Abstract
Artificial intelligence (AI) algorithms, particularly deep learning, have demonstrated remarkable progress in image-recognition tasks. Methods ranging from convolutional neural networks to variational autoencoders have found myriad applications in the medical image analysis field, propelling it forward at a rapid pace. Historically, in radiology practice, trained physicians visually assessed medical images for the detection, characterization and monitoring of diseases. AI methods excel at automatically recognizing complex patterns in imaging data and providing quantitative, rather than qualitative, assessments of radiographic characteristics. In this O pinion article, we establish a general understanding of AI methods, particularly those pertaining to image-based tasks. We explore how these methods could impact multiple facets of radiology, with a general focus on applications in oncology, and demonstrate ways in which these methods are advancing the field. Finally, we discuss the challenges facing clinical implementation and provide our perspective on how the domain could be advanced.
Artificial intelligence (AI) has recently made substantial strides in perception (the interpretation of sensory information), allowing machines to better represent and interpret complex data. This has led to major advances in applications ranging from web search and self-driving vehicles to natural language processing and computer vision — tasks that until a few years ago could be done only by humans1. Deep learning is a subset of machine learning that is based on a neural network structure loosely inspired by the human brain. Such structures learn discriminative features from data automatically, giving them the ability to approximate very complex nonlinear relationships (BOX 1). While most earlier AI methods have led to applications with subhuman performance, recent deep learning algorithms are able to match and even surpass humans in task-specific applications2–5 (FIG. 1). This is owing to recent advances in AI research, the massive amounts of digital data now available to train algorithms and modern, powerful computational hardware. Deep learning methods have been able to defeat humans in the strategy board game of Go, an achievement that was previously thought to be decades away given the highly complex game space and massive number of potential moves6. Following the trend towards a human-level general AI, researchers predict that AI will automate many tasks, including translating languages, writing best-selling books and performing surgery — all within the coming decades7.
Box 1 |. Artificial intelligence methods in medical imaging
Machine learning algorithms based on predefined engineered features
Traditional artificial intelligence (AI) methods rely largely on predefined engineered feature algorithms (Fig. 2a) with explicit parameters based on expert knowledge. Such features are designed to quantify specific radiographic characteristics, such as the 3D shape of a tumour or the intratumoural texture and distribution of pixel intensities (histogram). A subsequent selection step ensures that only the most relevant features are used. Statistical machine learning models are then fit to these data to identify potential imaging-based biomarkers. Examples of these models include support vector machines and random forests.
Deep learning algorithms
Recent advances in AI research have given rise to new, non-deterministic, deep learning algorithms that do not require explicit feature definition, representing a fundamentally different paradigm in machine learning111–113. The underlying methods of deep learning have existed for decades. However, only in recent years have sufficient data and computational power become available. Without explicit feature predefinition or selection, these algorithms learn directly by navigating the data space, giving them superior problem-solving capabilities. While various deep learning architectures have been explored to address different tasks, convolutional neural networks (CNNs) are the most prevalent deep learning architecture typologies in medical imaging today14. A typical CNN comprises a series of layers that successively map image inputs to desired end points while learning increasingly higher-level imaging features (Fig. 2b). Starting from an input image, ‘hidden layers’ within CNNs usually include a series of convolution and pooling operations extracting feature maps and performing feature aggregation, respectively. These hidden layers are then followed by fully connected layers providing high-level reasoning before an output layer produces predictions. CNNs are often trained end-to-end with labelled data for supervised learning. Other architectures, such as deep autoencoders96 and generative adversarial networks95, are more suited for unsupervised learning tasks on unlabelled data. Transfer learning, or using pre-trained networks on other data sets, is often utilized when dealing with scarce data114.
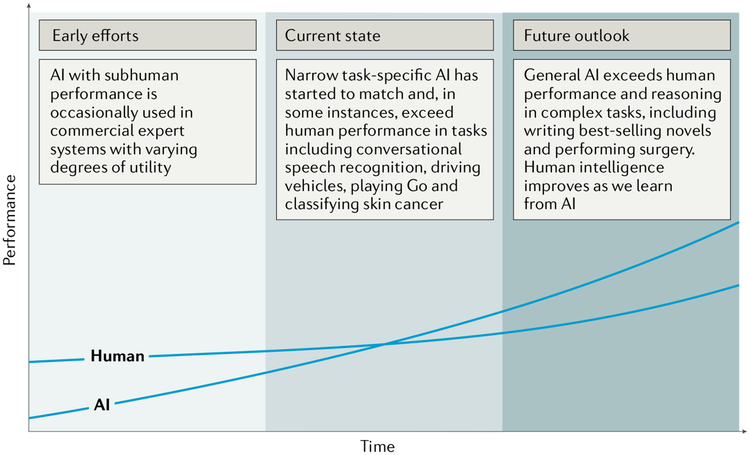
Fig. 1 |. Artificial versus human intelligence.
This plot outlines the performance levels of artificial intelligence (AI) and human intelligence starting from the early computer age and extrapolating into the future. Early AI came with a subhuman performance and varying degrees of success. Currently, we are witnessing narrow task-specific AI applications that are able to match and occasionally surpass human intelligence4–6,9. It is expected that general AI will surpass human performance in specific applications within the coming years. Humans will potentially benefit from the human-AI interaction, bringing them to higher levels of intelligence.
Within health care, AI is becoming a major constituent of many applications, including drug discovery, remote patient monitoring, medical diagnostics and imaging, risk management, wearables, virtual assistants and hospital management. Many domains with big data components such as the analysis of DNA and RNA sequencing data8 are also expected to benefit from the use of AI. Medical fields that rely on imaging data, including radiology, pathology, dermatology9 and ophthalmology10, have already begun to benefit from the implementation of AI methods (Box 2). Within radiology, trained physicians visually assess medical images and report findings to detect, characterize and monitor diseases. Such assessment is often based on education and experience and can be, at times, subjective. In contrast to such qualitative reasoning, AI excels at recognizing complex patterns in imaging data and can provide a quantitative assessment in an automated fashion. More accurate and reproducible radiology assessments can then be made when AI is integrated into the clinical workflow as a tool to assist physicians.
Box 2 |. Examples of clinical application areas of artificial intelligence in oncology
Radiology-based
Thoracic imaging.
Lung cancer is one of the most common and deadly tumours. Lung cancer screening can help identify pulmonary nodules, with early detection being lifesaving in many patients. Artificial intelligence (AI) can help in automatically identifying these nodules and categorizing them as benign or malignant.
Abdominal and pelvic imaging.
With the rapid growth in medical imaging, especially computed tomography (CT) and magnetic resonance imaging (MRI), more incidental findings, including liver lesions, are identified. AI may aid in characterizing these lesions as benign or malignant and prioritizing follow-up evaluation for patients with these lesions.
Colonoscopy.
Colonic polyps that are undetected or misclassified pose a potential risk of colorectal cancer. Although most polyps are initially benign, they can become malignant over time115. Hence, early detection and consistent monitoring with robust AI-based tools are critical.
Mammography.
Screening mammography is technically challenging to expertly interpret. AI can assist in the interpretation, in part by identifying and characterizing microcalcifications (small deposits of calcium in the breast).
Brain imaging.
Brain tumours are characterized by abnormal growth of tissue and can be benign, malignant, primary or metastatic; AI could be used to make diagnostic predictions116.
Radiation oncology.
Radiation treatment planning can be automated by segmenting tumours for radiation dose optimization. Furthermore, assessing response to treatment by monitoring over time is essential for evaluating the success of radiation therapy efforts. AI is able to perform these assessments, thereby improving accuracy and speed.
Non-radiology-based
Dermatology.
Diagnosing skin cancer requires trained dermatologists to visually inspect suspicious areas. With the large variability in sizes, shades and textures, skin lesions are rather challenging to interpret9. The massive learning capacity of deep learning algorithms qualifies them to handle such variance and detect characteristics well beyond those considered by humans.
Pathology.
