Abstract
Fine particulate matter (PM2.5) has been associated in humans with inflammation, oxidative stress and cancer. Studies had shown that curcumin could potentially inhibit these effects; however, there had been no in vivo or in vitro reports about the effects of curcumin on organisms exposed to PM2.5. This predictive study explored the possible biological functions and pathways involved in the mechanism of curcumin inhibition of the hazardous effects of PM2.5. For predictive analysis, microarray data were used to investigate the effect of PM2.5 on human bronchial epithelial cells (HBEC), and human target proteins of curcumin were retrieved from PubChem. Two protein-protein interaction (PPI) networks were established based upon differential genes and target proteins, respectively, and the common network of these two networks was found. Functional and pathway analysis of the common network was performed using the Ingenuity Pathways Analysis (IPA) software. The results suggested that the predictive effects of curcumin on HBEC exposed to PM2.5 were involved in bio-functions, including inflammatory response of airway, cancerogenesis, and apoptosis, and in pathways such as cancer, glucocorticoid receptor signaling, and NF-kappaB signaling. This study predicted for the first time that curcumin could be a potential therapeutic agent for protecting the human airway from the hazardous effects of PM2.5.
Keywords: curcumin, fine particulate matter, protein-protein interaction network, bioinformatics prediction, pathway analysis
1. Introduction
Air pollution had long been considered a hazard to human health. Ambient airborne particulate matter (PM), an important environmental pollutant, had been associated with multiple cardiopulmonary diseases and cancers [1]. In the past few decades, many studies had highlighted the role of the size and surface area of PM in determining the potential to elicit inflammatory injury, oxidative damage, and other biological effects [2]. These effects were stronger for fine particles (diameter < 2.5 μm, known as PM2.5), because they could penetrate deeper into the airways of the respiratory tract and reach the alveoli, where 50% of the PM2.5 were retained in the lung parenchyma [3]. In recent years, the hazardous effects of PM2.5 had captured more and more public attention. However, do we have other methods to protect us from the hazardous of PM2.5 in addition to reducing the discharge of PM2.5 into the atmosphere? Furthermore, can certain food or herbal additives intake actively defend the body against the damaging effects of PM2.5?
Curcumin, a yellow pigment extracted from the rhizome of the plant Curcuma longa (turmeric), had been widely used as a spice, food additive, and herbal medicine in Asia [4]. In recent years, extensive in vitro and in vivo studies had suggested that curcumin had anticancer, antiviral, antiarthritic, anti-amyloid, antioxidant, anti-inflammatory, and anti-aging properties [5]. Interestingly, these therapeutic effects of curcumin were in direct opposition to the detrimental effects of PM2.5. Therefore, we speculated that curcumin as a therapeutic agent might control or decrease the damage induced by PM2.5. In the present study, we predicted the underlying protective mechanism of curcumin on human airway epithelial cells (HBEC) exposed to PM2.5 based on gene expression profiling in Gene Expression Omnibus (GEO) and target protein data in PubChem.
2. Results and Discussion
2.1. Results
Using a t-test, we identified 89 genes differentially expressed between HBEC exposed to PM2.5 and vehicle control (Table 1). These genes could clearly distinguish primary HBEC exposed to PM2.5 from the HBEC in control. Of the 89 genes, 38 genes were significantly up-regulated and 51 genes were remarkably down-regulated.
Table 1.
Differentially expressed genes in HBEC exposed to PM2.5 versus control.
| Probe Set ID | RefSeq ID | Gene Symbol | p-value | Fold Change | Regulation |
|---|---|---|---|---|---|
| 203665_at | NM_002133 | HMOX1 | 0.0045 | 24.58 | up |
| 209921_at | NM_014331 | SLC7A11 | 0.0004 | 10.45 | up |
| 202436_s_at | NM_000104 | CYP1B1 | 0.0015 | 7.02 | up |
| 201266_at | NM_003330 | TXNRD1 | 0.0014 | 6.96 | up |
| 205749_at | NM_000499 | CYP1A1 | 0.0185 | 5.54 | up |
| 203925_at | NM_002061 | GCLM | 0.0006 | 3.71 | up |
| 202923_s_at | NM_001498 | GCLC | 0.0110 | 3.29 | up |
| 201468_s_at | NM_000903 | NQO1 | 0.0139 | 2.92 | up |
| 204151_x_at | NM_001353 | AKR1C1 | 0.0042 | 2.91 | up |
| 206172_at | NM_000640 | IL13RA2 | 0.0110 | 2.72 | up |
| 211653_x_at | NM_001354 | AKR1C2 | 0.0083 | 2.70 | up |
| 209387_s_at | NM_014220 | TM4SF1 | 0.0472 | 2.47 | up |
| 210845_s_at | NM_002659 | PLAUR | 0.0104 | 2.18 | up |
| 206683_at | NM_003447 | ZNF165 | 0.0090 | 2.14 | up |
| 212907_at | NM_021194 | SLC30A1 | 0.0357 | 2.14 | up |
| 214211_at | NM_002032 | FTH1 | 0.0270 | 2.08 | up |
| 208963_x_at | NM_013402 | FADS1 | 0.0202 | 2.03 | up |
| 205767_at | NM_001432 | EREG | 0.0160 | 1.98 | up |
| 219475_at | NM_182981 | OSGIN1 | 0.0103 | 1.98 | up |
| 207675_x_at | NM_057091 | ARTN | 0.0313 | 1.97 | up |
| 202842_s_at | NM_012328 | DNAJB9 | 0.0309 | 1.96 | up |
| 202266_at | NM_016614 | TDP2 | 0.0001 | 1.95 | up |
| 201625_s_at | NM_005542 | INSIG1 | 0.0446 | 1.93 | up |
| 209882_at | NM_006912 | RIT1 | 0.0114 | 1.93 | up |
| 201489_at | NM_005729 | PPIF | 0.0093 | 1.92 | up |
| 213112_s_at | NM_003900 | SQSTM1 | 0.0191 | 1.91 | up |
| 204420_at | NM_005438 | FOSL1 | 0.0323 | 1.80 | up |
| 202284_s_at | NM_000389 | CDKN1A | 0.0324 | 1.77 | up |
| 206907_at | NM_003811 | TNFSF9 | 0.0032 | 1.74 | up |
| 219697_at | NM_006043 | HS3ST2 | 0.0291 | 1.72 | up |
| 204970_s_at | NM_002359 | MAFG | 0.0032 | 1.69 | up |
| 213187_x_at | NM_000146 | FTL | 0.0471 | 1.68 | up |
| 212717_at | NM_014798 | PLEKHM1 | 0.0319 | 1.66 | up |
