Nero TL, Dhagat U, Kan WL, Hercus TR, Tvorogov D, Lopez AF, Parker MW (2015) The betac receptor family - Structural insights and their functional implications. Cytokine 74:247–258. [DOI] [PubMed] [Google Scholar]
- 7.Chamorro ME, Wenker SD, Vota DM, Vittori DC, Nesse AB (2013) Signaling pathways of cell proliferation are involved in the differential effect of erythropoietin and its carbamylated derivative. Biochimica et biophysica acta 1833:1960–1968. [DOI] [PubMed] [Google Scholar]
- 8.Choi M, Ko SY, Lee IY, Wang SE, Lee SH, Oh DH, Kim YS, Son H (2014) Carbamylated erythropoietin promotes neurite outgrowth and neuronal spine formation in association with CBP/p300. Biochemical and biophysical research communications 446:79–84. [DOI] [PubMed] [Google Scholar]
- 9.Coleman TR, Westenfelder C, Togel FE, Yang Y, Hu Z, Swenson L, Leuvenink HG, Ploeg RJ, d’Uscio LV, Katusic ZS, Ghezzi P, Zanetti A, Kaushansky K, Fox NE, Cerami A, Brines M (2006) Cytoprotective doses of erythropoietin or carbamylated erythropoietin have markedly different procoagulant and vasoactive activities. Proc Natl Acad Sci U S A 103:5965–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrenreich H et al. (2007) Improvement of cognitive functions in chronic schizophrenic patients by recombinant human erythropoietin. Molecular psychiatry 12:206–220. [DOI] [PubMed] [Google Scholar]
- 11.El-Kordi A, Radyushkin K, Ehrenreich H (2009) Erythropoietin improves operant conditioning and stability of cognitive performance in mice. BMC biology 7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkbeiner S, Tavazoie SF, Maloratsky A, Jacobs KM, Harris KM, Greenberg ME (1997) CREB: a major mediator of neuronal neurotrophin responses. Neuron 19:1031–1047. [DOI] [PubMed] [Google Scholar]
- 12.Girgenti MJ, Hunsberger J, Duman CH, Sathyanesan M, Terwilliger R, Newton SS (2009) Erythropoietin induction by electroconvulsive seizure, gene regulation, and antidepressant-like behavioral effects. Biol Psychiatry 66:267–274. [DOI] [PubMed] [Google Scholar]
- 12.Gouveia A, Hsu K, Niibori Y, Seegobin M, Cancino GI, He L, Wondisford FE, Bennett S, Lagace D, Frankland PW, Wang J (2016) The aPKC-CBP Pathway Regulates Adult Hippocampal Neurogenesis in an Age-Dependent Manner. Stem Cell Reports 7:719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassouna I et al. (2016) Revisiting adult neurogenesis and the role of erythropoietin for neuronal and oligodendroglial differentiation in the hippocampus. Molecular psychiatry 21:1752–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunsberger JG, Newton SS, Bennett AH, Duman CH, Russell DS, Salton SR, Duman RS (2007) Antidepressant actions of the exercise-regulated gene VGF. Nat Med 13:1476–1482. [DOI] [PubMed] [Google Scholar]
- 15.Leist M et al. (2004) Derivatives of erythropoietin that are tissue protective but not erythropoietic. Science (New York, NY 305:239–242. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Atalaya JP, Ciccarelli A, Viosca J, Valor LM, Jimenez-Minchan M, Canals S, Giustetto M, Barco A (2011) CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. The EMBO journal 30:4287–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR (2007) Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci 27:10445–10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miskowiak KW, Vinberg M, Christensen EM, Bukh JD, Harmer CJ, Ehrenreich H, Kessing LV (2014) Recombinant human erythropoietin for treating treatment-resistant depression: a double-blind, randomized, placebo-controlled phase 2 trial. Neuropsychopharmacology 39:1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miskowiak KW, Vinberg M, Macoveanu J, Ehrenreich H, Koster N, Inkster B, Paulson OB, Kessing LV, Skimminge A, Siebner HR (2015) Effects of Erythropoietin on Hippocampal Volume and Memory in Mood Disorders. Biol Psychiatry 78:270–277. [DOI] [PubMed] [Google Scholar]