The quantification of digital whole-slide images of biopsy samples is vital in the accurate diagnosis of many types of cancers. With the large variation in imaging hardware, slide preparation, magnification and staining techniques, traditional AI methods often require considerable tuning to address this problem. More robust AI is able to more accurately perform mitosis detection, segment histologic primitives (such as nuclei, tubules and epithelium), count events and characterize and classify tissue117–120.
DNA and RNA sequencing.
The ever-increasing amount of available sequencing data continues to provide opportunities for utilizing genomic end points in cancer diagnosis and care. AI-based tools are able to identify and extract high-level features correlating somatic point mutations and cancer types121 as well as predict the effect of mutations on sequence specificities of RNA-binding and DNA-binding proteins122.
As imaging data are collected during routine clinical practice, large data sets are — in principle — readily available, thus offering an incredibly rich resource for scientific and medical discovery. Radiographic images, coupled with data on clinical outcomes, have led to the emergence and rapid expansion of radiomics as a field of medical research11–13. Early radiomics studies were largely focused on mining images for a large set of predefined engineered features that describe radiographic aspects of shape, intensity and texture. More recently, radiomics studies have incorporated deep learning techniques to learn feature representations automatically from example images14, hinting at the substantial clinical relevance of many of these radiographic features. Within oncology, multiple efforts have successfully explored radiomics tools for assisting clinical decision making related to the diagnosis and risk stratification of different cancers15,16. For example, studies in non-small-cell lung cancer (NSCLC) used radiomics to predict distant metastasis in lung adenocarcinoma17 and tumour histological subtypes18 as well as disease recurrence19, somatic mutations20, gene-expression profiles21 and overall survival22. Such findings have motivated an exploration of the clinical utility of AI-generated biomarkers based on standard-of-care radiographic images23 — with the ultimate hope of better supporting radiologists in disease diagnosis, imaging quality optimization, data visualization, response assessment and report generation.
In this Opinion article, we start by establishing a general understanding of AI methods particularly pertaining to image-based tasks. We then explore how up-and-coming AI methods will impact multiple radiograph-based practices within oncology. Finally, we discuss the challenges and hurdles facing the clinical implementation of these methods.
AI in medical imaging
The primary driver behind the emergence of AI in medical imaging has been the desire for greater efficacy and efficiency in clinical care. Radiological imaging data continues to grow at a disproportionate rate when compared with the number of available trained readers, and the decline in imaging reimbursements has forced health-care providers to compensate by increasing productivity24. These factors have contributed to a dramatic increase in radiologists’ workloads. Studies report that, in some cases, an average radiologist must interpret one image every 3–4 seconds in an 8-hour workday to meet workload demands25. As radiology involves visual perception as well as decision making under uncertainty26, errors are inevitable — especially under such constrained conditions.
A seamlessly integrated AI component within the imaging workflow would increase efficiency, reduce errors and achieve objectives with minimal manual input by providing trained radiologists with pre-screened images and identified features. Therefore, substantial efforts and policies are being put forward to facilitate technological advances related to AI in medical imaging. Almost all image-based radiology tasks are contingent upon the quantification and assessment of radiographic characteristics from images. These characteristics can be important for the clinical task at hand, that is, for the detection, characterization or monitoring of diseases. The application of logic and statistical pattern recognition to problems in medicine has been proposed since the early 1960s27,28. As computers became more prevalent in the 1980s, the AI-powered automation of many clinical tasks has shifted radiology from a perceptual subjective craft to a quantitatively computable domain29,30. The rate at which AI is evolving radiology is parallel to that in other application areas and is proportional to the rapid growth of data and computational power.
There are two classes of AI methods that are in wide use today (BOX 1; FIG. 2). The first uses handcrafted engineered features that are defined in terms of mathematical equations (such as tumour texture) and can thus be quantified using computer programs31. These features are used as inputs to state-ofthe-art machine learning models that are trained to classify patients in ways that can support clinical decision making. Although such features are perceived to be discriminative, they rely on expert definition and hence do not necessarily represent the most optimal feature quantification approach for the discrimination task at hand. Moreover, predefined features are often unable to adapt to variations in imaging modalities, such as computed tomography (CT), positron emission tomography (PET) and magnetic resonance imaging (MRI), and their associated signal-to-noise characteristics.
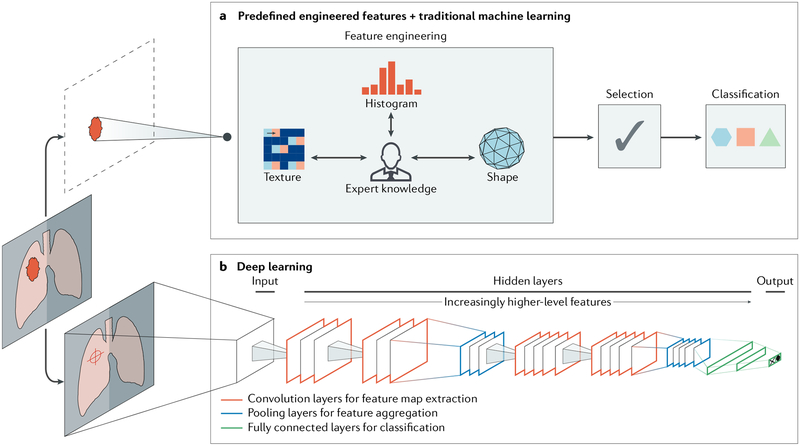
Fig. 2 |. Artificial intelligence methods in medical imaging.
This schematic outlines two artificial intelligence (AI) methods for a representative classification task, such as the diagnosis of a suspicious object as either benign or malignant. a | The first method relies on engineered features extracted from regions of interest on the basis of expert knowledge. Examples of these features in cancer characterization include tumour volume, shape, texture, intensity and location. The most robust features are selected and fed into machine learning classifiers. b | The second method uses deep learning and does not require region annotation — rather, localization is usually sufficient. It comprises several layers where feature extraction, selection and ultimate classification are performed simultaneously during training. As layers learn increasingly higher-level features (Box 1), earlier layers might learn abstract shapes such as lines and shadows, while other deeper layers might learn entire organs or objects. Both methods fall under radiomics, the data-centric, radiology-based research field.
The second method, deep learning, has gained considerable attention in recent years. Deep learning algorithms can automatically learn feature representations from data without the need for prior definition by human experts. This data-driven approach allows for more abstract feature definitions, making it more informative and generalizable. Deep learning can thus automatically quantify phenotypic characteristics of human tissues32, promising substantial improvements in diagnosis and clinical care. Deep learning has the added benefit of reducing the need for manual preprocessing steps. For example, to extract predefined features, accurate segmentation of diseased tissues by experts is often needed33. Because deep learning is data driven (Box 1), with enough example data, it can automatically identify diseased tissues and hence avoid the need for expert-defined segmentations. Given its ability to learn complex data representations, deep learning is also often robust against undesired variation, such as the inter-reader variability, and can hence be applied to a large variety of clinical conditions and parameters. In many ways, deep learning can mirror what trained radiologists do, that is, identify image parameters but also weigh up the importance of these parameters on the basis of other factors to arrive at a clinical decision.
Given the growing number of applications of deep learning in medical imaging14, several efforts have compared deep learning methods with their predefined feature-based counterparts and have reported substantial performance improvements with deep learning34,35. Studies have also shown that deep learning technologies are on par with radiologists’ performance for both detection36 and segmentation37 tasks in ultrasonography and MRI, respectively. For the classification tasks of lymph node metastasis in PET-CT, deep learning had higher sensitivities but lower specificities than radiologists38. As these methods are iteratively refined and tailored for specific applications, a better command of the sensitivity:specificity trade-off is expected. Deep learning can also enable faster development times, as it depends solely on curated data and the corresponding metadata rather than domain expertise. On the other hand, traditional predefined feature systems have shown plateauing performance over recent years and hence do not generally meet the stringent requirements for clinical utility. As a result, only a few have been translated into the clinic39. It is expected that high-performance deep learning methods will surpass the threshold for clinical utility in the near future and can therefore be expeditiously translated into the clinic.