| 206498_at | NM_000275 | OCA2 | 0.0221 | 1.66 | up |
| 202672_s_at | NM_001674 | ATF3 | 0.0041 | 1.57 | up |
| 202021_x_at | NM_005801 | EIF1 | 0.0460 | 1.55 | up |
| 202067_s_at | NM_000527 | LDLR | 0.0128 | 1.54 | up |
| 204958_at | NM_004073 | PLK3 | 0.0153 | 1.50 | up |
| 202207_at | NM_005737 | ARL4C | 0.0139 | 3.24 | down |
| 202887_s_at | NM_019058 | DDIT4 | 0.0123 | 2.66 | down |
| 201890_at | NM_001034 | RRM2 | 0.0097 | 2.26 | down |
| 211450_s_at | NM_000179 | MSH6 | 0.0486 | 2.26 | down |
| 201849_at | NM_004052 | BNIP3 | 0.0293 | 2.20 | down |
| 219250_s_at | NM_013281 | FLRT3 | 0.0425 | 2.10 | down |
| 209120_at | NM_021005 | NR2F2 | 0.0056 | 2.01 | down |
| 202464_s_at | NM_004566 | PFKFB3 | 0.0289 | 1.99 | down |
| 208808_s_at | NM_002129 | HMGB2 | 0.0462 | 1.99 | down |
| 203344_s_at | NM_002894 | RBBP8 | 0.0280 | 1.97 | down |
| 218718_at | NM_016205 | PDGFC | 0.0044 | 1.97 | down |
| 207173_x_at | NM_001797 | CDH11 | 0.0432 | 1.95 | down |
| 201669_s_at | NM_002356 | MARCKS | 0.0448 | 1.92 | down |
| 207826_s_at | NM_002167 | ID3 | 0.0285 | 1.84 | down |
| 204967_at | NM_001649 | SHROOM2 | 0.0141 | 1.80 | down |
| 202628_s_at | NM_000602 | SERPINE1 | 0.0486 | 1.77 | down |
| 212599_at | NM_015570 | AUTS2 | 0.0053 | 1.77 | down |
| 203274_at | NM_012151 | F8A1 | 0.0118 | 1.76 | down |
| 208673_s_at | NM_003017 | SRSF3 | 0.0188 | 1.76 | down |
| 203476_at | NM_006670 | TPBG | 0.0400 | 1.75 | down |
| 209189_at | NM_005252 | FOS | 0.0366 | 1.72 | down |
| 209784_s_at | NM_002226 | JAG2 | 0.0032 | 1.70 | down |
| 203625_x_at | NM_005983 | SKP2 | 0.0040 | 1.70 | down |
| 222036_s_at | NM_005914 | MCM4 | 0.0439 | 1.66 | down |
| 202219_at | NM_005629 | SLC6A8 | 0.0253 | 1.65 | down |
| 205449_at | NM_013299 | SAC3D1 | 0.0328 | 1.65 | down |
| 212168_at | NM_006047 | RBM12 | 0.0031 | 1.64 | down |
| 209286_at | NM_006449 | CDC42EP3 | 0.0060 | 1.63 | down |
| 204334_at | NM_003709 | KLF7 | 0.0105 | 1.63 | down |
| 208579_x_at | NM_017445 | H2BFS | 0.0173 | 1.62 | down |
| 204069_at | NM_002398 | MEIS1 | 0.0281 | 1.60 | down |
| 203797_at | NM_003385 | VSNL1 | 0.0172 | 1.58 | down |
| 203764_at | NM_014750 | DLGAP5 | 0.0181 | 1.58 | down |
| 213051_at | NM_020119 | ZC3HAV1 | 0.0104 | 1.58 | down |
| 208051_s_at | NM_006451 | PAIP1 | 0.0321 | 1.57 | down |
| 203405_at | NM_003720 | PSMG1 | 0.0304 | 1.57 | down |
| 211744_s_at | NM_001779 | CD58 | 0.0273 | 1.57 | down |
| 206277_at | NM_002564 | P2RY2 | 0.0179 | 1.56 | down |
| 204715_at | NM_015368 | PANX1 | 0.0375 | 1.56 | down |
| 201312_s_at | NM_003022 | SH3BGRL | 0.0383 | 1.55 | down |
| 213088_s_at | NM_015190 | DNAJC9 | 0.0253 | 1.55 | down |
| 203803_at | NM_016297 | PCYOX1 | 0.0350 | 1.54 | down |
| 201624_at | NM_001349 | DARS | 0.0225 | 1.54 | down |
| 214214_s_at | NM_001212 | C1QBP | 0.0468 | 1.54 | down |
| 212320_at | NM_178014 | TUBB | 0.0185 | 1.53 | down |
| 208405_s_at | NM_006016 | CD164 | 0.0465 | 1.51 | down |
| 213019_at | NM_012416 | RANBP6 | 0.0002 | 1.51 | down |
| 212922_s_at | NM_020197 | SMYD2 | 0.0002 | 1.50 | down |
| 209025_s_at | NM_006372 | SYNCRIP | 0.0481 | 1.50 | down |
| 201163_s_at | NM_001553 | IGFBP7 | 0.0458 | 1.50 | down |
| 214800_x_at | NM_001207 | BTF3 | 0.0036 | 1.50 | down |
57 human target proteins of curcumin (CID: 969516) were obtained from the PubChem database by PubChem Promiscuity online and identified by UniProt protein IDs (Table 2).
Table 2.
Human target proteins of curcumin in PubChem.
| GI | UniProtKB ID |
|---|---|
| 4507949 | 1433B_HUMAN |
| 31542303 | ABHD5_HUMAN |
| 37622910 | ACM1_HUMAN |
| 21361176 | AL1A1_HUMAN |
| 4885057 | APJ_HUMAN |
| 47132611 | ATG4B_HUMAN |
| 6683500 | BAZ2B_HUMAN |
| 53832009 | CAC1H_HUMAN |
| 4502601 | CBR3_HUMAN |
| 37187860 | CCR6_HUMAN |
| 67551261 | CLK1_HUMAN |
| 153791372 | CLK3_HUMAN |
| 13435386 | CP3A4_HUMAN |
| 32307159 | CRFR2_HUMAN |
| 30219 | CRHBP_HUMAN |
| 4503383 | DRD1_HUMAN |
| 4503385 | DRD2_HUMAN |
| 10835013 | ESR2_HUMAN |
| 4885263 | GEM_HUMAN |
| 122921310 | HCD2_HUMAN |
| 155969707 | IDE_HUMAN |
| 98986450 | KC1G1_HUMAN |
| 153791733 | KC1G2_HUMAN |
| 325651834 | KCNH2_HUMAN |
| 221046486 | KD4DL_HUMAN |
| 22035600 | M4K2_HUMAN |
| 11386165 | MCL1_HUMAN |
| 89993689 | MDM2_HUMAN |
| 88702791 | MDM4_HUMAN |
| 20986531 | MK01_HUMAN |
| 4505209 | MMP13_HUMAN |
| 66911845 | MRGX1_HUMAN |
| 34577122 | NFKB1_HUMAN |
| 222080095 | OX1R_HUMAN |
| 32307152 | OXYR_HUMAN |
| 4505587 | PA1B3_HUMAN |
| 5031975 | PAK4_HUMAN |
| 31881630 | PE2R2_HUMAN |
| 31542939 | PGDH_HUMAN |
| 4505811 | PIM1_HUMAN |
| 42821112 | PIM2_HUMAN |
| 223718196 | PLIN1_HUMAN |
| 116734717 | PPBT_HUMAN |
| 4826962 | RAC3_HUMAN |
| 41281453 | SLK_HUMAN |
| 23943882 | STK33_HUMAN |
| 8400711 | TAU_HUMAN |
| 223468676 | TF65_HUMAN |
| 4507533 | TLR4_HUMAN |
| 8394456 | TLR9_HUMAN |
| 4507615 | TNNC1_HUMAN |
| 151101270 | TNNI3_HUMAN |
| 48255881 | TNNT2_HUMAN |
| 4507681 | TRFR_HUMAN |
| 118600387 | UBP1_HUMAN |
| 4502331 | V1AR_HUMAN |
| 4507883 | VDR_HUMAN |
Based on the differentially expressed genes in Table 1 and human target proteins in Table 2, two biological networks showing protein-protein interactions were constructed. The two protein-protein interaction (PPI) networks were visualized using Cytoscape. The nodes represented proteins in the PPI network and the edges represented the biological relationship between two nodes. There were 1,962 nodes and 15,455 edges in the PPI network of HBEC exposed to PM2.5 (Supplementary Material Figure S1), and 1,284 nodes and 11,541 edges in the PPI network of human target proteins of curcumin (Supplementary Material Figure S2). Appling the function “Intersection” of the Advanced Network Merge plugin in Cytoscape, we found the common proteins and relationships (common network) in the two PPI networks. The common network had 1,197 nodes and 9,521 edges (Figure 1).