- 20.Mun KC, Golper TA (2000) Impaired biological activity of erythropoietin by cyanate carbamylation. Blood purification 18:13–17. [DOI] [PubMed] [Google Scholar]
- 21.Nadam J, Navarro F, Sanchez P, Moulin C, Georges B, Laglaine A, Pequignot JM, Morales A, Ryvlin P, Bezin L (2007) Neuroprotective effects of erythropoietin in the rat hippocampus after pilocarpine-induced status epilepticus. Neurobiol Dis 25:412–426. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa S, Kim JE, Lee R, Malberg JE, Chen J, Steffen C, Zhang YJ, Nestler EJ, Duman RS (2002) Regulation of neurogenesis in adult mouse hippocampus by cAMP and the cAMP response element-binding protein. J Neurosci 22:3673–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton SS, Collier EF, Hunsberger J, Adams D, Terwilliger R, Selvanayagam E, Duman RS (2003) Gene profile of electroconvulsive seizures: induction of neurotrophic and angiogenic factors. J Neurosci 23:10841–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmusson AM, Shi L, Duman R (2002) Downregulation of BDNF mRNA in the hippocampal dentate gyrus after re-exposure to cues previously associated with footshock. Neuropsychopharmacology 27:133–142. [DOI] [PubMed] [Google Scholar]
- 25.Renzi MJ, Farrell FX, Bittner A, Galindo JE, Morton M, Trinh H, Jolliffe LK (2002) Erythropoietin induces changes in gene expression in PC-12 cells. Brain research Molecular brain research 104:86–95. [DOI] [PubMed] [Google Scholar]
- 26.Sathyanesan M, Haiar JM, Watt MJ, Newton SS (2017) Restraint stress differentially regulates inflammation and glutamate receptor gene expression in the hippocampus of C57BL/6 and BALB/c mice. Stress (Amsterdam, Netherlands) 20:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sathyanesan M, Watt MJ, Haiar JM, Scholl JL, Davies SR, Paulsen RT, Wiederin J, Ciborowski P, Newton SS (2018) Carbamoylated erythropoietin modulates cognitive outcomes of social defeat and differentially regulates gene expression in the dorsal and ventral hippocampus. Translational psychiatry 8:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SHEPHERD JD, Bear MF (2011) New views of Arc, a master regulator of synaptic plasticity. Nature neuroscience 14:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirayama Y, Chen AC, Nakagawa S, Russell DS, Duman RS (2002) Brain-derived neurotrophic factor produces antidepressant effects in behavioral models of depression. J Neurosci 22:3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakker-Varia S, Krol JJ, Nettleton J, Bilimoria PM, Bangasser DA, Shors TJ, Black IB, Alder J (2007) The neuropeptide VGF produces antidepressant-like behavioral effects and enhances proliferation in the hippocampus. J Neurosci 27:12156–12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai PT, Ohab JJ, Kertesz N, Groszer M, Matter C, Gao J, Liu X, Wu H, Carmichael ST (2006) A critical role of erythropoietin receptor in neurogenesis and post-stroke recovery. J Neurosci 26:1269–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Um M, Gross AW, Lodish HF (2007) A “classical” homodimeric erythropoietin receptor is essential for the antiapoptotic effects of erythropoietin on differentiated neuroblastoma SH- SY5Y and pheochromocytoma PC-12 cells. Cellular signalling 19:634–645. [DOI] [PubMed] [Google Scholar]
- 33.Vo N, Goodman RH (2001) CREB-binding protein and p300 in transcriptional regulation. J Biol Chem 276:13505–13508. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y, Shang Y, Sun S, Liang H, Liu R (2007) Erythropoietin prevents PC12 cells from 1-methyl-4- phenylpyridinium ion-induced apoptosis via the Akt/GSK-3beta/caspase-3 mediated signaling pathway. Apoptosis : an international journal on programmed cell death 12:1365–1375. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF (2015) Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron 86:490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]


