Abstract
Investigating the effect of lipid hydrophobicity on the activity of amphiphilic neomycin B conjugates, six polycationic amphiphiles (PAs) were created. Four of the new compounds incorporated either palmitic or arachidic di-lipid lysine tails, while two had single fluorinated undecanoic acid tails. The basicity of half of the compounds was increased through the incorporation of six guanidine moieties, in order to assess the effect of base strength on antimicrobial activity. A panel of ten bacteria was used for the testing, with seven strains obtained from the American Type Culture Collection series and three clinical isolates from Canadian Intensive Care Units. When compared to previous results with hydrocarbon monolipids the PAs all compounds were found to have reduced activity, though the hemolytic activity of the compounds with fluorinated tails was sharply reduced, with only a moderate reduction in antimicrobial activity.
Keywords: aminoglycosides, cationic amphiphiles, carbohydrates, antibacterials
1. Introduction
Much of our current antibiotic arsenal is derived from scaffolds developed forty or more years ago, allowing bacterial resistance to slowly accrue [1]. To combat tomorrow’s “superbugs” we will need new scaffolds [2], with antimicrobials designed specifically to combat the emergence of drug resistance. Polycationic lipids are one such scaffold, as agents such as chlorhexidine and benzalkonium chloride have been used as disinfectants for decades with little resistance development [3]. Unfortunately, these compounds are toxic to mammalian cells, limiting their use to topical applications [3,4]. Extensive research around analogues of cationic antimicrobial peptides such a magainin, mellitin and LL-37 has explored their potential to act as less toxic amphiphiles [4,5], but interest is beginning to move from the membrane-interacting behaviour of the peptides to their immunomodulatory properties and specific peptide-target interactions [6,7].
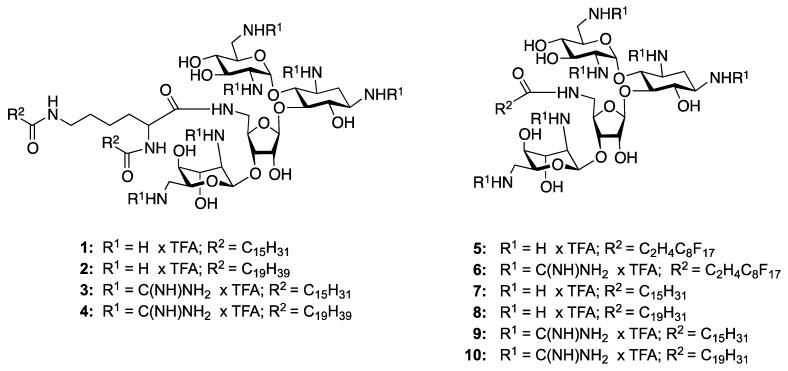
Starting from a class of compounds with known antibiotic action and limited ability to disrupt mammalian membranes, we have previously demonstrated the potential for a model aminoglycoside, neomycin, to act as a polycationic amphiphile (PA) [8,9,10,11,12]. In one study, coupling a single unit of palmitic acid to a C5"-amine restored good activity against a normally resistant strain of MRSA [12]. Reducing the size of the lipid tail to twelve carbons eliminated activity, confirming that the gain was due to an increase in aminoglycoside hydrophobicity. In this work we attempted to increase PA self-association in both the bacterial membrane and solution, through the incorporation of either a fluorocarbon tails or two hydrophobic tails (Figure 1). Self-association in the membrane was postulated to decrease the concentration required for membrane disruption, by forming pockets of high PA concentration, while self-association in solution could lead to micelle formation and general exclusion of the hydrophobic portion of the amphiphiles from circulation.
Figure 1.
Structures of neomycin B-based bilipids 1–4, the fluorinated monolipids 5 and 6, and the previously reported hydrocarbon monolipid-neomycin-B amphiphiles 7–10 [9,12].
2. Results and Discussion
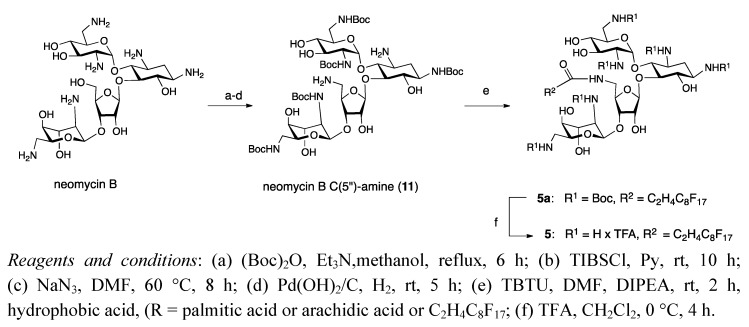
While an appealing scaffold for PA design, the polyfunctional nature of neomycin necessitates multiple protection/deprotection steps for selective modifications. This is often perceived as a serious limitation to drug development, but we have developed an efficient series of reactions for modification of the 5"-OH moiety. The four step synthesis begins with protection of the amine functionalities, followed by conversion of the 5"-OH to a primary amine for standard amide bond formation (Scheme 1). Crystal structures have confirmed that this site is not required for tight binding to the ribosome [13], and in the past this set of reactions was used to create a number of monolipid analogues [8,12], including some with the standard amine groups converted into more basic guanidines [9].