Figure 1.
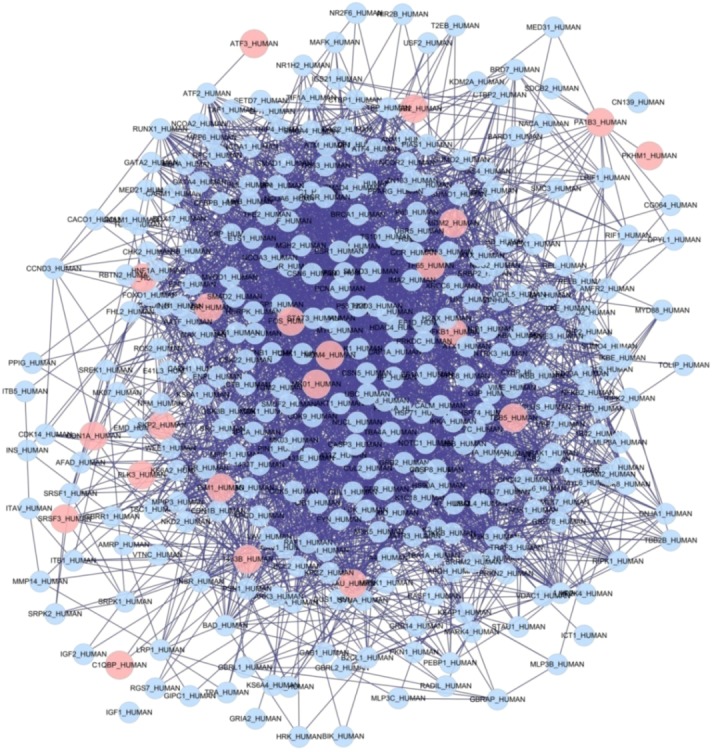
Common network of two PPI networks based on differentially expressed genes of HBEC exposed to PM2.5 and human target proteins of curcumin. Red cycles represent seed nodes, and blue cycles represent neighbor nodes. All edges represent interactions between the nodes.
The top five functions of the common network and the number of proteins associated with each function were found using Ingenuity Pathways Analysis (IPA). The most significant biological functions were grouped into three categories: (1) Diseases and Disorders, (2) Molecular and Cellular Functions, and (3) Physiological System Development and Function (Table 3).
Table 3.
Key functions associated with the common network using IPA.
| Top Bio Functions | p-value | Number of Molecules |
|---|---|---|
| Diseases and Disorders | ||
| Infectious Disease | 1.26E−12–4.25E−02 | 35 |
| Cancer | 3.45E–3.01E−02 | 8 |
| Genetic Disorder | 1.33E–3.01E−02 | 5 |
| Respiratory Disease | 1.33E–3.01E−02 | 6 |
| Inflammatory Response | 2.79E–2.79E−02 | 1 |
| Molecular and Cellular Functions | ||
| Cell Death | 9.91E−20–3.01E−02 | 31 |
| Cellular Growth and Proliferation | 5.64E−15–2.79E−02 | 32 |
| Cellular Development | 1.56E−08–2.79E−02 | 17 |
| Cell Cycle | 1.84E−07–2.79E−02 | 12 |
| Cellular Movement | 1.01E−04–2.79E−02 | 10 |
| Physiological System Development and Function | ||
| Organismal Survival | 2.02E−03–2.02E−03 | 4 |
| Respiratory System Development and Function | 2.28E−03–2.28E−03 | 2 |
| Tissue Development | 2.28E−03–2.79E−02 | 2 |
| Connective Tissue Development and Function | 1.94E−02–1.94E−02 | 2 |
| Tissue Morphology | 2.79E−02–2.79E−02 | 1 |
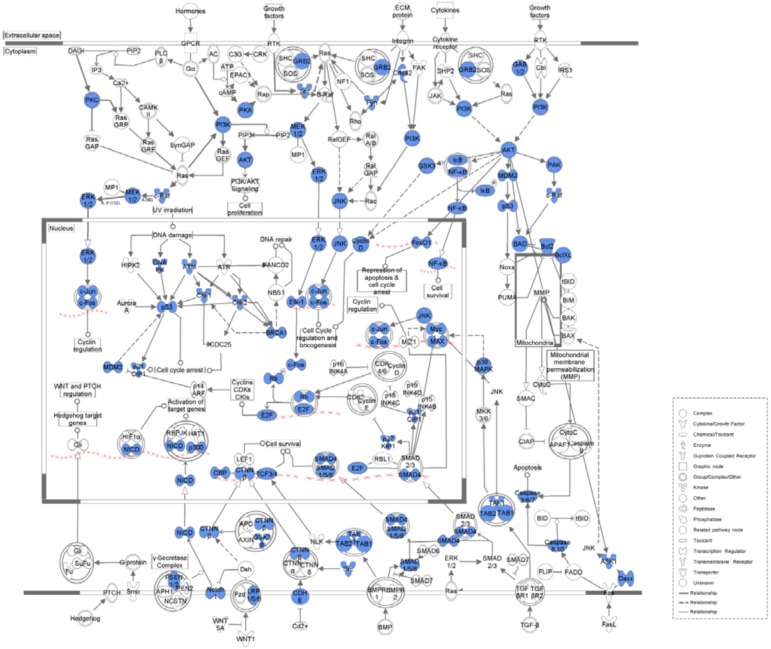
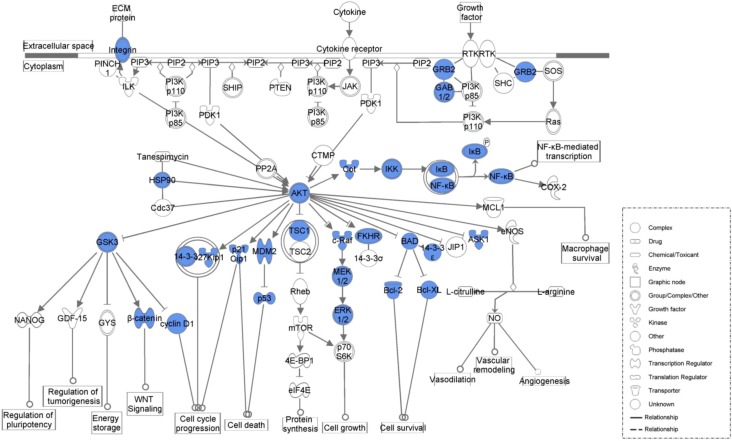
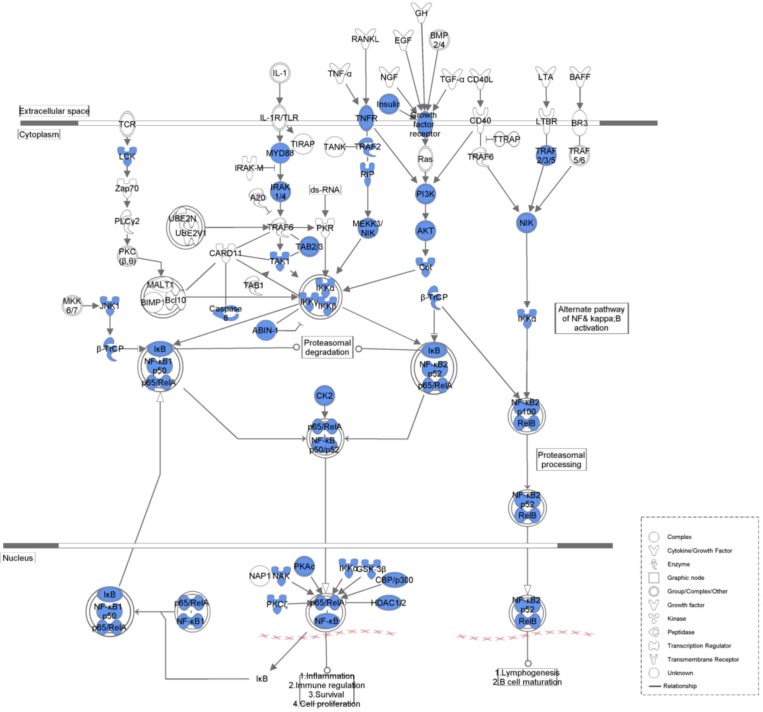
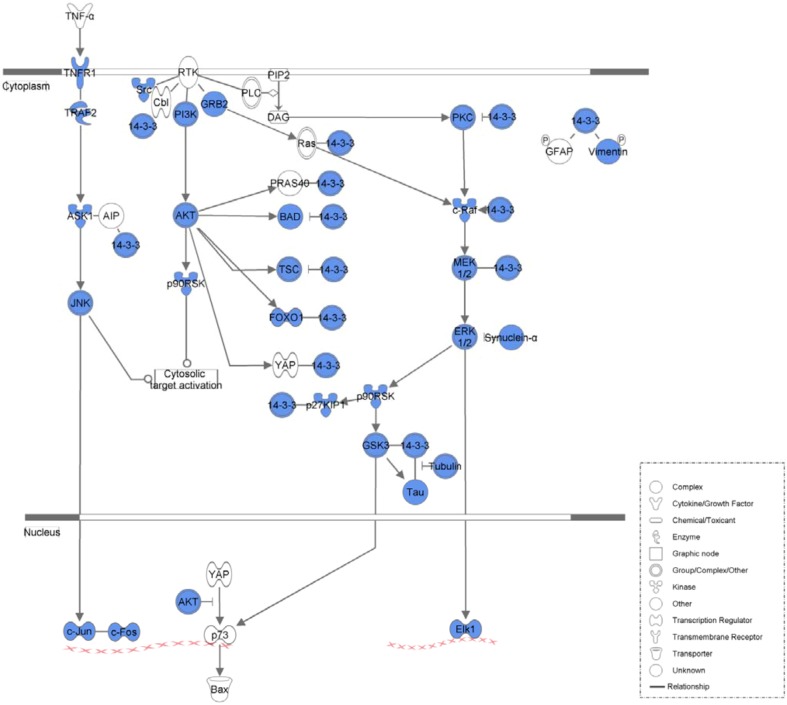
Table 4 lists the top five canonical pathways associated with the common network as calculated by IPA (Figure 2, Figure 3, Figure 4 and Figure 5, Supplementary Material 3). Calculation was either according to ratio (the number of genes from the data set that map to the canonical pathway in question divided by the total number of proteins that map to the same canonical pathway) or significance (Fischer’s exact test was used to calculate a P-value determining the probability that the association between the proteins in the dataset and the canonical pathway was explained by chance alone).
Table 4.
Key canonical pathways associated with the common network using IPA.
| Canonical Pathways | p-value | Ratio |
|---|---|---|
| Glucocorticoid Receptor Signaling | 2.57E−42 | 61/238 (0.256) |
| Molecular Mechanisms of Cancer | 6.68E−39 | 65/314 (0.207) |
| PI3K/AKT Signaling | 6.87E−36 | 41/110 (0.373) |
| NF-kappaB Signaling | 1.33E−30 | 41/143 (0.287) |
| 14-3-3-mediated Signaling | 1.37E−30 | 36/102 (0.353) |
Figure 2.
Molecular mechanisms of cancer associated with the common network. Blue legends represent proteins contained in the common network.
