Abstract
Pestalafuranones A–E (compounds 1–5), five new 2(5H)-furanones, have been isolated from cultures of an isolate of Pestalotiopsis besseyi. The structures of these compounds were elucidated mainly by analysis of their NMR spectroscopic data and HRESIMS experiments. Pestalafuranones A–C (compounds 1–3) displayed weak inhibitory effects against HIV-1 replication in C8166 cells, whereas pestalafuranones D (4) and E (5) showed moderate antifungal activity against the plant pathogens Verticillium dahiae and Alternaria longipes.
Keywords: pestalafuranones A–E, endophytes, Pestalotiopsis besseyi, bioactivity
1. Introduction
Plant endophytic fungi have been demonstrated to be rich sources of bioactive secondary metabolites [1,2], and Pestalotiopsis spp. are especially well-known as producers of bioactive natural products [3,4,5,6]. In the course of our search for new bioactive secondary metabolites from Pestalotiopsis spp., an isolate of P. besseyi (1009) which was grown in a solid-substrate fermentation culture was investigated. The organic solvent extract displayed inhibitory effect on HIV-1 replication in C8166 cells, and antifungal activity against the plant pathogens Verticillium dahiae (CGMCC 3758) and Alternaria longipes (CGMCC 2875). Bioassay-guided fractionation of this extract led to the isolation of five new 2(5H)-furanones namely pestalafuranones A–E (compounds 1–5). Detailed isolation, structure elucidation, and bioactivities of these compounds are presented herein.
2. Results and Discussion
2.1. Structural Elucidation
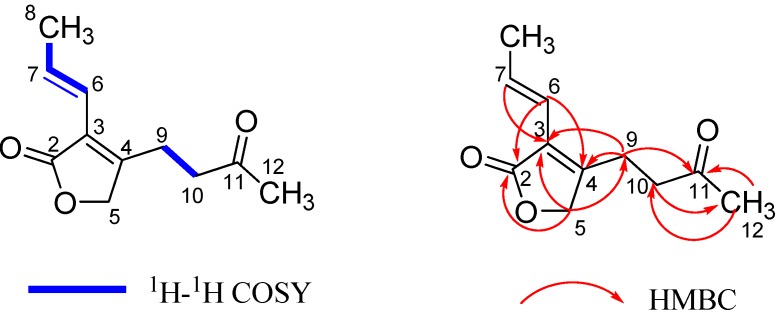
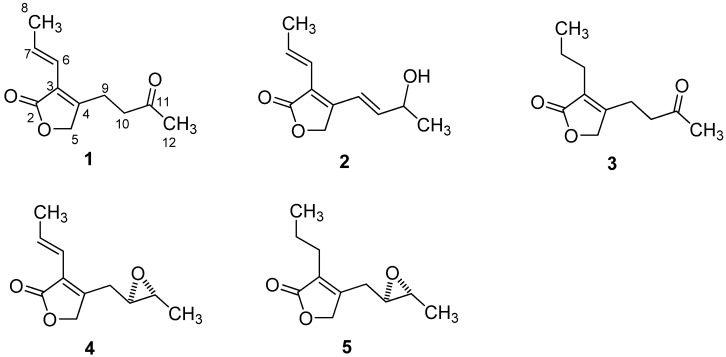
The molecular formula of pestalafuranone A (1) was established as C11H14O3 (five degrees of unsaturation) by analysis of its HRESIMS [m/z 217.0840 (M+Na)+; Δ +0.5 mmu] and NMR data (Table 1 and Table 2). Interpretation of the 1H-, 13C-, and HMQC-NMR spectroscopic data of 1 indicated the presence of two methyl groups, three methylene units (one oxygenated), one disubstituted and one tetrasubstituted olefin, and two carbonyl carbons, leaving one degree of unsaturation and this suggesting 1 to be a monocyclic compound. Analysis of the 1H–1H COSY NMR data led to the identification of two isolated proton spin-systems corresponding to the C-6–C-8 and C-9–C-10 subunits of structure 1. HMBC correlations from H-7 to C-3 and from H-6 to C-2, C-3, and C-4 led to the connection of C-3 to C-6, whereas correlations from H2-10 to C-4, H2-9 to C-3, C-4, C-5, and from the oxygenated methylene protons H2-5 to C-3, C-4 and C-9 indicated that C-3, C-5, and C-9 were all connected to C-4. Key HMBC correlation of H2-5 with C-2 established the α,β-unsaturated γ-lactone subunit. Cross peaks from Me-12 to C-10, C-11 and from CH2-10 to C-11, C-12 in the HMBC spectra (Figure 1) confirmed that C-11 was connected to C-10 and C-12. Thus the structure of 1 was determined as shown in Figure 2.