Scheme 1.
Strategy for the synthesis of neomycin B C(5")-modified analog 5.
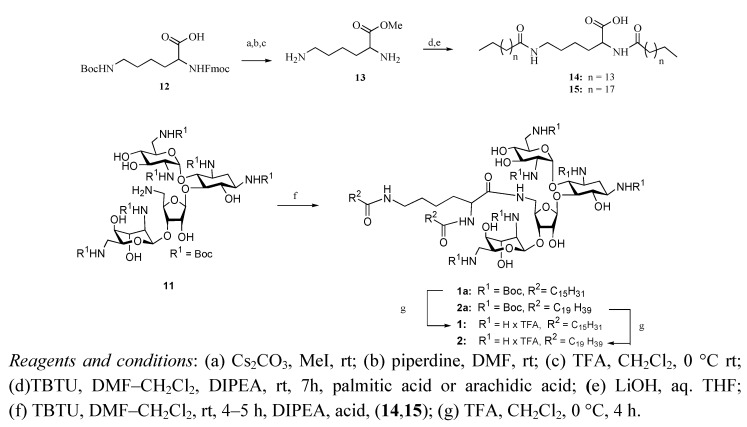
Incorporation of two hydrophobic tails required the use of a specially modified carboxylic acid. Due to its ready availability we began with the protected amino acid Fmoc-Lys(Boc)-OH (Scheme 2), first converting the carboxylic acid to a methyl ester via cesium carbonate and methyl iodide.
Scheme 2.
Synthesis of neomycin B C(5")-modified bilipids 1 and 2.
The amino acid protecting groups were then sequentially cleaved under standard conditions and either palmitic or arachidic acid was coupled to both free amines, with TBTU as the coupling agent. A mixture of DMF and DCM was required to effect solubilization during this reaction, due to the highly hydrophobic nature of arachidic acid. Following the coupling the methyl ester was cleaved by lithium hydroxide in aqueous THF to furnish dilipids 14 and 15.
Interested in the effect of guanidinylation on PA activity, two different types of lipid conjugated neomycin B amphiphiles were produced. Synthesis of both types began with protection of the amine moieties with Boc anhydride in a refluxing mixture of TEA and MeOH. The primary hydroxyl group was then activated as a triisopropylbenzenesulfonate ester, and the sulfonate was then displaced via SN2 nucleophilic attack by sodium azide. The bulky sulfonyl chloride was chosen for its preference for unhindered primary alcohols, which reduced substitution at the six secondary alcohols [14].
Reduction of the azide was accomplished via catalytic hydration by palladium hydroxide on charcoal and hydrogen gas, affording the free 5"-amino hexa-Boc protected neomycin B, 11. This compound was then coupled to dilipids 14 and 15, and following purification by column chromatography the protecting groups were cleaved with 95:5 TFA: H2O. Trituration in 98:2 Et2O/MeOH removed any residual hydrophobic impurities, providing dilipid neomycin salts 1 and 2 in good yield.
Compound 11 was also used to produce a monolipid analogue with a fluorinated tail, compound 5, again via standard peptide coupling reagents. A fluorinated undecanoic acid analogue was selected, due to a mix of commercial availability and stability of the final amide. Previous work with perfluorinated acids has shown that the resulting amide bond is not stable to hydrolysis, due to the strong acidity of the conjugate ester [15]. The addition of two methylene spacers between the fluorocarbon region and the carboxylic acid eliminated this hydrolysis, but limited commercial availability to tails that were a total of eleven carbons long or shorter. As this work was intended to explore the effect of self-associating tails on PA activity the longest commercially available lipid was selected. While significantly shorter the fluorinated undecanoic acid tail is nearly twice the molecular weight of palmitic acid, and so compounds 1 and 5 were similar in final molecular weight.
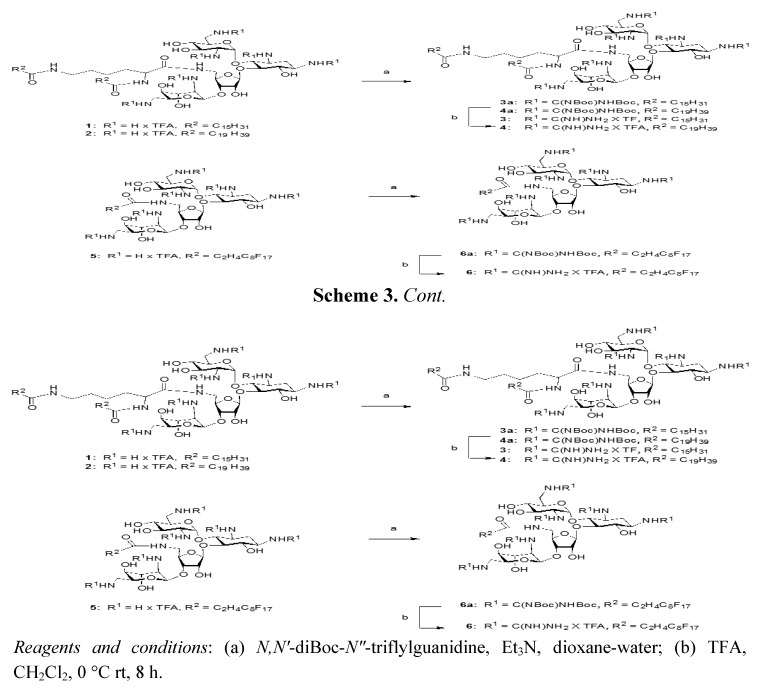
The second set of PAs was constructed by replacing the amines of neomycin B with guanidine groups, through the use of the commercially available N,N'-Diboc-N"-triflylguanidine [16] (Scheme 3).