Figure 3.
PI3K/AKT signaling associated with the common network. Blue legends represent proteins contained in the common network.
Figure 4.
NF-kappaB signaling associated with the common network. Blue legends represent proteins contained in the common network.
Figure 5.
14-3-3-mediated signaling associated with the common network. Blue legends represent proteins contained in the common network.
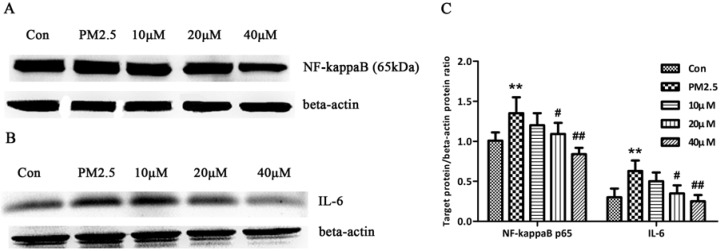
To partially validate the pathways listed in Table 4, we measured the expression of NF-kappaB p65 and IL-6 in human bronchial epithelial cells (16HBE) exposed to PM2.5. 16HBE were pre−treated with 10, 20, 40 μM curcumin for 30 min followed by exposure to PM2.5 (250 μg/mL) for 24 h in the presence or absence of curcumin. After 24 h, cells were collected and measured for NF-kappaB p65 and IL-6 expression by Western blot. Notably, NF-kappaB p65 or IL-6 expression level was markedly increased in 16HBE exposed to PM2.5 as compared with the control cells. However, curcumin treatment could attenuate the high expression of NF-kappaB p65 or IL-6 in cells induced by PM2.5 (Figure 6).
Figure 6.
The effect of curcumin on the NF-kappaB p65 and IL-6 of 16HBE exposed to PM2.5. Cells were pre−treated with 10, 20, 40 μM curcumin for 30 min followed by exposure to PM2.5 (250 μg/mL) for 24 h in the presence or absence of curcumin. After 24 h, cells were collected and measured for NF-kappaB p65 and IL-6 expression by Western blot. (A) Expression of NF-kappaB p65. (B) Expression of IL-6. (C) Bar graphs showing the quantification of Western blot bands. Beta-actin was used as an internal control. ** p < 0.01, compared with the control group, # p < 0.05, ## p < 0.01, compared with the PM2.5 group.
2.2. Discussion
Predictive analysis was a general method for predicting the accuracy of quantitative experiments. The use of predictive analysis allowed the designer of an experiment to estimate the accuracy that should be obtained from the experiment before the experimental setup was finalized [6]. Until now, there had been no in vivo or in vitro reports about the effects of curcumin on organisms exposed to PM2.5; therefore, we collected limited data associated with PM2.5 or curcumin available from online databases such as GEO and PubChem. Because the aim of our study was to outline the potential biofunctions and pathways associated with the effect of curcumin on HBEC exposed to PM2.5 predictively, we did not restrict all data reanalyzed at identical molecular level.
PubChem [7] is a public repository for biological properties of small molecules hosted by the US National Institutes of Health (NIH). The PubChem BioAssay database contained biological test results for more than 700,000 compounds. From the PubChem BioAssay database, we could retrieve the target proteins of compounds [8]. In our study, 57 human target proteins of curcumin (CID: 969516) were obtained.