Table 1.
1H-NMR Data for 1–3, 5 in CDCl3 and 4 in acetone-d6.
| Position | 1 a | 2 a | 3 a | 4 a | 5 a |
|---|---|---|---|---|---|
| 5 | 4.65, s | 4.85, s | 4.63, s | 4.78, s | 4.78, d (17) |
| 4.71, d (17) | |||||
| 6 | 6.10, d (16) | 6.26, d (16) | 2.45, t (7.5) | 6.24, d (16) | 2.61, t (7.5) |
| 7 | 6.86, m | 6.97, m | 1.52, m | 6.85, m | 1.55, m |
| 8 | 1.86, d (7.0) | 1.89, d (6.7) | 0.93, t (7.5) | 1.81, d (6.5) | 0.94, t (7.5) |
| 9 | 2.70, m | 6.77, d (16) | 2.67, br s | 2.90, dd (15, 3.6) | 2.86, dd (12, 4.0) |
| 2.65, dd (15, 6.0) | 2.44, dd (12, 7.0) | ||||
| 10 | 2.70, m | 6.04, dd (16, 7.7) | 2.67, br s | 2.77, m | 2.75, ddd (7.0, 4.0, 2.0) |
| 11 | 4.52, m | 2.85, dq (4.5, 2.0) | 2.81, dq (7.5, 2.0) | ||
| 11-OH | 1.75, br s | ||||
| 12 | 2.18, s | 1.68, d (7.0) | 2.19, s | 1.24, d (4.5) | 0.93, d (7.5) |
a Recorded at 500 MHz. J values are given in Hz.
Table 2.
13C-NMR data for 1–3, 5 in CDCl3 and 4 in acetone-d6.
| Position | 1 a | 2 a | 3 a | 4 a | 5 a | ||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 172.9, qC | 173.0, qC | 177.5, qC | 173.0, qC | 174.5, qC | ||||
| 3 | 123.3, qC | 122.7, qC | 127.9, qC | 123.9, qC | 128.7, qC | ||||
| 4 | 156.6, qC | 149.4, qC | 158.9, qC | 156.3, qC | 158.5, qC | ||||
| 5 | 70.8, CH2 | 68.6, CH2 | 71.2, CH2 | 71.7, CH2 | 71.8, CH2 | ||||
| 6 | 118.1, CH | 118.7, CH | 25.6 CH2 | 119.9, CH | 25.6, CH2 | ||||
| 7 | 133.0, CH | 134.3, CH | 20.6 CH2 | 132.3, CH | 21.4, CH2 | ||||
| 8 | 19.3, CH3 | 19.6, CH3 | 13.9, CH3 | 17.4, CH3 | 13.9, CH3 | ||||
| 9 | 20.4, CH2 | 118.6, CH | 21.3, CH2 | 30.2, CH2 | 29.9 CH2 | ||||
| 10 | 41.0, CH2 | 140.8, CH | 41.3, CH2 | 57.2, CH | 56.8, CH | ||||
| 11 | 206.1, qC | 68.3, CH | 206.0, qC | 54.7, CH | 54.5, CH | ||||
| 12 | 29.8, CH3 | 23.5, CH3 | 29.8, CH3 | 19.1, CH3 | 17.2, CH3 |
a Recorded at 125 MHz.
Figure 1.
1H-1H COSY and Key HMBC correlations for pestalafuranone A (1).
Figure 2.
Structures of compounds 1–5.
The molecular formula of pestalafuranone B (2) was determined to be C11H14O3 by analysis of its HRESIMS (m/z 217.0842 [M+Na]+; Δ +0.7 mmu) and NMR data (Table 1 and Table 2). Analysis of the 1H and 13C-NMR data of 2 revealed the presence of structural features similar to those found in 1, except the signal for the ketone group (δC 206.1) present in 1 was replaced by an oxymethine (δH 4.52; δC 68.3), and the two methylene units CH2-9 and CH2-10 were replaced by two olefinic methines (δH 6.77 and 6.04). The 1H–1H COSY correlations for pestalafuranone B finally confirmed the above observations, leading to the assignment of the planar structure of 2.
Pestalafuranone C (3) was assigned a molecular formula of C11H16O3 (four degrees of unsaturation) on the basis of HRESIMS analysis [m/z 219.0998 (M+Na)+; Δ +0.6 mmu] and NMR data (Table 1 and Table 2), thus having two mass units more than 1. Analysis of the 1H- and 13C-NMR data for 3 indicated that the C-6/C-7 olefinic bond observed in 1 was reduced to give two methylene units, which was further confirmed by analysis of its 1H–1H COSY NMR data.