Scheme 3.
Synthesis of neomycin B C(5")-modified guanidinylated analogs 3, 4 and 6.
PAs 1–3 were reacted over a period of three to four days, followed by extraction and column chromatography to give compounds 3a, 4a, and 6a. Treatment with 1:1 TFA: DCM for four h cleaved the Boc protecting groups to give compounds 3, 4, and 6; which were purified via trituration in 98:2 Et2O : MeOH.
Polycationic amphiphiles in hand, we then determined their antimicrobial activity against a panel of both American Type Culture Collection (ATCC) and clinically relevant bacterial strains. Pathogenic isolates were obtained during the CAN-ICU surveillance study [17], and represent “clinically significant” strains of Escherichia coli and Pseudomonas aeruginosa. ATCC strains used in this testing allow for comparison to previous PA studies [18], with gentamicin and neomycin serving as positive controls.
We were especially interested in comparing the activity of our dilipid and fluorinated lipid aminoglycosides to the activity of PAs 7–10 (Figure 1), which have been reported previously [9,12]. In line with previous results involving monolipid tails [9,12], PAs 1–6 demonstrated higher activity against Gram positive bacteria than Gram negative (Table 1), with the palmitoyl guanidylated dilipid 3 being the most active compound overall. Guanidinylation increased the activity of the amphiphiles against Gram positive bacteria 2–8 fold, with both strains of Staphylococcus epidermidis highly susceptible, in line with the activity of neomycin against these strains. The greatest improvement in activity was against the Gram negative bacteria P. aeruginosa, where the MIC decreased from 512 ug/mL for neomycin to 32 ug and 16 ug for compound 3 (against ATCC strain 27853 and CAN-ICU strain 62308, respectively). While compound 3 was more active than the PAs with fluorinated monolipids, both 5 and 6 had improved activity over the remaining dilipids, perhaps due to the potential for the dilipids to form either insoluble aggregates.
Table 1.
Antibacterial and hemolytic activity (MIC) in µg/mL of various neomycin B-based mono- and bilipids 1–6.
| Control Organism | Genta-mycin | Neo- mycin | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|---|
| S. aureus ATCC29213 | 1 | 1 | 16 | 32 | 4 | 64 | 16 | 8 |
| MRSA ATCC33592 | 2 | 256 | 16 | 128 | 4 | 128 | 64 | 8 |
| S. epidermidis ATCC14990 | 0.25 | 0.25 | 4 | 16 | 2 | 16 | 8 | 8 |
| MRSE CAN-ICU 61589 | 32 | 0.5 | 4 | 16 | 4 | 64 | 4 | 4 |
| E. faecalis ATCC29212 | n.d. | 16 | 16 | 128 | 8 | 128 | 64 | 16 |
| E. facium ATCC27270 | n.d. | 4 | 16 | 128 | 4 | 128 | 32 | 2 |
| S. pneumoniae ATCC49619 | 4 | 32 | 128 | 128 | 64 | >128 | 64 | 32 |
| E. coli ATCC25922 | 1 | 4 | 32 | 32 | 64 | >128 | 32 | 16 |
| E. coli CAN-ICU 61714 | 128 | 8 | 32 | 64 | 32 | >128 | 32 | 32 |
| E. coli CAN-ICU 63074 | 8 | n.d. | 64 | 64 | 32 | >128 | 64 | 64 |
| P. aeruginosa ATCC27853 | 8 | 512 | 128 | >256 | 32 | 128 | 128 | 64 |
| P. aeruginosa CAN-ICU 62308 | 128 | 512 | 64 | 128 | 16 | 128 | 32 | 8 |
| % hemolysis at 100 µg/mL/ (500 µg/mL) | <0.4 (<0.4) | <0.4 (<0.4) | n.d. (n.d.) | n.d. (n.d.) | n.d. (n.d.) | n.d. (n.d.) | 1.1 (9.4) | 1.8 (11.9) |
Moving from a monolipid to a dilipid scaffold appeared to reduce the activity in the context of palmitic and arachidic tails. In general there was a 2–4 fold reduction in activity against most bacterial species, with no significant difference in reduction between Gram positive and Gram negative strains. PAs with fluorinated lipid tails were less active than the palmitoyl monolipids, which had a similar mass, but demonstrated significantly reduced toxicity towards eukaryotic cells. While hemolysis data was not obtained for PAs 1–4, we are encouraged by this broadening of the therapeutic window.