PPI were extremely important cellular events that affected many of the most important molecular processes in the cell, such as DNA replication. They formed the basis for many signal transduction pathways and transcriptional regulatory networks. The availability of complete and annotated genome sequences of several organisms had led to a paradigm shift from the study of individual proteins in a cell to proteome−wide analysis in an organism. The whole proteome analysis had illustrated that PPI affected cellular biological functions through many orchestrating networks such as metabolic, signaling and regulatory pathways in an organism [9].
Within the airway, the epithelium forms the mucosal immune barrier, the first structural cell defense against common environmental insults such as microorganisms and particulate matter. Hence, respiratory infectious diseases share similar pathologic processes such as the inflammatory response or oxidative stress with bronchial diseases induced by PM2.5 [10,11,12]. The inflammatory response was the main acute effect induced by PM2.5 in the respiratory tract, a target organ of PM2.5. In vitro studies had shown that airway epithelial cells responded to PM2.5 exposure by the release of inflammatory cytokines such as IL-1beta, TNF-alpha, and IL-6 [13], chemokines such as IL-8 [14], and erythropoietic cytokines such as G-CSF and GM-CSF [15,16]. Because curcumin was observed to inhibit secretion of the pro-inflammatory cytokines NF-kappaB mediating in HBEC exposed to pollutants [17,18], we predicted that curcumin might also have an anti-inflammatory effect on HBEC exposed to PM2.5.
Some researchers conducting large epidemiological cohort studies in the United States and Europe had comfirmed the relationship between long-term exposure to particulate air pollution (PM10 and PM2.5) and increased mortality from lung cancer, especially in combination with other known risk factors, such as smoking, passive smoking, and occupational exposure [19,20]. By contrast, curcumin, a natural antitumor compound, had been shown to have the effect of inhibiting lung cancer cell invasion and metastasis in several studies [21,22,23] and have promising potential as a diet-derived cancer chemopreventive agent [24]. Thus, we inferred that curcumin could inhibit the carcinogenesis of airway epithelial cells resulting from PM2.5 exposure.
Generally, PM2.5 led to the proliferation inhibition and apoptosis of HBEC [25,26,27]. PM2.5 could induce cell cycle arrest in G1 phase, inhibit DNA synthesis, and block airway epithelial cell proliferation [28]. The P53 pathway, tumor necrosis factor-alpha (TNF-alpha) pathway, and mitochondrial pathway played critical roles in the apoptosis processes induced by PM2.5 [29,30]. However, as a dietary antioxidant, curcumin had been proven to have preventive potential against apoptosis induced by peroxide or cigarette smoke extract in HBEC though inhibition of NF-kappaB [17,31]. Moreover, curcumin was a selective apoptosis modulator. For most noncancerous cells, curcumin was a protector and prevents cells from apoptosis induced by various adverse factors, but for cancer cells, curcumin was a killer and arrested cell cycle, inhibited cell proliferation, and/or caused apoptosis. For example, when mammary epithelial cells and breast cancer cells accumulated a similar amount of curcumin, a significantly higher percentage of apoptotic cells was induced in cancer cells compared to epithelial cells [32]. Similarly, we speculated that curcumin might have a two-way regulating effect on HBEC when exposed to PM2.5.
Glucocorticoids (GCs) could control airway inflammation in respiratory diseases such as chronic obstructive pulmonary disease (COPD) and asthma, and the airway epithelium was a primary target of GC anti-inflammatory actions [33]. GC effects were mediated through the GC receptor (GR). Previous studies had indicated that cultured HBEC from smokers possess GRs with a lower binding affinity, and this might result from the inflammation found in the airways in smokers [34]. Although there had been no studies involving the effect of PM2.5 on GRs in HBEC, cigarette smoke and PM2.5 shared a similar inflammatory effect on HBEC, and we could speculate that PM2.5 might decrease GR signaling. In addition, GR action was shown to be tightly regulated by histone deacetylase 2 (HDAC2), which suppressed inflammatory gene expression in inflammatory airway disease [35]. Acting as an HDAC activator, curcumin was found to restore HDAC2 activity, thereby restoring the function of the GR [36]. In summary, regulation of the GR pathway was a possible mechanism by which curcumin inhibits the hazardous effects of PM2.5.
Recent studies have suggested that numerous components of phosphoinositide 3-kinase (PI3K)-dependent signaling, mediated by Akt kinase, played a crucial role in the expression and activation of inflammatory mediators, inflammatory cell recruitment, immune cell function, airway remodeling, and corticosteroid insensitivity in chronic inflammatory respiratory disease [37], especially in COPD and asthma [38]. PM2.5 or cigarette smoke could induce activation of the PI3K/Akt pathway in HBEC and promote transcription of downstream inflammatory mediators [39,40,41]. However, studies had proved that curcumin could inhibit PI3K/Akt/NF-kappaB signals in human lung epithelial cells [42], block Akt translocation to the nucleus and further decrease inflammation in human tracheal smooth muscle cells [43]. Therefore, we predicted that curcumin also might have potential to prevent HBEC from the toxicity effects of PM2.5 by modulating PI3K/Akt signaling.
Recent research indicated that the NF-kappaB/IkappaB pathway played an important role in the inflammatory response induced by PM2.5 in the lung [44]. The activation of the NF-kappaB/IkappaB complex preceded cytotoxicity or inflammation in PM2.5-exposed human bronchial or lung epithelial cells through the reactive oxygen species (ROS)-dependent NF-kappaB pathway [45,46]. As an inhibitor of NF-kappaB, curcumin exhibited a potent anti-inflammatory effect, and could decrease the airway epithelial cell inflammatory cytokine response to the pollutant cadmium or cigarette smoke extract [17,18]. Like cadmium and cigarette smoke, PM2.5 was also a pollutant in the environment, so we hypothesized that curcumin might perform its anti-inflammatory effect on PM2.5 by inhibiting the NF-kappaB pathway.
14-3-3 family members tightly regulated cell fate through interaction with a wide spectrum of proteins that were targeted by various classes of protein kinases [47]. 14-3-3 proteins played particularly important roles in coordinating the progression of cells through the cell cycle, regulating their response to DNA damage and influencing life−death decisions [48]. Studies reported that 14-3-3 might contribute to lung tumorigenesis. In H322 cells, over-expression of 14-3-3 protein resulted in abnormal DNA replication and polyploidization [49], and in A549 cells, 14-3-3 promoted cellular proliferation [50]. Other studies found that curcumin could induce the typical features of apoptosis and inhibited the expression of 14-3-3 in HT-29 cells [51]. Based on the evidence mentioned above, we predicted that curcumin might prevent HBEC exposed to PM2.5 from carcinogenesis by inhibiting the 14-3-3 pathway.
In the NF-kappaB signaling pathway, NF-kappaB played a pivotal role as inflammatory response regulator, and IL-6 was an important inflammatory factor regulated by NF-kappaB and caused the damage response of PM2.5 [52]. Therefore, in a validating experiment, we selected NF-kappaB p65 and IL-6 as validated molecules and found that curcumin treatment could attenuate the high expression of NF-kappaB p65 or IL-6 in cells induced by PM2.5. These results supported our part prediction.