Impact on oncology imaging
In this section, we focus on three main clinical radiology tasks that specifically pertain to oncology: abnormality detection, followed by characterization and subsequent monitoring of change (FIG. 3). These tasks require a diversified set of skills: medical, in terms of disease diagnosis and care, as well as technical, for capturing and processing radiographic images. Both these skills hint at the ample opportunities where up-and-coming AI technologies can positively impact clinical outcomes by identifying phenotypic characteristics in images. In addition to being used in radiographic cancer images, such as in thoracic imaging and mammography, these tasks are also commonly used in other oncology subspecialties with non-radiographic images (BOX 2). For each of these tasks, we investigate technologies currently being utilized in the clinic and provide highlights of research efforts aimed at integrating state-of-the-art AI developments in these practices.
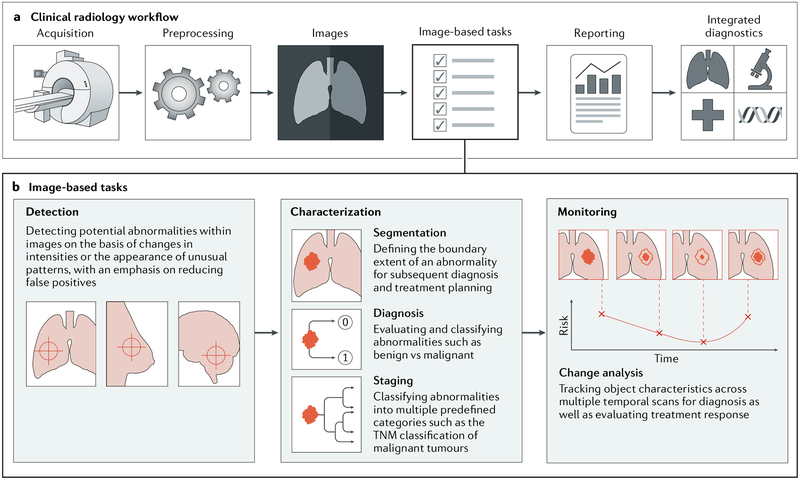
Fig. 3 |. Artificial intelligence impact areas within oncology imaging.
This schematic outlines the various tasks within radiology where artificial intelligence (AI) implementation is likely to have a large impact. a | The workflow comprises the following steps: preprocessing of images after acquisition, image-based clinical tasks (which usually involve the quantification of features either using engineered features with traditional machine learning or deep learning), reporting results through the generation of textual radiology reports and, finally, the integration of patient information from multiple data sources. b | AI is expected to impact image-based clinical tasks, including the detection of abnormalities; the characterization of objects in images using segmentation, diagnosis and staging; and the monitoring of objects for diagnosis and assessment of treatment response. TNM, tumour–node–metastasis.
Detection.
Within the manual detection workflow, radiologists rely on manual perceptive skills to identify possible abnormalities, followed by cognitive skills to either confirm or reject the findings.
Radiologists visually scan through stacks of images while periodically adjusting viewing planes and window width and level settings. Relying on education, experience and an understanding of the healthy radiograph, radiologists are trained to identify abnormalities on the basis of changes in imaging intensities or the appearance of unusual patterns. These criteria, and many more, fall within a somewhat subjective decision matrix that enables reasoning in problems ranging from detecting lung nodules to breast lesions and colon polyps. As dependence on computers has increased, automated methods for the identification and processing of these predefined features — collectively known as computer-aided detection (CADe) — have long been proposed and occasionally utilized in the clinic31. Radiologist-defined criteria are distilled into a pattern-recognition problem where computer vision algorithms highlight conspicuous objects within the image40. However, these algorithms are often task-specific and do not generalize across diseases and imaging modalities. Additionally, the accuracy of traditional predefined feature-based CADe systems remains questionable, with ongoing efforts to reduce false positives. It is often the case that outputs have to be assessed by radiologists to decide whether a certain automated annotation merits further investigation, thereby making it labour intensive. In examining mammograms, some studies have reported that radiologists rarely altered their diagnostic decisions after viewing results from predefined, feature-based CADe systems and that their clinical integration had no statistical significance on the radiologists’ performance41,42. This is owing, in part, to the subhuman performance of these systems. Recent efforts have explored deep learning-based CADe to detect pulmonary nodules in CT43 and prostate cancer in multiparametric imaging, specifically multiparametric MRI44. In detecting lesions in mammograms, early results show that utilizing convolutional neural networks (CNNs; deep learning algorithms; Box 1) in CADe outperforms traditional CADe systems at low sensitivity while performing comparably at high sensitivity and shows similar performance compared with human readers45. These findings hint at the utility of deep learning in developing robust, high-performance CADe systems.
Characterization.
Characterization is an umbrella term referring to the segmentation, diagnosis and staging of a disease. These tasks are accomplished by quantifying radiological characteristics of an abnormality, such as the size, extent and internal texture. While handling routine tasks of examining medical images, humans are simply not capable of accounting for more than a handful of qualitative features. This is exacerbated by the inevitable variability across human readers, with some performing better than others. Automation through AI can, in principle, consider a large number of quantitative features together with their degrees of relevance while performing the task at hand in a reproducible manner every time. For instance, it is difficult for humans to accurately predict the status of malignancy in the lung owing to the similarity between benign and malignant nodules in CT scans. AI can automatically identify these features, and many others, while treating them as imaging biomarkers. Such biomarkers could hence be used to predict malignancy likelihood among other clinical end points including risk assessment, differential diagnosis, prognosis and response to treatment.
Within the initial segmentation step, while non-diseased organs can be segmented with relative ease, identifying the extent of diseased tissue is potentially orders of magnitude more challenging. Typical practices of tumour segmentation within clinical radiology today are often limited to high-level metrics such as the largest inplane diameter. However, in other clinical cases, a higher specificity and precision are vital. For instance, in clinical radiation oncology, the extents of both tumour and non-tumour tissues have to be accurately segmented for radiation treatment planning. Attempts at automating segmentation have made their way into the clinic, with varying degrees of success46. Segmentation finds its roots in earlier computer vision research carried out in the 1980s47, with continued refinement over the past decades. Simpler segmentation algorithms used clustered imaging intensities to isolate different areas or utilized region growing, where regions are expanded around user-defined seed points within objects until a certain homogeneity criterion is no longer met48. A second generation of algorithms saw the incorporation of statistical learning and optimization methods to improve segmentation precision, such as the watershed algorithm, where images are transformed into topological maps with intensities representing heights49. More advanced systems incorporate previous knowledge into the solution space, as in the use of a probabilistic atlas — often an attractive option when objects are ill-defined in terms of their pixel intensities. Such atlases have enabled more accurate automated segmentations, as they contain information regarding the expected locations of tumours across entire patient populations46. Applications of probabilistic atlases include segmenting brain MRI for locating diffuse low-grade glioma50, prostate MRI for volume estimation51 and head and neck CT for radiotherapy treatment planning52, to name a few.
Recently proposed deep learning architectures for segmentation include fully convolutional networks, which are networks comprising convolutional layers only, that output segmentation probability maps across entire images53. Other architectures, such as the U-net54, have been specifically designed for medical images. Studies have reported that a single deep learning system is able to perform diverse segmentation tasks across multiple modalities and tissue types, including brain MRI, breast MRI and cardiac CT angiography (CTA), without task-specific training55. Others describe deep learning methods for brain MRI segmentation that completely eliminate the need for image registration, a required preprocessing step in atlas-based methods56.