3. Experimental
3.1. General Methods
NMR spectra were recorded on a Bruker Avance 300 spectrometer (300 MHz for 1H-NMR, 75 MHz for 13C) or AMX 500 spectrometer (500 MHz for 1H-NMR). Optical rotation was measured at a concentration of g/100 mL, with a Perkin-Elmer polarimeter (accuracy 0.002°). GC-MS analyses were performed on a Perkin-Elmer Turbomass-Autosystem XL. Analytical thin-layer chromatography was performed on precoated silica gel plates, with spot visualization via ultraviolet light and/or by staining with ninhydrin solution in ethanol. Chromatographic separations were performed on a silica gel column by flash chromatography (Kiesel gel 40, 0.040–0.063 mm; Merck). Yields are given after purification, unless explicitly stated. Reactions requiring anhydrous conditions were performed under nitrogen or argon gas.
3.2. Synthesis of Lysine-Dilipid Conjugates 14 and 15
Commercially available side chain protected Fmoc-Lys(NHBoc)-COOH was converted to methyl ester using TBTU in DMF followed by removal of Fmoc- and Boc-protecting groups followed by condensation to palmitic acid/arachidic. Ester hydrolysis was performed with LiOH in a mixture of THF/H2O (2:1).
N,N-di-Hexadecanoyl-Lys-acid (14). Yield = 65%; 1H-NMR (300 MHz, CD3OD and CDCl3): δ 6.22 (d, 1H, J = 7.9 Hz), 5.76 (t, 1H, J = 5.0 Hz), 4.55 (m, 1H), 3.22 (t, 2H, J = 6.3 Hz), 2.19 (m, 4H), 1.84 (m, 2H), 1.61 (m, 4H), 1.41 (m, 4H), 1.24 (s, 48H), 0.87 (t, 6H, J = 6.6 Hz); EIMS: calcd for C38H74NaN2O4+ 645.56 Found: 645.88 [M+Na]+.
N,N-di-Nonadecanoyl-Lys-acid (15). Yield = 61%; 1H-NMR (300 MHz, CD3OD and CDCl3): δ 6.22 (d, 1H, J = 7.9 Hz), 5.76 (t, 1H, J = 5.0 Hz), 4.55 (m, 1H), 3.32 (t, 2H, J = 6.3 Hz), 2.19 (m, 4H), 1.84 (m, 2H), 1.73–1.41 (m, 8H), 1.24 (s, 64H), 0.87 (t, 6H, J = 6.6 Hz); EIMS: calcd for C46H90N2NaO4+ 775.69 Found: 775.97 [M+Na]+.
3.3. General Procedure for Coupling Reaction of Neomycin B-based Amine 11
To a solution of neomycin amine 11 (1 equiv.) [12] in dry DMF, TBTU (2 equiv.), the lipid of choice (1 equiv.) and DIPEA (3 equiv.) were added. The mixture was stirred at room temperature for 2 h, then extracted with water and ethyl acetate. The ethyl acetate layer was washed with water, brine, dried over sodium sulfate and concentrated. The crude residue was purified using flash silica gel (CH2Cl2/MeOH). Spectroscopic data is provided below.
3.4. General Procedure for Final Deprotection
All Boc-protected lipids were treated with 95:5 TFA–H2O at 0 °C for 3 min, following which the solvent was removed at reduced pressure. To the residue 2% methanol in ether was added and the solvent was decanted to get the solid neomycin-lipid conjugate as salt. The spectroscopic data is given below.
5"-N-(N,N-di-Hexadecanoyl-Lys)-1,3,2',6',2''',6'''-hexa-N-(tert-butoxycarbonyl)-5''-deoxy-neomycin (1a). Yield = 78%; Rf 0.35 (MeOH/CH2Cl2 1:9); 1H-NMR (300 MHz, CD3OD): δ 5.48 (br s, 1H), 5.09 (br s, 1H), 4.88 (s, 1H), 4.31 (d, 1H, J = 3.4 Hz), 4.07 (t, 1H, J = 4.4 Hz), 3.96–3.80 (m, 5H), 3.76 (m, 2H), 3.56 (m, 4H), 3.48 (br t, 3H, J = 3.9 Hz), 3.39 (m, 4H), 3.27 (d, 2H, J = 7.6 Hz), 3.20 (m, 2H), 3.12 (m, 1H), 2.34 (m, 3H), 1.97 (dt, 1H, J = 4.3, 11.1 Hz), 1.64 (m, 4H), 1.51–144 (m, 56H), 1.43 (s, 9H), 1.29 (s, 45H), 0.90 (t, 6H, J = 6.1 Hz); 13C-NMR (75 MHz, CD3OD): δ 176.6 (amide C=O), 176.4 (×2) (peptide amide C=O), 158.9–158.7 (Boc-CO), 112.1, 100.4, 98.8 (anomeric carbons), 88.4, 80.8–80.3, 79.4, 76.2, 75.6, 74.5, 74.1, 73.4, 71.7, 69.0, 69.3, 56.8, 53.6, 51.3, 52.5, 43.9, 42.7, 42.5, 38.9, 37.0–23.7 (aliphatic CH2), 14.5 (aliphatic CH3); [α]D25 = +32.0 (c 0.53, MeOH); EIMS: calcd for C91H167N9NaO27+ 1841.22 Found: 1841.45 [M+Na]+; Anal. Calcd. for C91H167N9O27 C, 60.08; H, 9.25; N, 6.93; Found: C, 60.33; H, 9.45; N, 7.03.