3. Experimental
3.1. Microarray Data Analysis
A microarray dataset (accession number GSE7010) [53] was downloaded from the GEO [54], and analyzed it based on the Affymetrix Human Genome U133A Array. This dataset was derived from a study observing global gene expression in HBEC and identifying cellular pathways associated with coarse, fine and ultrafine particulate matter exposure. Ambient PM was collected in three different size fractions from Chapel Hill air; particles were extracted from foam or filter matrices and lyophilized. Primary HBEC were exposed to PM2.5 at 250 μg/mL or vehicle control for 6 h in culture [55]. In this study, we used three samples from the control group (GSM161787, GSM161793, GSM161798) and three samples from the fine particulate matter (PM2.5) group (GSM161790, GSM161796, GSM161801). Probes showing differential expression were extracted by volcano plot analysis with the filtering criteria of a 1.5-fold change using GeneSpring GX version 11.0 after per chip and per gene normalization.
3.2. Target Proteins of Curcumin
The human target proteins of curcumin (CID: 969516) in PubChem [56] were retrieved using PubChem Promiscuity [57] online [58] with the filtering criteria of “not less than one Active Bioassay”.
3.3. Construction of PPI Networks and Detection of Common Network
PPI represented a basic blueprint for the analysis of self-organization and homeostasis in living organisms [59]. In this study, a Cytoscape [60] plugin, BisoGenet [61], was applied for assembling the PPI network. Information on human PPI networks involving relevant genes was obtained from various databases, including HPRD (Human Protein Reference Database), BIND (Biomolecular Interaction Network Database), BioGRID (The General Repository for Interaction Datasets), DIP (Database of Interacting Proteins), IntAct (Database system and analysis tools for protein interaction data), and MINT (Molecular Interactions Database). Two PPI networks were constructed based on the differential expression of genes from microarray data analysis and the target proteins of curcumin from PubChem. Another Cytoscape plugin, Advanced Network Merge, was used to find the common proteins and relations (common network) in the two PPI networks.
3.4. Functional and Pathway Analysis of Common Network
For further analysis, a data file was uploaded into IPA (Ingenuity® Systems, www.ingenuity.com, Redwood City, CA, USA). This file contained the proteins in the common network. Each protein identifier was mapped to its corresponding protein object in the Ingenuity Pathways Knowledge Base (IPKB). The functional analysis identified the biological functions and/or diseases that were most significant to the data set. Proteins from the data set that met the P-value threshold of 0.05 (Fisher’s exact test) and were associated with biological functions and/or diseases in the IPKB were kept for analysis. Canonical pathway analysis identified the pathways most significant to the data set, based on two parameters: (1) a ratio of the number of proteins from the data set that map to the pathway divided by the total number of proteins that map to the canonical pathway and (2) a p-value calculated with Fisher’s exact test determining the probability that the association between the proteins in the dataset and the canonical pathway is explained by chance alone.
3.5. Validating Experiment
3.5.1. Chemicals
All reagents used in this validating experiment including curcumin (purity: 70%) were purchased from Sigma (Sigma-Aldrich, St. Louis, MO, USA) unless specified.
3.5.2. Cell Culture
Human bronchial epithelial cells 16HBE were purchased from American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained at 37 °C and 5% CO2 in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum, 10 U/mL of penicillin and 10 U/mL of streptomycin.
3.5.3. Preparation of Particles
Urban atmospheric PM2.5 was kindly provided by Prof. Xiaohong Zhao of College of Arts and Sciences of Beijing Union University. PM2.5 was collected on 150 mm diameter nitrocellulose filters (HAWP, Sartorius, La Fert’esous-Jouarre, France) with a high volume sampler machine (DA-80 Digitel, Cugy, Switzerland, flowrate: 30 m3/h) during the winter of 2008 on the roof of a five−story building in Xueyuan Road, Haidian District, Beijing. Particles were processed as previously described [55].
3.5.4. Treatment of Cells with Curcumin and PM2.5
The cells were pretreated with curcumin (10, 20, 40 μM) for 30 min followed by exposure to PM2.5 (250 μg/mL) for 24 h in the presence or absence of curcumin. After 24 h, total cell lysates were prepared and 30 μg protein was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblot analysis.
3.5.5. Western Blot
Rabbit polyclonal anti-NF-kappaB p65, anti-IL-6 antibody and mouse monoclonal anti-beta-actin antibody were purchased from Cell Signaling Technology, Abcam Inc. and Applygen Technologies Inc., respectively. Goat anti-rabbit horseradish peroxidase−conjugated immunoglobulin G (IgG-HRP; Santa Cruz Biotechnology) and goat anti-mouse IgG-HRP (Santa Cruz Biotechnology) were used as secondary antibodies for the rabbit and mouse primary antibodies, respectively. Western blot was performed following the standard protocol. Precision Plus ProteinTM Dual Color Standards (Bio-Rad Laboratories) and PageRulerTM Plus Prestained Protein Ladder (Fermentas) were used as molecular weight markers. The immunoblot was finally visualized by exposure on film with ECL Plus Western Blotting Detection Reagents (Amersham & Pharmacia Biotech). Each experiment was independently repeated in triplicate.
4. Conclusions
In this study, we predicted for the first time that the anticancer and anti-inflammatory effects of curcumin might play a key role in protecting human airway from the hazardous effect of PM2.5. Curcumin had the potential to be an airway-protective agent against PM2.5. The current findings were based on bioinformatic studies and require further investigation to confirm.
Acknowledgments
This work is supported by National Natural Science Foundation of China (No. 81102680) and China Postdoctoral Science Foundation (No. 20100470524, No. 20110490548).
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/17/10/12406/s1.
Footnotes
Sample Availability: Not Available.
References
- 1.Valavanidis A., Fiotakis K., Vlachogianni T. Airborne particulate matter and human health: toxicological assessment and importance of size and composition of particles for oxidative damage and carcinogenic mechanisms. J. Environ. Sci. Heal. C. 2008;26:339–362. doi: 10.1080/10590500802494538. [DOI] [PubMed] [Google Scholar]
- 2.Sacks J.D., Stanek L.W., Luben T.J., Johns D.O., Buckley B.J., Brown J.S., Ross M. Particulate matter-induced health effects: who is susceptible? Environ. Health Perspect. 2011;119:446–454. doi: 10.1289/ehp.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polichetti G., Cocco S., Spinali A., Trimarco V., Nunziata A. Effects of particulate matter (PM(10), PM(2.5) and PM(1)) on the cardiovascular system. Toxicology. 2009;261:1–8. doi: 10.1016/j.tox.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 4.Hatcher H., Planalp R., Cho J., Torti F.M., Torti S.V. Curcumin: From ancient medicine to current clinical trials. Cell. Mol. Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H., Beevers C.S., Huang S. The targets of curcumin. Curr. Drug Targets. 2011;12:332–347. doi: 10.2174/138945011794815356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolberg J.R. Designing Quantitative Experiments : Prediction Analysis. Springer; Berlin, Germany: 2010. [Google Scholar]
- 7.PubChem. [(accessed on 16 October 2012)]. Available online: http://pubchem.ncbi.nlm.nih.gov.