Multiple radiographic characteristics are also employed in subsequent diagnosis tasks. These are critical to determine, for instance, whether a lung nodule is solid or whether it contains non-solid areas, also known as ground-glass opacity (GGO) nodules. GGO nodules are rather challenging to diagnose and often require special management protocols, mainly owing to the lack of associated characteristics of malignancy or invasiveness in radiographs57. Generally, tumour radiographic characteristics may include information regarding size, maximum diameter, sphericity, internal texture and margin definition. The logic for diagnosis is based on these, often subjective, characteristics, enabling the stratification of objects into classes indicative of being benign or malignant. Methods to automate diagnoses are collectively referred to as computer-aided diagnosis (CADx) systems. Similar to CADe, they often rely on predefined engineered discriminative features. Several systems are already in clinical use, as is the case with screening mammograms58. They usually serve as a second opinion in complementing a radiologist’s assessment59, and their perceived successes have led to the development of similar systems for other imaging modalities, including ultrasonography and MRI60. For instance, traditional CADx systems have been used on ultrasonography images to diagnose cervical cancer in lymph nodes, where they have been found to improve the performance of particularly inexperienced radiologists as well as reduce variability among them61. Other application areas include prostate cancer in multiparametric MRI, where a malignancy probability map is first calculated for the entire prostate, followed by automated segmentation for candidate detection62.
The accuracy of traditional predefined feature-based CADx systems is contingent upon several factors, including the accuracy of previous object segmentations. It is often the case that errors are magnified as they propagate through the various image-based tasks within the clinical oncology workflow. We also find that some traditional CADx methods fail to generalize across different objects. For instance, while the measurement of growth rates over time is considered a major factor in assessing risk, pulmonary nodule CADx systems designed around this criterion are often unable to accurately diagnose special nodules such as cavity and GGO nodules63. Such nodules require further descriptors for accurate detection and diagnosis — descriptors that are not discriminative when applied to the more common solid nodules64. This eventually leads to multiple solutions that are tailored for specific conditions with limited generalizability. Without explicit predefinition of these discriminative features, deep learning-based CADx is able to automatically learn from patient populations and form a general understanding of variations in anatomy — thus allowing it to capture a representation of common and uncommon cases alike.
Architectures such as CNNs are well suited for supervised diagnostic classification tasks (FIG. 2b). For both the breast lesion and lung nodule classification tasks, studies report a substantial performance gain of deep learning-based CADx methods — specifically those utilizing stacked denoising autoencoders — over their traditional state-of-the-art counterparts. This is mainly owing to the automatic feature exploration mechanism and higher noise tolerance of deep learning. Such performance gain is assessed using multiple metrics, including the area under receiver operating characteristic curve (AUC), accuracy, sensitivity and specificity, to name a few35.
Staging systems, such as tumour-node-metastasis (TNM) in oncology, rely on preceding information gathered through segmentation and diagnosis to classify patients into multiple predefined categories65. This enables a well-informed choice of the type of treatment and aids in predicting survival likelihood and prognosis. Staging has generally seen little to no automation because it relies on qualitative descriptions that are often difficult to quantitatively measure. The automated staging of primary tumour size, nearby lymph nodes and distant metastasis requires different feature sets and approaches. While traditional machine learning might have relied on ensemble methods where multiple distinct models are combined, deep learning has the ability to learn joint data representations simultaneously66 — making it well suited for such multi-faceted classification problems. Most deep learning efforts to detect lymph node involvement and distant metastasis — and ultimately obtain an accurate staging — have been carried out on pathology images67,68. However, more work on radiographic images is expected to appear in the near future.
Monitoring.
Disease monitoring is essential for diagnosis as well as for evaluation of treatment response. The workflow involves an image registration preprocess where the diseased tissue is aligned across multiple scans, followed by an evaluation of simple metrics on them using predefined protocols — which is very similar to diagnosis tasks on single time-point images. A simple data comparison protocol follows and is used to quantify change. In oncology, for instance, these protocols define information regarding tumor size. Examples include the Response Evaluation Criteria in Solid Tumours (RECIST) and those created by the World Health Organization (WHO)69. Here, we find that the main goal behind such simplification is reducing the amount of effort and data a human reader must interact with while performing the task. However, this simplification is often based on incorrect assumptions regarding isotropic tum our growth. Whereas some change characteristics are directly identifiable by humans, such as moderately large variations in object size, shape and cavitation, others are not. These could include subtle variations in texture and heterogeneity within the object. Poor image registration, dealing with multiple objects and physiological changes over time all contribute to more challenging change analyses. Moreover, the inevitable interobserver variability70 remains a major weakness in the process. Computer-aided change analysis is considered a relatively younger field than CADe and CADx systems and has not yet achieved as much of a widespread adoption71. Early efforts in automating change analysis workflows relied on the automated registration of multiple images followed by subtraction of one from another, after which changed pixels are highlighted and presented to the reader. Other more sophisticated methods perform a pixel-by-pixel classification — on the basis of predefined discriminative features — to identify changed regions and hence produce a more concise map of change72. As the predefined features used for registration differ from those used for the subsequent change analysis, a multistep procedure combining different feature sets is required. This could compromise the change analysis step, as it becomes highly sensitive to registration errors. With computer-aided change analysis based on deep learning, feature engineering is eliminated and a joint data representation can be learned. Deep learning architectures, such as recurrent neural networks, are very well suited for such temporal sequence data formats and are expected to find ample applications in monitoring tasks.
Other opportunities.
In addition to the three primary clinical tasks mentioned above, AI is expected to impact other image-based tasks within the clinical radiology workflow. These include the preprocessing steps following image acquisition as well as subsequent reporting and integrated diagnostics (FIG. 3a).
Starting at the outset of the workflow, the first of these tasks to be improved is reconstruction. We find a widening gap between advancements in image acquisition hardware and image-reconstruction software, a gap that can potentially be addressed by new deep learning methods for suppressing artefacts and improving overall quality. For instance, CT reconstruction algorithms have seen little to no change in the past 25 years73. Additionally, many filtered back-projection image-reconstruction algorithms are computationally expensive, signifying that a trade-off between distortions and runtime is inevitable74. Recent efforts report the flexibility of deep learning in learning reconstruction transformations for various MRI acquisition strategies, which is achieved by treating the reconstruction process as a supervised learning task where a mapping between the scanner sensors and resultant images is derived75. Other efforts employ novel AI methods to correct for artefacts as well as address certain imaging modality-specific problems such as the limited angle problem in CT76 — a missing data problem where only a portion of the scanned space can be reconstructed owing to the scanner’s inability to perform full 180° rotations around objects. Studies have also utilized CNNs and synthetically generated artefacts to combine information from original and corrected images as a means to suppress metal artefacts77. More work is needed to investigate the accuracy of deep learning-based reconstruction algorithms and their ability to recreate rare, unseen structures, as initial errors propagated throughout the radiology workflow can have adverse effects on patient outcome.
Another preprocessing task to be improved is registration, as touched upon previously in the monitoring section. This process is often based on predefined similarity criteria such as landmark and edge-based measures. In addition to the computational power and time consumed by these predefined feature-based methods, some are sensitive to initializations, chosen similarity features and the reference image78. Deep learning methods could handle complex tissue deformations through more advanced non-rigid registration algorithms while providing better motion compensation for temporal image sequences. Studies have shown that deep learning leads to generally more consistent registrations and is an order of magnitude faster than more conventional methods79. Additionally, deep learning is multimodal in nature where a single shared representation among imaging modalities can be learned80. Multimodal images in cancer have enabled the association of multiple quantitative functional measurements, as in the PET hybrids PET-MRI and PET-CT, thus improving the accuracy of tumour characterization and assessment81. With robust registration algorithms based on deep learning, the utility of multimodal imaging can be further explored without concerns regarding registration accuracy.