1,3,2',6',2''',6'''-Hexaammonium-5"-deoxy-5"-N-(N,N-di-hexadecanoyl-Lys)-neomycin hexakis-(trifluoroacetate) (1). Yield = 89%; Rf 0.10 (NH4OH/MeOH/CH2Cl2 2:9:4); 1H-NMR (300 MHz, CD3OD): δ 5.86 (d, 1H, J = 3.1 Hz), 5.40 (d, 1H, J = 4.4 Hz), 5.30 (s, 1H), 4.41 (t, 1H, J = 5.4 Hz), 4.30 (m, 2H), 4.22 (m, 1H), 4.16 (m, 2H), 4.08 (t, 2H, J = 5.9 Hz), 3.99 (t, 2H, J = 3.6 Hz), 3.89 (t, 2H, J = 8.8 Hz), 3.68 (m, 4H), 3.52 (dd, 2H, J = 2.6, 11.1 Hz), 3.45 (m, 3H), 3.41 (t, 1H, J = 3.6 Hz), 3.37 (d, 1H, J = 1.6 Hz), 3.24 (m, 1H), 3.20 (m, 1H), 2.48 (d, 2H, J = 11.8 Hz), 2.26 (d, 3H, J = 7.8 Hz), 2.08 (d, 2H, J = 11.8 Hz), 1.61 (m, 3H), 1.29 (s, 52H), 0.90 (t, 6H, J = 6.1 Hz); 13C-NMR (75 MHz, CD3OD): δ 177.4 (amide C=O), 177.3 (×2) (peptide amide C=O), 164.0–162.7 (TFA, q with J 2CF ~ 34.8 Hz), 124.1–112.4 (q with J 1CF ~ 292.0 Hz), 109.5, 97.2, 96.7 (anomeric carbons), 86.6, 83.4, 78.1, 76.6, 76.4, 75.6, 74.1, 74.0, 72.9, 72.5, 72.0, 71.9, 71.8, 69.4, 67.6, 64.9, 55.4, 52.9, 51.1, 50.3, 41.9, 41.7, 37.2–23.7 (aliphatic CH2), 14.7 (CH3); [α]D25 = +24.0 (c 1.58, MeOH); EIMS: calcd for C61H120N9O15+1218.88 Found: 1219.09 [M+H]+; Anal. Calcd. for C73H125F18N9O27 C, 46.08; H, 6.62; F, 17.97; N, 6.63; Found: C, 46.23; H, 6.76; F, 18.01; N, 6.46.
5"N-(N,N-di-Nonadecane-Lys)-1,3,2',6',2''',6'''-hexa-N-(tert-butoxycarbonyl)-5"-deoxy-neomycin (2a). Yield = 70%; Rf 0.33 (MeOH/CH2Cl2 1:9); 1H-NMR (300 MHz, 2:1 mixture of CD3OD and CDCl3): δ 5.08 (m, 2H), 4.74 (m, 1H), 4.36 (m, 2H), 4.22 (m, 2H), 4.03 (t, 1H, J = 7.3 Hz), 3.99 (m, 4H), 3.79 (m, 2H), 3.70 (m, 2H), 3.55 (m, 3H), 3.46 (m, 4H), 3.43 (m, 2H), 3.25 (m, 2H), 3.13 (m, 1H), 2.34 (m, 2H), 2.24 (t, 2H, J = 7.9 Hz), 1.99 (m, 2H), 1.69 (m, 2H), 1.55–1.40 (m, 72H), 1.28 (s, 54H), 0.89 (t, 6H, J = 7.1 Hz); 13C-NMR (75 MHz, CD3OD): δ 176.2 (amide C=O), 175.7 (×2) (peptide amide C=O), 158.3–157.2 (Boc-CO), 101.6, 100.3, 97.6 (anomeric carbons), 87.5, 81.4–80.6, 78.4, 76.4, 75.5, 75.3, 74.5, 73.5, 73.3, 72.4, 71.4, 68.9, 57.1, 56.1, 53.4, 52.3, 44.2, 42.8, 40.0, 38.0, 37.7–24.0 (aliphatic CH2), 14.4(CH3); [α]D25 = +55.0 (c 0.73, MeOH); EIMS: C99H183N9NaO27+ 1953.32 Found: 1953.67 [M+Na]+; Anal. Calcd. for C99H183N9O27 C, 61.56; H, 9.55; N, 6.53; Found: C, 62.22; H, 10.09; N, 6.53.