- 8.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Zhou Z., Han L., Karapetyan K., Dracheva S., Shoemaker B.A., et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raman K. Construction and analysis of protein-protein interaction networks. Autom. Exp. 2010;2:2. doi: 10.1186/1759-4499-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phung T.T., Sugamata R., Uno K., Aratani Y., Ozato K., Kawachi S., Thanh Nguyen L., Nakayama T., Suzuki K. Key role of regulated upon activation normal T-cell expressed and secreted, nonstructural protein1 and myeloperoxidase in cytokine storm induced by influenza virus PR-8 (A/H1N1) infection in A549 bronchial epithelial cells. Microbiol. Immunol. 2011;55:874–884. doi: 10.1111/j.1348-0421.2011.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dergham M., Lepers C., Verdin A., Billet S., Cazier F., Courcot D., Shirali P., Garcon G. Prooxidant and proinflammatory potency of air pollution particulate matter (PM(2).(5)(-)(0).(3)) produced in rural, urban, or industrial surroundings in human bronchial epithelial cells (BEAS-2B) Chem. Res. Toxicol. 2012;25:904–919. doi: 10.1021/tx200529v. [DOI] [PubMed] [Google Scholar]
- 12.Koarai A., Sugiura H., Yanagisawa S., Ichikawa T., Minakata Y., Matsunaga K., Hirano T., Akamatsu K., Ichinose M. Oxidative stress enhances toll-like receptor 3 response to double-stranded RNA in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2010;42:651–660. doi: 10.1165/rcmb.2008-0345OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfaro-Moreno E., Torres V., Miranda J., Martinez L., Garcia-Cuellar C., Nawrot T.S., Vanaudenaerde B., Hoet P., Ramirez-Lopez P., Rosas I., et al. Induction of IL-6 and inhibition of IL-8 secretion in the human airway cell line Calu-3 by urban particulate matter collected with a modified method of PM sampling. Environ. Res. 2009;109:528–535. doi: 10.1016/j.envres.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Veranth J.M., Moss T.A., Chow J.C., Labban R., Nichols W.K., Walton J.C., Watson J.G., Yost G.S. Correlation of in vitro cytokine responses with the chemical composition of soil-derived particulate matter. Environ. Health Perspect. 2006;114:341–349. doi: 10.1289/ehp.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baulig A., Sourdeval M., Meyer M., Marano F., Baeza-Squiban A. Biological effects of atmospheric particles on human bronchial epithelial cells. Comparison with diesel exhaust particles. Toxicol. In Vitro. 2003;17:567–573. doi: 10.1016/S0887-2333(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 16.Reibman J., Hsu Y., Chen L.C., Kumar A., Su W.C., Choy W., Talbot A., Gordon T. Size fractions of ambient particulate matter induce granulocyte macrophage colony-stimulating factor in human bronchial epithelial cells by mitogen-activated protein kinase pathways. Am. J. Respir. Cell Mol. Biol. 2002;27:455–462. doi: 10.1165/rcmb.2001-0005OC. [DOI] [PubMed] [Google Scholar]
- 17.Liu X., Togo S., Al-Mugotir M., Kim H., Fang Q., Kobayashi T., Wang X., Mao L., Bitterman P., Rennard S. NF-kappaB mediates the survival of human bronchial epithelial cells exposed to cigarette smoke extract. Respir. Res. 2008;9:66. doi: 10.1186/1465-9921-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rennolds J., Malireddy S., Hassan F., Tridandapani S., Parinandi N., Boyaka P.N., Cormet-Boyaka E. Curcumin regulates airway epithelial cell cytokine responses to the pollutant cadmium. Biochem. Biophys. Res. Commun. 2012;417:256–261. doi: 10.1016/j.bbrc.2011.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner M.C., Krewski D., Pope C.A., 3rd, Chen Y., Gapstur S.M., Thun M.J. Long-term ambient fine particulate matter air pollution and lung cancer in a large cohort of never-smokers. Am. J. Respir. Crit. Care Med. 2011;184:1374–1381. doi: 10.1164/rccm.201106-1011OC. [DOI] [PubMed] [Google Scholar]
- 20.Pope C.A., 3rd., Burnett R.T., Thun M.J., Calle E.E., Krewski D., Ito K., Thurston G.D. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H.W., Lee J.Y., Huang J.Y., Wang C.C., Chen W.J., Su S.F., Huang C.W., Ho C.C., Chen J.J., Tsai M.F., et al. Curcumin inhibits lung cancer cell invasion and metastasis through the tumor suppressor HLJ1. Cancer Res. 2008;68:7428–7438. doi: 10.1158/0008-5472.CAN-07-6734. [DOI] [PubMed] [Google Scholar]
- 22.Lin S.S., Lai K.C., Hsu S.C., Yang J.S., Kuo C.L., Lin J.P., Ma Y.S., Wu C.C., Chung J.G. Curcumin inhibits the migration and invasion of human A549 lung cancer cells through the inhibition of matrix metalloproteinase-2 and -9 and Vascular Endothelial Growth Factor (VEGF) Cancer Res. 2009;285:127–133. doi: 10.1016/j.canlet.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Alexandrow M.G., Song L.J., Altiok S., Gray J., Haura E.B., Kumar N.B. Curcumin: A novel Stat3 pathway inhibitor for chemoprevention of lung cancer. Eur. J. Cancer Prev. 2012;21:407–412. doi: 10.1097/CEJ.0b013e32834ef194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gescher A.J., Sharma R.A., Steward W.P. Cancer chemoprevention by dietary constituents: a tale of failure and promise. Lancet Oncol. 2001;2:371–379. doi: 10.1016/S1470-2045(00)00392-2. [DOI] [PubMed] [Google Scholar]
- 25.Agopyan N., Head J., Yu S., Simon S.A. TRPV1 receptors mediate particulate matter-induced apoptosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;286:L563–l572. doi: 10.1152/ajplung.00299.2003. [DOI] [PubMed] [Google Scholar]
- 26.Nel A.E., Diaz-Sanchez D., Li N. The role of particulate pollutants in pulmonary inflammation and asthma: Evidence for the involvement of organic chemicals and oxidative stress. Curr. Opin. Pulm. Med. 2001;7:20–26. doi: 10.1097/00063198-200101000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Gualtieri M., Ovrevik J., Mollerup S., Asare N., Longhin E., Dahlman H.J., Camatini M., Holme J.A. Airborne urban particles (Milan winter-PM2.5) cause mitotic arrest and cell death: Effects on DNA, mitochondria, AhR binding and spindle organization. Mutat. Res. 2011;713:18–31. doi: 10.1016/j.mrfmmm.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J., Ghio A.J., Gao M., Wei K., Rosen G.D., Upadhyay D. Ambient particulate matter induces alveolar epithelial cell cycle arrest: role of G1 cyclins. FEBS Lett. 2007;581:5315–5320. doi: 10.1016/j.febslet.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soberanes S., Panduri V., Mutlu G.M., Ghio A., Bundinger G.R., Kamp D.W. p53 mediates particulate matter-induced alveolar epithelial cell mitochondria-regulated apoptosis. Am. J. Respir. Crit. Care Med. 2006;174:1229–1238. doi: 10.1164/rccm.200602-203OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dagher Z., Garcon G., Billet S., Gosset P., Ledoux F., Courcot D., Aboukais A., Shirali P. Activation of different pathways of apoptosis by air pollution particulate matter (PM2.5) in human epithelial lung cells (L132) in culture. Toxicology. 2006;225:12–24. doi: 10.1016/j.tox.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 31.Siddiqui M.A., Ahamed M., Ahmad J., Majeed Khan M.A., Musarrat J., Al-Khedhairy A.A., Alrokayan S.A. Nickel oxide nanoparticles induce cytotoxicity, oxidative stress and apoptosis in cultured human cells that is abrogated by the dietary antioxidant curcumin. Food Chem. Toxicol. 2012;50:641–647. doi: 10.1016/j.fct.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Ramachandran C., You W. Differential sensitivity of human mammary epithelial and breast carcinoma cell lines to curcumin. Breast Cancer Res. Treat. 1999;54:269–278. doi: 10.1023/A:1006170224414. [DOI] [PubMed] [Google Scholar]