The molecular formula of pestalafuranone D (4) was established as C11H14O3 (five degrees of unsaturation) by analysis of its HRESIMS [m/z 217.0846 (M+Na)+; Δ +1.1 mmu] and NMR data (Table 1 and Table 2). The 1H- and 13C-NMR data of 4 suggested the presence of the same α,β-unsaturated γ-lactone subunit with a propenyl side chain attached to C-3, as that appearing in compounds 1 and 3. Analysis of the 1H–1H COSY NMR data of 4 led to the identification of the C-9–C-12 partial structure. Considering the 13C chemical shifts for C-10 (δC 57.2) and C-11 (δC 54.7) as well as the unsaturation index, C-10 and C-11 must be attached to the remaining oxygen to form an epoxide moiety. Therefore, the planar structure of pestalafuranone D was elucidated as depicted in 4.
The molecular formula of pestalafuranone E (5) was determined to be C11H16O3 on the basis of HRESIMS analysis [m/z 219.0995 (M+Na)+; Δ +0.3 mmu] and NMR data (Table 1 and Table 2). Examination of the 1H- and 13C-NMR data revealed the close resemblance of 5 to 4, except for the reduction of the C-6/C-7 olefinic bond in 4 giving the structure of 5, which was further supported by relevant 1H–1H COSY correlations.
The C-6/C-7 double bond in compounds 1, 2 and 4, and the C-9/C-10 double bond in 2, were assigned an E-geometry on the basis of the coupling constants observed for the corresponding olefinic protons (J = 16 Hz). The vicinal coupling constant of J = 2.0 Hz between H-10 and H-11 in compounds 4 and 5 suggested a cis relationship between these two protons [7]. Due to the limited amount available of compound 2, its absolute configuration was not determined.
2.2. Anti-HIV Activity
Pestalafuranones A–E (compounds 1–5) were tested for in vivo activity against HIV-1. Pestalafuranones A–C (compounds 1–3) showed weak inhibitory effects on HIV-1 replication in C8166 cells, with EC50 values of 10.52, 24.32 and 36.74 μg/mL, respectively (the EC50 values of positive control zidovudine (AZT) and indinavir sulfate (IDV) were 2.17 ng/mL and 3.92 ng/mL, respectively).
2.3. Antifungal Activity
In the antifungal bioassay, pestalafuranone D (4) showed weak activity against the plant pathogenic fungi Verticillium dahiae (CGMCC 3758) with IC50 values of 24.5 μg/mL, whereas pestalafuranone E (5) displayed moderate activity against Alternaria longipes (CGMCC 2875) with IC50 values of 10.3 μg/mL (positive control fluconazole showed IC50 value of 0.50 and 0.30 μg/mL).
3. Experimental
3.1. General
Optical rotations were measured on a Perkin-Elmer 241 polarimeter, and UV data were recorded on Shimadzu Biospec-1601 spectrophotometer. IR data were recorded using a Nicolet Magna-IR 750 spectrophotometer. 1H- and 13C-NMR data were acquired with Varian Mercury-400/500 spectrometers using solvent signals (CDCl3; δH 7.26/δC 77.7; acetone-d6; δH 2.05/δC 29.8, 206.0) as references. The HMQC and HMBC experiments were optimized for 145.0 and 8.0 Hz, respectively. ESIMS data were recorded on a Bruker Esquire 3000plus spectrometer. HRESI MS data were obtained using a Bruker APEX III 7.0 T spectrometer.
3.2. Fungal Material
The culture of P. besseyi was isolated by Professor Guo from Dongling Mountain, Beijing, in May 2005. The isolate was identified by Prof. Guo, and assigned the accession number 1,009 in his culture collection at the Institute of Microbiology, Chinese Academy of Sciences, Beijing. The isolate was subcultured on PDA slants at 25 °C for 15 days. The agar plugs were used to inoculate 250-mL Erlenmeyer flasks, each containing 50 mL of media (0.4% glucose, 1% malt extract, and 0.4% yeast extract), and the final pH of the media was adjusted to 6.5 before sterilization. Flask cultures were incubated at 25 °C on a rotary shaker at 170 rpm for five days. Twenty 500-mL Erlenmeyer flasks, each containing 150 mL of liquid media (6% dextrin, 2% maltose, 0.75% cotton-seed meal, 0.7% peptone, 0.25% CaCO3, 0.25% gSO4·7H2O, 0.1% FeSO4·7H2O, 0.001% ZnSO4; final pH 6.0) and 30 g of vermiculite were individually inoculated with 15 mL of the seed culture, and incubated at 25 °C for 10 days.