1,3,2',6',2"',6"'-Hexaammonium-5"-deoxy-5"-N-(N,N-di-nonadecanoyl-Lys)-neomycin hexakis-(trifluoroacetate) (2). Yield = 87%; Rf 0.10 (NH4OH/MeOH/CH2Cl2 2:9:4); 1H-NMR (300 MHz, 2:1 mixture of CD3OD and CDCl3): δ 5.40 (d, 1H, J = 3.1 Hz), 5.25 (d, 1H, J = 4.4 Hz), 5.20 (s, 1H), 4.79 (m, 1H), 4.62 (m, 1H), 4.40–4.00 (m, 9H), 3.89 (t, 1H, J = 5.9 Hz), 3.68 (m, 2H), 3.60 (m, 1H), 3.44 (m, 6H), 3.22 (m, 4H), 2.45 (m, 1H), 2.21 (d, 2H, J = 8.1 Hz), 2.19 (d, 2H, J = 7.1 Hz), 2.00 (d, 2H, J = 6.4 Hz), 1.60 (m, 5H), 1.22 (s, 70H), 0.90 (t, 6H, J = 7.2 Hz); 13C-NMR (75 MHz, CD3OD): δ 177.2 (amide C=O), 176.1 (×2) (peptide amide C=O), 164.0–162.7 (TFA, q with J 2CF ~ 34.8 Hz), 124.1–112.4 (q with J 1CF ~ 292.0 Hz), 109.5, 97.2, 96.3 (anomeric carbons), 86.3, 83.0, 79.2, 78.2, 75.9, 75.5, 73.8, 72.9, 72.4, 72.1, 71.9, 71.7, 69.4, 69.0, 61.9, 55.9, 52.9, 51.8, 50.3, 41.9, 41.7, 40.6, 40.5, 37.5–23.7 (aliphatic CH2) 14.7 (CH3); [α]D25 = +55.0 (c 0.71, MeOH); EIMS: calcd for C69H136N9O15+ 1331.01 Found: 1331.45 [M+H]+; Anal. Calcd for C81H141F18N9O27 C, 48.28; H, 7.05; F, 16.97; N, 6.26 Found: C, 48.28; H, 7.05; F, 16.97; N, 6.26.
5"-N-(Heptadecafluoro-decanoyl)-1,3,2',6',2"',6"'-hexa-N-(tert-butoxycarbonyl)-5"-deoxy-neomycin (5a). Yield = 88%; Rf 0.56 (MeOH/CH2Cl2 1:9); 1H-NMR (300 MHz, CD3OD): δ 5.54 (br s, 1H), 5.10 (br s, 1H), 4.89 (br s, 1H), 4.34 (d, J = 4.38, 1H), 4.11–4.04 (m, 2H), 3.97–3.85 (m, 4H), 3.79–3.72 (m, 2H), 3.59–3.47 (m, 6H), 3.44–3.36 (m, 3H), 3.28–3.18 (m, 2H), 3.08 (dd, J = 8.6, 13.6 Hz, 1H), 2.82–2.47 (m, 4H), 1.97–192 (d, J =12.6 Hz, 1H), 1.67–1.60 (m, 2H), 1.51–1.43 (m, 54H), 1.39–1.22 (m, 2H); 13C-NMR (75 MHz, CD3OD): δ 173.0 (amide-CO) 159.1, 158.9, 158.7, 158.6, 158.3, 157.9 (Boc-CO), 112.3, 100.3, 98.7 (anomeric carbons), 88.6, 81.0, 80.8, 80.7, 80.4, 80.3, 76.3, 75.5, 74.5, 73.4, 72.7, 72.5, 71.7, 69.1, 56.8, 53.6, 52.7, 51.3, 44.2, 42.7, 42.0, 35.7, 28.9–27.5 (Boc CH3 and aliphatic CH2); EIMS: calcd. for C64H99F17N7O25+ 1688.64. Found: 1688.45 [M+H]+.
1,3,2',6',2''',6"'-Hexaammonium-5"-deoxy-5"-N-(heptadecafluoro-decanoyl)-neomycin hexakis-(trifluoroacetate) (5). Yield = 89%; Rf 0.14 (NH4OH/MeOH/CH2Cl2 1:5:5); 1H-NMR (300 MHz, CD3OD): δ 5.85 (d, J = 3.7 Hz, 1H), 5.40 (d, J = 4.8 Hz, 1H), 5.29 (br s, 1H), 4.41–4.33 (m, 1H), 4.25 (m, 4H), 4.14 (m, 1H), 4.08 (m, 1H), 4.00 (m, 2H), 3.88 (dd, 1H, J = 9.0, 7.8 Hz), 3.73–3.66 (m, 2H), 3.64 (m, 1H), 3.55–3.34 (m, 8H), 3.26–3.21 (m, 1H), 3.16 (dd, 1H, J = 13.4, 8.1 Hz), 2.58 (m, 3H), 2.46 (m, 1H), 2.07 (m, 1H); 13C-NMR (75 MHz, CD3OD): δ 173.70 (NH-CO-), 163.3 (TFA), 123.93, 120.38, 120.09, 120.07, 119.86, 119.82 (F attached), 117.2 (TFA), 109.61, 97.33, 96.71 (anomeric carbons), 86.55, 83.19, 78.18, 76.50, 75.58, 73.99, 73.05, 72.04, 71.94, 69.48, 69.33, 69.18, 66.87, 55.03, 52.88, 51.14, 50.31, 42.21, 41.96, 41.68, 29.43, 27.51; EIMS: calcd. for C34H51F17N7O13+ 1088.32. Found: 1088.45 [M+H]+.
3.5. General Procedure for the Guanidinylation of Aminoglycosides
N,N'-DiBoc-N"-triflylguanidine was purchased from Fluka and used without further purification. To a solution of aminoglycoside (5 amines, 0.054 mmol) in H2O (0.5 mL) was added 1,4-dioxane (2.5 mL) and N,N'-diBoc-N"-triflylguanidine (0.82 mmol) in alternating portions so the solution remained relatively clear. After 5 min, NEt3 (0.82 mmol) was added at room temperature. After 3–4 days, the 1,4-dioxane was removed under reduced pressure. The remaining residue and H2O was extracted with CH2Cl2 (3 × 10 mL), washed with H2O and brine, and dried (MgSO4). The fully guanidinylated product can be isolated by flash column chromatography (FCC) on silica gel (CH2Cl2/MeOH).