Radiology reports lie at the intersection of radiology and multiple oncology subspecialties. However, the generation of these textual reports can be a laborious and routine time-consuming task. When compared with conventional dictation, even structured reporting systems with bulleted formatting have been shown not to improve attending physicians’ perceptions of report clarity82. As the report generation task falls towards the end of the radiology workflow, it is the most sensitive to errors from preceding steps. Additionally, the current radiologist-oncologist communication model has not been found to be optimally coordinated — especially with regard to monitoring lesions over time83. Owing to the often different formats in which data are recorded by medical professionals, Al-run, automatic, report-generation tools can pave the way for a more standardized terminology — an area that currently lacks stringent standards and an agreed-upon understanding of what constitutes a ‘good’ report84. Such tools could also replace the traditional qualitative text-based approach with a more interactive quantitative one, which has been shown to improve and promote collaboration between different parties85. Within lung cancer screening, this could include quantified information about the size and location of a nodule, probability of malignancy and associated confidence level. These well-structured reports are also immensely beneficial to population sciences and big data mining efforts. Following deep learning advances in the automatic caption generation from photographic images86, recent efforts have explored means to diagnose abnormalities in chest radiography scans and automatically annotate them in a textual format87.
After carrying out various clinical tasks and generating radiology reports (FIG. 3a), Al-based integrated diagnostics could potentially enable health-care-wide assimilation of data from multiple streams, thus capitalizing on all data types pertaining to a particular patient. In addition to radiology reports describing findings from medical images and their associated metadata, other data could be sourced from the clinic or from pathology or genomics testing. Data from wearables, social media and other lifestyle-quantifying sources could all potentially offer valid contributions to such a comprehensive analysis. This will be crucial in providing AI biomarkers with robust generalizability towards different end points. Such consolidation of standard medical data, using traditional AI methods, has already demonstrated the ability to advance clinical decision making in lung cancer diagnosis and care21.
AI challenges in medical imaging
We are currently witnessing a major paradigm shift in the design principles of many computer-based tools used in the clinic. There is great debate about the speed with which newer deep learning methods will be implemented in clinical radiology practice88, with speculations for the time needed to fully automate clinical tasks ranging from a few years to decades. The development of deep learning-based automated solutions will begin with tackling the most common clinical problems where sufficient data are available. These problems could involve cases where human expertise is in high demand or data are far too complex for human readers; examples of these include the reading of lung screening CTs, mammograms and images from virtual colonoscopy. A second wave of efforts is likely to address more complex problems such as multiparametric MRI. A common trait among current AI tools is their inability to address more than one task, as is the case with any narrow intelligence. A comprehensive AI system able to detect multiple abnormalities within the entire human body is yet to be developed.
Data continue to be the most central and crucial constituent for learning AI systems. With one out of four Americans receiving a CT examination89 and one out of ten receiving an MRI examination90 annually, millions of medical images are produced each year. Additionally, recent well-implemented advances in US-based digital health systems — such as the Picture Archiving and Communication System (PACS) — have ensured that medical images are electronically organized in a systematic manner91,92, with parallel efforts in Europe93 and developing countries94. It is clear that large amounts of medical data are indeed available and are stored in such a manner that enables moderate ease in access and retrieval. However, such data are rarely curated, and this represents a major bottleneck in attempting to learn any AI model. Curation can refer to patient cohort selection relevant for a specific AI task but can also refer to segmenting objects within images. Curation ensures that training data adheres to a defined set of quality criteria and is clear of compromising artefacts. It can also help avoid unwanted variance in data owing to differences in data-acquisition standards and imaging protocols, especially across institutions, such as the time between contrast agent administration and actual imaging. An example of data curation within oncology could include assembling a cohort of patients with specific stages of disease and tumour histology grades. Although photographic images can be labelled by nonexperts, using, for instance, crowdsourcing approaches, medical images do require domain knowledge. Hence, it is imperative that such curation is performed by a trained reader to ensure credibility — making the process expensive. It is also very time consuming, although utilizing recent deep learning algorithms promises to reduce annotation time substantially: meticulous slice-by-slice segmentation can potentially be substituted by single seed points within the object, from which full segmentations could be automatically generated. The amount of data requiring curation is another limiting factor and is highly dependent on the AI approach — with deep learning methods being more prone to overfitting and hence often requiring more data.
The suboptimal performance of many automated and semi-automated segmentation algorithms46 has hindered their utility in curating data, as human readers are almost always needed to verify accuracy. More complications arise with rare diseases, where automated labelling algorithms are non-existent. The situation is exacerbated when only a limited number of human readers have previous exposure and are capable of verifying these uncommon diseases. One solution that enables automated data curation is unsupervised learning. Recent advances in unsupervised learning, including generative adversarial networks95 and variational autoencoders96 among others, show great promise, as discriminative features are learned without explicit labelling. Recent studies have explored unsupervised domain adaptation using adversarial networks to segment brain MRI, leading to a generalizability and accuracy close to those of supervised learning methods97. Others employ sparse autoencoders to segment breast density and score mammographic texture in an unsupervised manner98. Self-supervised learning efforts have also utilized spatial context information as supervision for recognizing body parts in CT and MRI volumes through the use of paired CNNs99. Nevertheless, public repositories such as The Cancer Imaging Archive (TCIA)100 offer unparalleled open-access to labelled medical imaging data, allowing immediate AI model prototyping and thus eliminating lengthy data curation steps.
Albeit intuitively leading to higher states of intelligence, the recent paradigm shift from programs based on well-defined rules to others that learn directly from data has brought certain unforeseen concerns to the spotlight. A strong theoretical understanding of deep learning is yet to be established101 despite the reported successes across many fields — explaining why deep learning layers that lie between inputs and outputs are labelled as ‘hidden layers’ (Box 1; fig. 2b). Identifying specific features of an image that contribute to a predicted outcome is highly hypothetical, causing a lack of understanding of how certain conclusions are drawn by deep learning. This lack of transparency makes it difficult to predict failures, isolate the logic for a specific conclusion or troubleshoot inabilities to generalize to different imaging hardware, scanning protocols and patient populations. Not surprisingly, many uninterpretable AI systems with applications in radiology have been dubbed ‘black-box medicine’ (REF102).
From a regulatory perspective, discussions are underway regarding the legal right of regulatory entities to interrogate AI frameworks on the mathematical reasoning for an outcome103. While such questioning is possible with explicitly programmed mathematical models, new AI methods such as deep learning have opaque inner workings, as mentioned above. Sifting through hundreds of thousands of nodes in a neural network, and their respective associated connections, to make sense of their stimulation sequence is unattainable. An increased network depth and node count brings more complex decision making together with a much more challenging system to take apart and explore. On the other hand, we find that many safe and effective US Food and Drug Administration (FDA)-approved drugs have unknown mechanisms of action104,105. From that perspective and despite the degree of uncertainty surrounding many AI algorithms, the FDA has already approved high-performance software solutions, though they are known to have somewhat obscure working mechanisms. Regulatory bodies, such as the FDA, have been regulating CADe and CADx systems that rely on machine learning and pattern-recognition techniques since the earliest days of computing. However, it is the shift to deep learning that now poses new regulatory challenges and requires new guidance for submissions seeking approval. Even after going to market, deep learning methods evolve over time as more data are processed and learned from. Thus, it is crucial to understand the implications of such lifelong learning in these adaptive systems. Periodic testing over specific time intervals could potentially ensure that learning and its associated prediction performance are following forecasted projections. Additionally, such benchmarking tests need to adapt to AI specifics such as the sensitivity of prediction probabilities in CNNs.
Other ethical issues may arise from the use of patient data to train these AI systems. Data are hosted within networks of medical institutions, often lacking secure connections to state-of-the-art AI systems hosted elsewhere. More recently, Health Insurance Portability and Accountability Act (HIPAA)-compliant storage systems have paved the way for more stringent privacy preservation. Studies have explored systems that enable multiple entities to jointly train AI models without sharing their input data sets — sharing only the trained model106,107. Other efforts use a decentralized ‘federated’ learning approach108. During training, data remains local, while a shared model is learned by combining local updates. Inference is then performed locally on live copies of the shared model, eliminating data sharing and privacy concerns. ‘Cryptonets’ are deep learning networks trained on encrypted data, and they even make encrypted predictions that can be decrypted only by the owner of a decryption key — thus ensuring complete confidentiality throughout the entire process109. All these solutions, albeit still in early developmental stages, promise to create a sustainable ‘data to AI’ ecosystem — without undermining privacy and HIPAA compliance.