- 33.Pujolsa L., Mullol J., Picado C. Glucocorticoid receptor in human respiratory epithelial cells. Neuroimmunomodulation. 2009;16:290–299. doi: 10.1159/000216187. [DOI] [PubMed] [Google Scholar]
- 34.Verheggen M.M., Adriaansen-Soeting P.W., Berrevoets C.A., van Hal P.T., Brinkmann A.O., Hoogsteden H.C., Versnel M.A. Glucocorticoid receptor expression in human bronchial epithelial cells: effects of smoking and COPD. Mediat. Inflamm. 1998;7:275–281. doi: 10.1080/09629359890965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnes P.J. Histone deacetylase-2 and airway disease. Ther. Adv. Respir. Dis. 2009;3:235–243. doi: 10.1177/1753465809348648. [DOI] [PubMed] [Google Scholar]
- 36.Marwick J.A., Ito K., Adcock I.M., Kirkham P.A. Oxidative stress and steroid resistance in asthma and COPD: pharmacological manipulation of HDAC-2 as a therapeutic strategy. Expert Opin. Ther. Tar. 2007;11:745–755. doi: 10.1517/14728222.11.6.745. [DOI] [PubMed] [Google Scholar]
- 37.Ito K., Caramori G., Adcock I.M. Therapeutic potential of phosphatidylinositol 3-kinase inhibitors in inflammatory respiratory disease. J. Pharmacol. Exp. Ther. 2007;321:1–8. doi: 10.1124/jpet.106.111674. [DOI] [PubMed] [Google Scholar]
- 38.Barnes P.J. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009;6:693–696. doi: 10.1513/pats.200907-071DP. [DOI] [PubMed] [Google Scholar]
- 39.Watterson T.L., Hamilton B., Martin R.S., Coulombe R.A., Jr. Urban particulate matter activates Akt in human lung cells. Arch. Toxicol. 2012;86:121–135. doi: 10.1007/s00204-011-0739-5. [DOI] [PubMed] [Google Scholar]
- 40.Syed D.N., Afaq F., Kweon M.H., Hadi N., Bhatia N., Spiegelman V.S., Mukhtar H. Green tea polyphenol EGCG suppresses cigarette smoke condensate-induced NF-kappaB activation in normal human bronchial epithelial cells. Oncogene. 2007;26:673–682. doi: 10.1038/sj.onc.1209829. [DOI] [PubMed] [Google Scholar]
- 41.Yu H., Li Q., Kolosov V.P., Perelman J.M., Zhou X. Regulation of cigarette smoke-induced mucin expression by neuregulin1beta/ErbB3 signalling in human airway epithelial cells. Basic Clin. Pharmacol. Toxicol. 2011;109:63–72. doi: 10.1111/j.1742-7843.2011.00686.x. [DOI] [PubMed] [Google Scholar]
- 42.Moriyuki K., Sekiguchi F., Matsubara K., Nishikawa H., Kawabata A. Curcumin Inhibits the proteinase-activated receptor-2-triggered prostaglandin E2 production by suppressing cyclooxygenase-2 upregulation and Akt-dependent activation of nuclear factor-kappaB in human lung epithelial cells. J. Pharmacol. Sci. 2010;114:225–229. doi: 10.1254/jphs.10126SC. [DOI] [PubMed] [Google Scholar]
- 43.Lee C.W., Lin C.C., Lin W.N., Liang K.C., Luo S.F., Wu C.B., Wang S.W., Yang C.M. TNF-alpha induces MMP-9 expression via activation of Src/EGFR, PDGFR/PI3K/Akt cascade and promotion of NF-kappaB/p300 binding in human tracheal smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;292:L799–l812. doi: 10.1152/ajplung.00311.2006. [DOI] [PubMed] [Google Scholar]
- 44.Mantecca P., Farina F., Moschini E., Gallinotti D., Gualtieri M., Rohr A., Sancini G., Palestini P., Camatini M. Comparative acute lung inflammation induced by atmospheric PM and size-fractionated tire particles. Toxicol. Lett. 2010;198:244–254. doi: 10.1016/j.toxlet.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Dagher Z., Garcon G., Billet S., Verdin A., Ledoux F., Courcot D., Aboukais A., Shirali P. Role of nuclear factor-kappa B activation in the adverse effects induced by air pollution particulate matter (PM2.5) in human epithelial lung cells (L132) in culture. J. Appl. Toxicol. 2007;27:284–290. doi: 10.1002/jat.1211. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y., Usatyuk P.V., Gorshkova I.A., He D., Wang T., Moreno-Vinasco L., Geyh A.S., Breysse P.N., Samet J.M., Spannhake E.W., et al. Regulation of COX-2 expression and IL-6 release by particulate matter in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2009;40:19–30. doi: 10.1165/rcmb.2008-0105OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freeman A.K., Morrison D.K. 14–3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011;22:681–687. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardino A.K., Yaffe M.B. 14–3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin. Cell Dev. Biol. 2011;22:688–695. doi: 10.1016/j.semcdb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qi W., Liu X., Chen W., Li Q., Martinez J.D. Overexpression of 14–3-3gamma causes polyploidization in H322 lung cancer cells. Mol. Carcinog. 2007;46:847–856. doi: 10.1002/mc.20314. [DOI] [PubMed] [Google Scholar]
- 50.Kawamoto S., Iemura N., Inoue Y., Katakura Y., Shirahata S. Effect of 14–3-3 protein induction on cell proliferation of A549 human lung adenocarcinoma. Cytotechnology. 2000;33:253–257. doi: 10.1023/A:1008129020728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J.B., Qi L.L., Zheng S.D., Wang H.Z., Wu T.X. Curcumin suppresses PPARdelta expression and related genes in HT-29 cells. World J. Gastroenterol. 2009;15:1346–1352. doi: 10.3748/wjg.15.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyata R., van Eeden S.F. The innate and adaptive immune response induced by alveolar macrophages exposed to ambient particulate matter. Toxicol. Appl. Pharmacol. 2011;257:209–226. doi: 10.1016/j.taap.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 53.GSE7010. [(accessed on 16 October 2012)]. Available online: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7010.
- 54.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y.C., Karoly E.D., Dailey L.A., Schmitt M.T., Silbajoris R., Graff D.W., Devlin R.B. Comparison of gene expression profiles induced by coarse, fine, and ultrafine particulate matter. J. Toxicol. Environ. Health A. 2011;74:296–312. doi: 10.1080/15287394.2010.516238. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic acids Res. 2009;37:W623–w633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canny S.A., Cruz Y., Southern M.R., Griffin P.R. PubChem promiscuity: A web resource for gathering compound promiscuity data from PubChem. Bioinformatics. 2012;28:140–141. doi: 10.1093/bioinformatics/btr622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.PubChem Promiscuity. [(Accessed on 16 October 2012)]. Available online: http://chemutils.florida.scripps.edu:8080/pcpromiscuity.
- 59.Real-Chicharro A., Ruiz-Mostazo I., Navas-Delgado I., Kerzazi A., Chniber O., Sanchez-Jimenez F., Medina M.A., Aldana-Montes J.F. Protopia: A protein-protein interaction tool. BMC Bioinformatics. 2009;10(Suppl. 12):S17. doi: 10.1186/1471-2105-10-S12-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin A., Ochagavia M.E., Rabasa L.C., Miranda J., Fernandez-de-Cossio J., Bringas R. BisoGenet: A new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.