3.3. Extraction and Isolation
The fermented material was freeze-dried and extracted with methyl ethyl ketone (3 × 500 mL), and the organic solvent was evaporated to dryness under vacuum to afford 5.0 g of crude extract. The extract was fractionated by silica gel column chromatography (5 × 40 cm) using CHCl3–CH3OH gradient elution. The fraction (50 mg) that was eluted with 1% MeOH was further separated by semipreparative reversed-phase HPLC (Kramosil C18 column; 10-µm; 10 × 250 mm, 2 mL/min) to afford pestalafuranone B (2; 1.9 mg, tR 15.3 min; 50% MeOH in H2O over 2 min, 50–72% over 10 min, 72–74% over 13 min). The fraction (180 mg) that was eluted with 2% MeOH was then separated by Sephadex LH-20 column chromatography (CH2Cl2–n-C6H14 = 4:1) to afford six fractions. Fraction 3 (60 mg) was successively separated by semipreparative reversed-phase HPLC (32% MeOH in H2O over 2 min, 32–55% over 60 min) afforded pestalafuranones A (1; 1.6 mg, tR 25.7 min), C (3; 6.0 mg, tR 26.6 min), D (4; 1.5 mg, tR 30.0 min), and E (5; 3.5 mg, tR 31.5 min).
Pestalafuranone A (1): colorless oil; UV (CH3OH) λmax 272 (ε 7800), 265 (ε 6600) nm; IR (Neat) νmax 2962, 1748, 1717, 1448, 1364 cm−1; 1H- and 13C-NMR data, see Table 1 and Table 2; HRESIMS obsd. m/z 217.0840 [M+Na]+ (calcd for C11H14O3Na, 217.0835).
Pestalafuranone B (2): pale yellow oil; [α]D −16 (c 0.05, CH3OH); UV (CH3OH) λmax 270 (ε 3300) nm; IR (Neat) νmax 3403 (br), 2932, 1749, 1446 cm−1; 1H- and 13C-NMR data, see Table 1 and Table 2; HRESIMS obsd. m/z 217.0842 [M+Na]+ (calcd for C11H14O3Na, 217.0835).
Pestalafuranone C (3): pale yellow oil; UV (CH3OH) λmax 265 (ε 6600), 235 (ε 4300) nm; IR (Neat) νmax 2919, 1752, 1717, 1445, 1365 cm−1; 1H- and 13C-NMR data, see Table 1 and Table 2; HRESIMS obsd. m/z 219.0998 [M+Na]+ (calcd for C11H16O3Na, 219.0992).
Pestalafuranone D (4): colorless oil; [α]D +32 (c 0.05, CH3OH); UV (CH3OH) λmax 275 (ε 4460) nm; IR (Neat) νmax 2963, 1751, 1450, 1381 cm−1; 1H- and 13C-NMR data, see Table 1 and Table 2; HRESIMS obsd. m/z 217.0846 [M+Na]+ (calcd for C11H14O3Na, 217.0835).
Pestalafuranone D (5): colorless oil; [α]D +16 (c 0.05, CH3OH); UV (CH3OH) λmax 275 (ε 6270) nm; IR (Neat) νmax 2931, 1753, 1446, 1346 cm−1; 1H- and 13C-NMR data, see Table 1 and Table 2; HRESIMS obsd. m/z 219.0995 [M+Na]+ (calcd for C11H16O3Na, 219.0992).
3.4. Anti-HIV Assay
Anti-HIV assays were conducted in triplicate by the following methods: the inhibition of HIV replication, represented by the reduction of p24 antigen was determined by p24 antigen capture ELISA [8]. C8166 cells were exposed to HIV-1LAI (MOI = 0.058) at 37 °C for 1 h, washed with PBS to remove free viruses, and then seeded into a 96-well microtitre plate at 3 × 104 cells per well in the absence or presence of test compounds. AZT and IDV (provided by the NIH AIDS Reagent Program) were used as positive controls. After 4 days, the supernatant was collected and inactivated by 0.5% Triton X-100. The supernatant was diluted three times, added to the plate coating with anti-p24 McAb (provided by Dr. Bin Yan), and incubated at 37 °C for 1 h. After washing 5 times with PBST, the HRP labeled anti-p24 antibody (provided by Dr. Bin Yan) was added and incubated at 37 °C for 1 h. The plate was washed 5 times with PBST, followed by adding OPD reaction mixture and termination buffer. The assay plate was read at 490 nm using a microplate reader within 30 min. The inhibition rate and the EC50 based on p24 antigen expression level were calculated.