Future perspectives
From the early days of X-ray imaging in the 1890s to more recent advances in CT, MRI and PET scanning, medical imaging continues to be a pillar of medical treatment. Current advances in imaging hardware — in terms of quality, sensitivity and resolution — enable the discrimination of minute differences in tissue densities. Such differences are, in some cases, difficult to recognize by a trained eye and even by some traditional AI methods used in the clinic. These methods are thus not fully on par with the sophistication of imaging instruments, yet they serve as another motivation to pursue this paradigm shift towards more powerful AI tools. Moreover, and in contrast to traditional methods based on predefined features, we find that deep learning algorithms scale with data, that is, as more data are generated every day and with ongoing research efforts, we expect to see relative improvements in performance. All these advances promise an increased accuracy and reduction in the number of routine tasks that exhaust time and effort.
Aligning research methodologies is crucial in accurately assessing the impact of AI on patient outcome. In addition to the undeniable importance of reproducibility and generalizability, utilizing agreed-upon benchmarking data sets, performance metrics, standard imaging protocols and reporting formats will level the experimentation field and enable unbiased indicators. It is also important to note that AI is unlike human intelligence in many ways; excelling in one task does not necessarily imply excellence in others. Therefore, the promise of up-and-coming AI methods should not be overstated. Almost all state-of-the-art advances in the field of AI fall under the narrow AI category, where AI is trained for one task and one task only — with only a handful exceeding human intelligence. While such advances excel in interpreting sensory perceptual information in a bottom-up fashion, they lack higher-level, top-down knowledge of contexts and fail to make associations the way a human brain does. Thus, it is evident that the field is still in its infancy, and overhyped excitement surrounding it should be replaced with rational thinking and mindful planning. It is also evident that AI is unlikely to replace radiologists within the near or even distant future. The roles of radiologists will expand as they become more connected to technology and have access to better tools. They are also likely to emerge as critical elements in the AI training process, contributing knowledge and overseeing efficacy. As different forms of AI exceed human performance, we expect it to evolve into a valuable educational resource. Human operators will not only oversee outcomes but also seek to interpret the reasoning behind them — as a means of validation and as a way to potentially discover hidden information that might have been overlooked (FIG. 1).
In contrast to traditional AI algorithms locked within proprietary commercial packages, we find that the most popular deep learning software platforms available today are open-source. This has fostered, and continues to foster, experimentation on a massive scale. In terms of data, AI efforts are expected to shift from processed medical images to raw acquisition data. Raw data are almost always downsampled and optimized for human viewers. This simplification and loss of information are both avoidable when the analyses are run by machines but are associated with caveats including reduced interpretability and impeded human validation. As more data are generated, more signal is available for training. However, more noise is also present. We expect the process of discerning signal from noise to become more challenging over time. With difficulties in curating and labelling data, we foresee a major push towards unsupervised learning techniques to fully utilize the vast archives of unlabelled data.
Open questions include the ambiguity of who controls AI and is ultimately responsible for its actions, the nature of the interface between AI and health care and whether implementation of a regulatory policy too soon will cripple AI application efforts. Enabling interoperability among the multitude of AI applications that are currently scattered across health care will result in a network of powerful tools. This AI web will function at not only the inference level but also the lifelong training level. We join the many calls110 that advocate for creating an interconnected network of de-identified patient data from across the world. Utilizing such data to train AI on a massive scale will enable a robust AI that is generalizable across different patient demographics, geographic regions, diseases and standards of care. Only then will we see a socially responsible AI benefiting the many and not the few.
Acknowledgements
The authors acknowledge financial support from the US National Institutes of Health (NIH-USA U24CA194354 and NIH-USA U01CA190234).
Glossary
- Area under receiver operating characteristic curve
(AUC). A sensitivity versus specificity metric for measuring the performance of binary classifiers that can be extended to multi-class problems. The area under the curve is equal to the probability that a randomly chosen positive sample ranks above a randomly chosen negative one or is regarded to have a higher probability of being positive.
- Artificial intelligence
(AI). A branch of computer science involved with the development of machines that are able to perform cognitive tasks that would normally require hum an intelligence.
- Caption generation
The often automated generation of qualitative text describing an illustration or image and its contents.
- Ground-glass opacity
(GGO). A visual feature of some subsolid pulmonary nodules that is characterized by focal areas of slightly increased attenuation on computed tomography. Underlying bronchial structures and vessels are often visually preserved (being even more recognizable owing to increased contrast), thus making the detection and diagnosis of such nodules somewhat challenging.
- Health Insurance Portability and Accountability Act
(HIPAA). A US act that sets provisions for protecting and securing sensitive patient medical data.
- Image registration
A process that involves aligning medical images either in terms of spatial or temporal characteristics, mostly intramodality and occasionally intermodality.
- Imaging modalities
A multitude of imaging methods that are used to non-invasively generate visualizations of the human anatomy. Examples of these include computed tomography (CT), computed tomography angiography (CTA), magnetic resonance imaging (MRI), mammography, ultrasonography (echocardiography) and positron emission tomography (PET).
- Initializations
Within optimization problems, constantly adjusted parameters during run time need to be initialized to some value before the start of the process. Good initialization techniques aid models in converging faster and hence speed up the iteration process.
- Machine learning
A branch of artificial intelligence and computer science that enables computers to learn without being explicitly programmed.
- Multiparametric imaging
Medical imaging in which two or more parameters are used to visualize differences between healthy and diseased tissue. In multiparametric magnetic resonance imaging (MRI), these parameters include T2-weighted MRI, diffusion-weighted MRI and dynamic contrastenhanced MRI, among others.
- Predefined engineered features
A set of context-based human-crafted features designed to represent knowledge regarding a specific data space.
- Probabilistic atlas
A single composite image formed by combining and registering pre-segmented images of multiple patients that thus contains knowledge on population variability.
- Radiomics
A data-centric field investigating the clinical relevance of radiographic tissue characteristics automatically quantified from medical images.
- Report generation
The communication of assessments and findings in both image and text formats among medical professionals.
- Segmentation
The partitioning of images to produce boundary delineations of objects of interest. Such a boundary is defined by pixels and voxels (3D pixels) when performed in 2D and 3D, respectively.
- Self-supervised learning
A type of supervised learning where labels are determined by the input data as opposed to being explicitly provided.
- Supervised learning
A type of machine learning where functions are inferred from labelled training data. Example data pairs consist of the input together with its desired output or label.
- Unsupervised learning
A type of machine learning where functions are inferred from training data without corresponding labels.