3.6. General Procedure for the Deprotection of Guanidinoglycosides
A solution of TFA: CH2Cl2 (1:1, 1 mL) was added to the protected guanidinoglycoside (0.041 mmol) at room temperature. After approximately 4 h, the solution was diluted with toluene, concentrated in vacuo, and dissolved in H2O. Subsequent lyophilization of H2O provided the deprotected guanidinoglycoside as a fluffy white powder.
5"-Deoxy-5"-N-(N,N-di-hexadecanoyl-Lys)-1,3,2',6',2"',6'''-hexa-N-(tert-butoxycarbonyl-guanidino)-5"-deoxy-neomycin (3a). Yield = 75%; Rf 0.33 (MeOH/CH2Cl2 1:9); 1H-NMR (300 MHz, CD3OD): δ 5.62 (s, 1H), 5.22 (s, 2H), 5.08 (s, 1H), 4.55 (m, 1H), 4.38 (m, 2H), 4.23 (m, 3H), 4.10 (m, 4H), 3.99 (m, 2H), 3.91–3.77 (m, 2H), 3.72 (m, 2H), 3.63–3.50 (m, 6H), 3.40 (t, 1H, J = 5.3 Hz), 3.20 (m, 1H), 2.26 (m, 4H), 1.72 (m, 2H), 1.50 (s, 128H), 1.30 (s, 36H), 0.89 (t, 6H, J = 7.1 Hz); 13C-NMR (75 MHz, CD3OD, HSQC): δ 176.5 (amide C=O), 176.2 (×2) (peptide amide C=O), 164.4–164.1 (Boc C=O), 158.0–157.4 (guanidine C=NH), 154.4–153.7 (Boc C=O), 111.3, 97.7, 95.8 (anomeric carbons), 88.3, 84.8–84.1, 82.6, 80.7–80.4, 79.6, 77.3, 76.5, 75.3, 74.2, 73.1, 72.6, 71.7, 71.2, 68.4, 56.4, 54.2, 52.9, 50.8, 51.7, 44.4, 43.6, 43.3, 41.8, 39.7–22.6 (aliphatic CH3) 13.4 (CH3); [α]D25 = +56.0 (c 0.56, MeOH); EIMS: calcd for C127H227N21NaO39+ 2693.64 Found: 2693.89 [M+Na]+; Anal. Calcd. for C127H227N21O39 C, 57.08; H, 8.56; N, 11.01; Found: C, 57.67; H, 9.11; N, 11.41.
1,3,2',6',2''',6'''-Hexaammonium-guanidinyl-5"-deoxy-5"-N-(N,N-di-hexadecanoyl-Lys)-neomycin hexakis(trifluoroacetate) (3). Yield = 81%; Rf 0.10 (NH4OH/MeOH/CH2Cl2 2:9:4); 1H-NMR (300 MHz, CD3OD): δ 5.64 (d, 1H, J = 2.7 Hz), 5.18 (m, 2H), 5.06 (m, 1H), 5.03 (d, 1H, J = 5.3 Hz), 4.77 (m, 1H), 4.23 (m, 1H), 4.15–4.08 (m, 3H), 4.01 (m, 2H), 3.80–3.77 (m, 3H), 3.67 (m, 3H), 3.57–3.48 (m, 6H), 3.42 (m, 2H), 3.37 (m, 1H), 3.19 (t, 1 H, J = 7.3 Hz), 2.24 (m, 3H), 2.19 (m, 2H), 1.72 (m, 2H), 1.61 (m, 4H), 1.46 (m, 2H), 1.29 (s, 49H), 0.91 (t, 6H, J = 6.5 Hz); 13C-NMR (75 MHz, CD3OD): δ 177.6 (amide C=O), 177.4 (×2) (peptide amide C=O), 159.6, 159.2 (×4), 158.6 (guanidine C=NH), 100.6, 97.9, 97.6 (anomeric carbons), 88.4, 86.6, 83.9, 82.4, 79.6, 79.4, 76.4, 75.2, 74.0, 72.7, 72.0, 71.4, 70.9, 69.8, 57.1, 55.4, 52.9, 51.7, 50.3, 43.6, 41.9, 41.7, 38.9–23.7 (aliphatic CH2), 14.7 (aliphatic CH3); [α]D25 = +44.0 (c 0.67, MeOH); EIMS: calcd for C67H131N21O15+ 1471.01 Found: 1471.22 [M+H]+; Anal. Calcd. for C79H137F18N21O27 C, 44.03; H, 6.41; F, 15.87; N, 13.65; Found: C, 44.22; H, 6.82; F, 16.11; N, 14.15.