3.5. Antifungal Bioassay
Antifungal bioassays were conducted in triplicate by following Clinical and Laboratory Standards Institute (CLSI) recommendations [9]. The plant pathogenic fungal strains were obtained from China General Microbial Culture Collection (CGMCC), and grown on potato dextrose agar (PDA). The fungal inocula were then prepared from broth cultures that were incubated at 25 °C for 48 h, and the final suspensions contained 104 hyphae/mL of corresponding plant pathogenic fungi. Test samples (10 mg/mL as stock solution in DMSO and serial dilutions) were transferred to 96-well clear plate in triplicate, and the suspensions of test organisms were added to each well achieving a final volume of 200 μL (fluconazole was used as positive control; 10 μL of 10% alamar blue solution was added to each well as indicator). After incubation (28 °C for 48 h), the fluorescence intensity was measured at Ex/Em = 544/590 nm. The inhibition rate was calculated and plotted versus test concentrations to afford the IC50 values.
4. Conclusions
This was the first report in which five new furanones pestalafuranones A–E (compounds 1–5), were characterized from the endophytic fungus P. besseyi, which would further provide the evidence that endophytes, inhabiting a unique environment, could produce diverse secondary metabolites with a wide range of bioactivities.
Acknowledgments
We gratefully acknowledged financial support from the National Natural Science Foundation of China (31000011, 31000008, 31101481), and the Chinese National S&T Special Project on Major New Drug Innovation (2011ZX09307-002-01).
Footnotes
Sample Availability: Samples of the compounds are not available from the authors because those compounds were used by bioactivity tests.
References
- 1.Suryanarayanan T.S., Thirunavukkarasu N., Govindarajulu M.B., Sasse F., Jansen R., Murali T.S. Fungal endophytes and bioprospecting. Fungal Biol. Rev. 2009;23:9–19. doi: 10.1016/j.fbr.2009.07.001. [DOI] [Google Scholar]
- 2.Aly A.H., Debbab A., Kjer J., Proksch P. Fungal endophytes from higher plants: A prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 2010;41:1–16. doi: 10.1007/s13225-010-0034-4. [DOI] [Google Scholar]
- 3.Yang X.L., Zhang J.Z., Luo D.Q. The taxonomy, Biology and chemistry of the fungal Pestalotiopsis genus. Nat. Prod. Rep. 2012;29:622–641. doi: 10.1039/c2np00073c. [DOI] [PubMed] [Google Scholar]
- 4.Wang K.W., Lei J.X., Wei J.G., Yao N. Bioactive natural compounds from the plant endophytic fungi Pestalotiopsis genus. Mini Rev. Med. Chem. 2012;12:1382–1393. doi: 10.2174/13895575112091382. [DOI] [PubMed] [Google Scholar]
- 5.Xu J., Ebada S.S., Proksch P. Pestalotiopsis a highly creative genus: Chemistry and bioactivity of secondary metabolites. Fungal Divers. 2010;44:15–31. doi: 10.1007/s13225-010-0055-z. [DOI] [Google Scholar]
- 6.Maharachchikumbura S.S.N., Guo L.D., Chukeatirote E., Bahkali A.H., Hyde K.D. Pestalotiopsis—Morphology, Phylogeny, Biochemistry and diversity. Fungal Divers. 2011;50:167–187. doi: 10.1007/s13225-011-0125-x. [DOI] [Google Scholar]
- 7.Tori M., Arbiyanti H., Taira Z., Asakawa Y. Terpenoids of the liverwort Frullanoides densifolia and Trocholejeunea sandvicensis. Phytochemistry. 1993;32:335–348. doi: 10.1016/S0031-9422(00)94991-4. [DOI] [Google Scholar]
- 8.Zhang G.H., Wang Q., Chen J.J., Zhang X.M., Tam S.C., Zheng Y.T. The anti-HIV-1 effect of scutellarin. Biochem. Biophys. Res. Commun. 2005;334:812–816. doi: 10.1016/j.bbrc.2005.06.166. [DOI] [PubMed] [Google Scholar]
- 9.Espinel-Ingroff A., Fothergill A., Ghannoum M.A., Manavathu E., Ostrosky-Zeichner L., Pfaller M., Rinaldi M., Schell W., Walsh T. Quality control and reference guidelines for CLSI broth micro-dilution method (M38-A document) for susceptibility testing of anidulafungin against moulds. J. Clin. Microbiol. 2007;45:2180–2182. doi: 10.1128/JCM.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]