- Wearables
A collective term describing health-monitoring devices, smartwatches and fitness trackers that have recently been integrated into the health-care ecosystem as a means to remotely track vitals and adhere to treatment plans.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Editors, N. Auspicious machine learning. Nat. Biomed. Engineer 1, 0036 (2017). [Google Scholar]
- 2.Mnih V et al. Human-level control through deep reinforcement learning. Nature 518, 529–533 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Moravcík M et al. DeepStack: Expert-level artificial intelligence in heads-up no-limit poker. Science 356, 508–513 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Xiong W et al. Toward human parity in conversational speech recognition. IEEE/ACM Trans. Audio Speech Language Process 25, 2410–2423 (2017). [Google Scholar]
- 5.Pendleton SD et al. Perception, planning, control, and coordination for autonomous vehicles. Machines 5, 6 (2017). [Google Scholar]
- 6.Silver D et al. Mastering the game of Go with deep neural networks and tree search. Nature 529, 484–489 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Grace K, Salvatier J, Dafoe A, Zhang B & Evans O When will AI exceed human performance? Evidence from AI experts. Preprint at arXiv, 1705.08807 (2017). [Google Scholar]
- 8.Rusk N Deep learning. Nat. Methods 13, 35–35 (2015). [Google Scholar]
- 9.Esteva A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulshan V et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Aerts HJWL The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2, 1636–1642 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Kumar V et al. Radiomics: the process and the challenges. Magn. Reson. Imag 30, 1234–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambin P et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48, 441–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litjens G et al. A survey on deep learning in medical image analysis. Med. Image Anal 42, 60–88 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Kolossváry M, Kellermayer M, Merkely B & Maurovich-Horvat P Cardiac computed tomography radiomics: a comprehensive review on radiomic techniques. J. Thorac. Imag 33, 26–34 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Aerts HJWL et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun 5, 4006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coroller TP et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol 114, 345–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu W et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front. Oncol 6, 71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh E et al. Associations of radiomic data extracted from static and respiratory-gated CT scans with disease recurrence in lung cancer patients treated with SBRT. PLoS ONE 12, e0169172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rios Velazquez E et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 77, 3922–3930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossmann P et al. Defining the biological basis of radiomic phenotypes in lung cancer. eLife 6, e23421 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmar C, Grossmann P, Bussink J, Lambin P & Aerts HJWL Machine learning methods for quantitative radiomic biomarkers. Sci. Rep 5, 13087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor JPB et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol 14, 169–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boland GWL, Guimaraes AS & Mueller PR The radiologist’s conundrum: benefits and costs of increasing CT capacity and utilization. Eur. Radiol 19, 9–12 (2009). [DOI] [PubMed] [Google Scholar]
- 25.McDonald RJ et al. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol 22, 1191–1198 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald R Error in radiology. Clin. Radiol 56, 938–946 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Ledley RS & Lusted LB Reasoning foundations of medical diagnosis; symbolic logic, probability, and value theory aid our understanding of how physicians reason. Science 130, 9–21 (1959). [DOI] [PubMed] [Google Scholar]
- 28.Lodwick GS, Keats TE & Dorst JP The coding of Roentgen images for computer analysis as applied to lung cancer. Radiology 81, 185–200 (1963). [DOI] [PubMed] [Google Scholar]
- 29.Ambinder EP A history of the shift toward full computerization of medicine. J. Oncol. Pract 1, 54–56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haug PJ Uses of diagnostic expert systems in clinical care. Proc. Annu. Symp. Comput. Appl. Med. Care, 379–383 (1993). [PMC free article] [PubMed] [Google Scholar]
- 31.Castellino RA Computer aided detection (CAD): an overview. Cancer Imag. 5, 17–19 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen D, Wu G & Suk H-I Deep learning in medical image analysis. Annu. Rev. Biomed. Eng 19, 221–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veeraraghavan H MO-A-207B-01: Radiomics: Segmentation & feature extraction techniques. Med. Phys 43, 3694–3694 (2016). [Google Scholar]
- 34.Paul R et al. Deep feature transfer learning in combination with traditional features predicts survival among patients with lung adenocarcinoma. Tomography 2, 388–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng J-Z et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci. Rep 6, 24454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Zheng Y, Park J-H, Heng P-A & Zhou SK in Medical Image Computing and Computer-Assisted Intervention — MICCAI 2016 487–495 (Athens, Greece, 2016). [Google Scholar]
- 37.Ghafoorian M et al. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci. Rep 7, 5110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. 7, 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Ginneken B, Schaefer-Prokop CM & Prokop M Computer-aided diagnosis: how to move from the laboratory to the clinic. Radiology 261, 719–732 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Nagaraj S, Rao GN & Koteswararao K The role of pattern recognition in computer-aided diagnosis and computer-aided detection in medical imaging: a clinical validation. Int. J. Comput. Appl 8, 18–22 (2010). [Google Scholar]
- 41.Cole EB et al. Impact of computer-aided detection systems on radiologist accuracy with digital mammography. AJR Am. J. Roentgenol 203, 909–916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehman CD et al. Diagnostic accuracy of digital screening mammography with and without computeraided detection. JAMA Intern. Med 175, 1828–1837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X, Shan J & Vaidya V in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 379–383 (Melbourne, Australia, 2017). [Google Scholar]
- 44.Tsehay YK et al. in Proceedings of SPIE https://doi.org/10.1117/12.2254423 (2017).
- 45.Kooi T et al. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal 35, 303–312 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Sharma N & Aggarwal LM Automated medical image segmentation techniques. J. Med. Phys 35, 3–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haralick RM & Shapiro LG Image segmentation techniques. Computer Vision Graph. Image Process 29, 100–132 (1985). [Google Scholar]
- 48.Pham DL, Xu C & Prince JL Current methods in medical image segmentation. Annu. Rev. Biomed. Eng 2, 315–337 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Grau V, Mewes AUJ, Alcañiz M, Kikinis R & Warfield SK Improved watershed transform for medical image segmentation using prior information. IEEE Trans. Med. Imag 23, 447–458 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Parisot S et al. A probabilistic atlas of diffuse WHO grade II glioma locations in the brain. PLoS ONE 11, e0144200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghose S et al. in 2012 19th IEEE International Conference on Image Processing 541–544 (Orlando, FL, USA, 2012). [Google Scholar]
- 52.Han X et al. Atlas-based auto-segmentation of head and neck CT images. Med. Image Comput. Comput. Assist. Interv 11, 434–441 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Long J, Shelhamer E & Darrell T in 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 3431–3440 (Boston, MA, USA, 2015). [Google Scholar]
- 54.Ronneberger O, Fischer P & Brox TU in Medical Image Computing and Computer-Assisted Intervention — MICCAI 2015 234–241 (Munich, Germany, 2015). [Google Scholar]
- 55.Moeskops P et al. in Medical Image Computing and Computer- Assisted Intervention — MICCAI 2016 478–486 (Athens, Greece, 2016). [Google Scholar]
- 56.de Brebisson A & Montana G in 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) 20–28 (Boston, MA, USA, 2015). [Google Scholar]
- 57.Cioffi U, Raveglia F, De Simone M & Baisi A Ground-glass opacities: a curable disease but a big challenge for surgeons. J. Thorac. Cardiovasc. Surg 154, 375–376 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Champaign JL & Cederbom GJ Advances in breast cancer detection with screening mammography. Ochsner J 2, 33–35 (2000). [PMC free article] [PubMed] [Google Scholar]
- 59.Shiraishi J, Li Q, Appelbaum D & Doi K Computeraided diagnosis and artificial intelligence in clinical imaging. Semin. Nucl. Med 41, 449–462 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Ayer T, Ayvaci MU, Liu ZX, Alagoz O & Burnside ES Computer-aided diagnostic models in breast cancer screening. Imag. Med 2, 313–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Wang Y, Yu B, Shi X & Zhang Y Application of computer-aided diagnosis to the sonographic evaluation of cervical lymph nodes. Ultrason. Imag 38, 159–171 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Giannini V et al. A fully automatic computer aided diagnosis system for peripheral zone prostate cancer detection using multi-parametric magnetic resonance imaging. Comput. Med. Imaging Graph 46, 219–226 (2015). [DOI] [PubMed] [Google Scholar]
- 63.El-Baz A et al. Computer-aided diagnosis systems for lung cancer: challenges and methodologies. Int. J. Biomed. Imag 2013, 942353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edey AJ & Hansell DM Incidentally detected small pulmonary nodules on CT. Clin. Radiol 64, 872–884 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Mirsadraee S, Oswal D, Alizadeh Y, Caulo A & van Beek E Jr. The7th lung cancer TNM classification and staging system: review of the changes and implications. World J. Radiol 4, 128–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sohn K, Shang W & Lee H in Advances in Neural Information Processing Systems 27 (NIPS 2014) (eds Ghahramani Z, Welling M, Cortes C, Lawrence ND & Weinberger KQ) 2141–2149 (Montreal, Canada, 2014). [Google Scholar]
- 67.Litjens G et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep 6, 26286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cruz-Roa A et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci. Rep 7, 46450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaffe CC Measures of response: RECIST, WHO, and new alternatives. J. Clin. Oncol 24, 3245–3251 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Thiesse P et al. Response rate accuracy in oncology trials: reasons for interobserver variability. Groupe Français d’Immunothérapie of the Fédération Nationale des Centres de Lutte Contre le Cancer. J. Clin. Oncol 15, 3507–3514 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Khorasani R, Erickson BJ & Patriarche J New opportunities in computer-aided diagnosis: change detection and characterization. J. Am. Coll. Radiol 3, 468–469 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Patriarche JW & Erickson BJ Part 1. Automated change detection and characterization in serial MR studies of brain-tumor patients. J. Digit. Imag 20, 203–222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pan X, Sidky EY & Vannier M Why do commercial CT scanners still employ traditional, filtered backprojection for image reconstruction? Inverse Probl 25, 1230009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pipatsrisawat T, Gacic A, Franchetti F, Puschel M & Moura JMF in Proceedings. (ICASSP ‘05). IEEE International Conference on Acoustics, Speech, and Signal Processing, 2005 v/153–v/156 (Philadelphia, PA, USA, 2005). [Google Scholar]
- 75.Zhu B, Liu JZ, Cauley SF, Rosen BR & Rosen MS Image reconstruction by domain-transform manifold learning. Nature 555, 487–492 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Hammernik K, Würfl T, Pock T & Maier AA in Bildverarbeitung für die Medizin 2017 (eds Maier-Hein K, Deserno T, Handels H & Tolxdorff T) 92–97 (Springer, Berlin, Heidelberg, 2017). [Google Scholar]
- 77.Gjesteby L et al. in Developments in X-Ray Tomography XI 10391–31 (San Diego, CA, USA, 2017). [Google Scholar]
- 78.El-Gamal FE-ZA, Elmogy M & Atwan A Current trends in medical image registration and fusion. Egypt. Informat. J 17, 99–124 (2016). [Google Scholar]
- 79.Yang X, Kwitt R, Styner M & Niethammer M Quicksilver: fast predictive image registration — a deep learning approach. Neuroimage 158, 378–396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ngiam J et al. in Proceedings of the 28th International Conference on Machine Learning 689–696 (Bellevue, WA, USA, 2011). [Google Scholar]
- 81.Yankeelov TE, Abramson RG & Quarles CC Quantitative multimodality imaging in cancer research and therapy. Nat. Rev. Clin. Oncol 11, 670–680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson AJ, Chen MYM, Zapadka ME, Lyders EM & Littenberg B Radiology report clarity: a cohort study of structured reporting compared with conventional dictation. J. Am. Coll. Radiol 7, 501–506 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Levy MA & Rubin DL Tool support to enable evaluation of the clinical response to treatment. AMIA Annu. Symp. Proc 2008, 399–403 (2008). [PMC free article] [PubMed] [Google Scholar]
- 84.European Society of Radiology (ESR). Good practice for radiological reporting. Guidelines from the European Society of Radiology (ESR). Insights Imag. 2, 93–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Folio LR et al. Quantitative radiology reporting in oncology: survey of oncologists and radiologists. AJR Am. J. Roentgenol 205, W233–W243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karpathy A & Fei-Fei L Deep visual-semantic alignments for generating image descriptions. IEEE Trans. Pattern Anal. Mach. Intell 39, 664–676 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Shin H-C et al. in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2497–2506 (Las Vegas, NV, USA, 2016). [Google Scholar]
- 88.Lee J-G et al. Deep learning in medical imaging: general overview. Kor. J. Radiol 18, 570–584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.OECD. Computed tomography (CT) exams. https://doi.org/10.1787/3c994537-en (2018).
- 90.OECD. Magnetic resonance imaging (MRI) exams. https://doi.org/10.1787/1d89353f-en (2018).
- 91.Bryan S et al. Radiology report times: impact of picture archiving and communication systems. AJR Am. J. Roentgenol 170, 1153–1159 (1998) [DOI] [PubMed] [Google Scholar]
- 92.Mansoori B, Erhard KK & Sunshine JL Picture Archiving and Communication System (PACS) implementation, integration and benefits in an integrated health system. Acad. Radiol 19, 229–235 (2012). [DOI] [PubMed] [Google Scholar]
- 93.Lemke HU PACS developments in Europe. Comput. Med. Imag. Graph 27, 111–120 (2003). [DOI] [PubMed] [Google Scholar]
- 94.Mendel JB & Schweitzer AL PACS for the developing world. J. Global Radiol 1, 5 (2015). [Google Scholar]
- 95.Goodfellow I et al. in Advances in Neural Information Processing Systems 27 (NIPS 2014) (eds Ghahramani Z, Welling M, Cortes C, Lawrence ND & Weinberger KQ) 2672–2680 (Montreal, Canada, 2014). [Google Scholar]
- 96.Kingma DP & Welling M Auto-encoding variational Bayes. Preprint at arXiv, 1312.6114 (2013). [Google Scholar]
- 97.Kamnitsas K et al. in Information Processing in Medical Imaging 597–609 (Springer, Cham, 2017). [Google Scholar]
- 98.Kallenberg M et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans. Med. Imag 35, 1322–1331 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Zhang P, Wang F & Zheng Y in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 578–582 (Melbourne, Australia, 2017). [Google Scholar]
- 100.Clark K et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imag 26, 1045–1057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang GA Perspective on deep imaging. IEEE Access 4, 8914–8924 (2016). [Google Scholar]
- 102.Ford RA, Price W & Nicholson II Privacy and accountability in black-box medicine. Mich. Telecomm. Tech. L. Rev 23, 1 (2016). [Google Scholar]
- 103.Selbst AD & Powles J Meaningful information and the right to explanation. Int. Data Privacy Law 7, 233–242 (2017). [Google Scholar]
- 104.Imming P, Sinning C & Meyer A Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov 5, 821–834 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Mehlhorn H et al. in Encyclopedia of Parasitology 3rd edn (ed. Mehlhorn H) 400–402 (Springer, Berlin, Heidelberg, 2008). [Google Scholar]
- 106.Shokri R & Shmatikov V in Proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security 1310–1321 (Denver, CO, USA, 2015). [Google Scholar]
- 107.Phong LT, Aono Y, Hayashi T, Wang L & Moriai S in Applications and Techniques in Information Security. 8th International Conference, ATIS 2017 (eds Batten L, Kim DS, Zhang X & Li G) 719, 100–110 (Auckland, New Zealand, 2017). [Google Scholar]
- 108.McMahan HB, Moore E, Ramage D, Hampson S & y Arcas BA in Proceedings of the 20th International Conference on Artificial Intelligence and Statistics (AISTATS) 1273–1282 (Fort Lauderdale, FL, USA, 2017). [Google Scholar]
- 109.Gilad-Bachrach R et al. in Proceedings of the 33rd International Conference on Machine Learning 201–210 (New York, NY, USA, 2016). [Google Scholar]
- 110.Cahan A & Cimino JJA Learning health care system using computer-aided diagnosis. J. Med. Internet Res 19, e54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 112.Miotto R, Wang F, Wang S, Jiang X & Dudley JT Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinform https://doi.org/10.1093/bib/bbx044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kevin Zhou S, Greenspan H & Shen D Deep Learning for Medical Image Analysis. (Academic Press, 2017). [Google Scholar]
- 114.Shin H-C et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imag 35, 1285–1298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shin Y & Balasingham I in 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 3277–3280 (Jeju Island, Korea, 2017). [DOI] [PubMed] [Google Scholar]
- 116.Orringer DA et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng 1, 0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Albarqouni S et al. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans. Med. Imag 35, 1313–1321 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Djuric U, Zadeh G, Aldape K & Diamandis P Precision histology: how deep learning is poised to revitalize histomorphology for personalized cancer care. Precision Oncol. 1, 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Janowczyk A & Madabhushi A Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inform 7, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bejnordi BE et al. in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 929–932 (Melbourne, Australia, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yuan Y et al. DeepGene: an advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinform. 17, 476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Alipanahi B, Delong A, Weirauch MT & Frey BJ Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]