5''-Deoxy-5"-N-((N,N-di-nonadecanoyl-Lys)-1,3,2',6',2''',6'''-hexa-N-(tert-butoxycarbonylguanidino)-5"-deoxyneomycin (4a). Yield = 71%; Rf 0.32 (MeOH/CH2Cl2 1:9); 1H-NMR (300 MHz, CD3OD and CDCl3): δ 5.74 (s, 1H), 5.05 (m, 2H), 4.74 (m, 1H), 4.55 (m, 1H), 4.38 (m, 2H), 4.18 (m, 3H), 4.03 (m, 1H), 3.99 (m, 2H), 3.83 (m, 3H), 3.72–3.35 (m, 6H), 3.23 (t, 2H, J = 9.3 Hz), 3.17 (d, 1H, J = 6.7 Hz), 3.43 (d, 3H, J = 13.4 Hz), 2.25 (t, 2H, J = 6.9 Hz), 2.17 (t, 2H, J = 7.3 Hz), 2.03 (m, 2H), 1.72 (m, 2H), 1.50 (s, 108H), 1.30 (s, 74H), 0.89 (t, 6H, J = 7.1 Hz); 13C-NMR (75 MHz, CD3OD, HSQC): δ 176.2 (amide C=O), 176.1 (×2) (peptide amide C=O), 164.6–163.9 (Boc C=O), 158.2–157.3 (guanidine C=NH), 154.4–153.7 (Boc C=O), 111.3, 97.7, 95.8 (anomeric carbons), 86.8, 85.1–84.0, 82.0, 80.9–80.3, 78.7, 78.5, 75.3, 74.2, 73.5, 72.6, 71.7, 71.2, 70.6, 69.4, 67.6, 67.3, 66.2, 63.3, 53.9, 50.1, 51.4, 48.1, 47.3, 42.6, 41.3, 40.0, 39.7–22.6 (aliphatic CH2) 13.4 (aliphatic CH3); [α]D25 = +48.0 (c 0.49, MeOH); EIMS: calcd for C135H243N21NaO39+ 2815.77 Found: 2816.45 [M+Na]+; Anal. Calcd for C135H243N21O39 C, 58.23; H, 8.80; N, 10.56; Found: C, 58.44; H, 9.10; N, 10.56.
1,3,2',6',2''',6'''-Hexaammonium-guanidinyl-5''-deoxy-5''-N-(N,N-di-nonadecanoyl-Lys)-neomycin hexakis(trifluoroacetate) (4). Yield = 84%; Rf 0.10 (NH4OH/MeOH/CH2Cl2 2:9:4); 1H-NMR (300 MHz, CD3OD and CDCl3): δ 5.06 (m, 2H), 4.74 (m, 2H), 4.08 (m, 2H), 3.81 (m, 3H), 3.60 (m, 1H), 3.50 (m, 5H), 3.43 (m, 3H), 3.42–3.26 (m, 10H), 2.91 (m, 1H), 1.95 (dt, 4H, J = 6.3 Hz), 1.33 (m, 3H), 1.03 (s, 74H), 0.89 (t, 6H, J = 7.1 Hz); 13C-NMR (75 MHz, CD3OD): δ 177.2 (amide C=O), 176.4 (×2) (peptide amide C=O), 164.0–162.7 (TFA, q with J 2CF ~ 34.8 Hz), 159. 6 (×2), 159.4 (×3), 159.2 (guanidine C=NH), 124.1–112.4 (q with J 1CF ~ 292.0 Hz), 112.1, 99.2, 97.3 (anomeric carbons), 83.0, 79.2, 78.2, 75.9, 74.0, 73.8, 72.9, 72.4, 72.1, 71.9, 71.7, 70.8, 68.8, 61.9, 56.9, 55.1, 53.2, 51.8, 43.4, 43.0, 40.8, 40.0, 37.5–23.7 (aliphatic CH3) 14.7 (aliphatic CH3); [α]D25 = +42.0 (c 0.65, MeOH); EIMS: calcd for C75H148N21O15+ 1583.14 Found: 1583.14 [M+H]+; Anal. Calcd for C87H153F18N21O27 C, 46.09; H, 6.80; F, 15.08; N, 12.97; Found: C, 46.54; H, 7.22; F, 15.28; N, 13.19.
5''-N-(Hexadecafluorodecanoyl)-1,3,2',6',2''',6'''-hexa-N-(tert-butoxycarbonyl-guanidino)-5''-deoxy-neomycin (6a). Yield = 86%; Rf 0.36 (MeOH/CH2Cl2 1:9); 1H-NMR (300 MHz, CD3OD): δ 5.88 (d, 1H, J = 3.68 Hz), 5.05 (dd, 2H, J =5.56, 1.70 Hz), 4.63–4.55 (m, 2H), 4.38 (m, 1H), 4.32–4.23 (m, 3H), 4.16 (m, 1H), 4.05–3.68 (m, 10H), 3.59–3.44 (m, 5H), 3.27 (m, 1H), 2.75–2.45 (m, 4H), 2.2 (m, 1H), 1.59–1.45 (m, 108H); 13C-NMR (75 MHz, CD3OD): δ 172.4,164.6.164.5, 164.3, 164.2, 164.1, 164.0, 158.8, 158.0, 157.9, 157.8, 157.6, 157.5, 154.7, 154.4, 154.2, 154.1, 154.0, 153.3, 153.2, 151.4, 140.5, 129.9, 127.7, 123.4, 120.4, 119.2, 114.9, 113.3, 99.3, 97.1, 89.1, 84.8, 84.7, 84.6, 84.5, 84.1, 84.0, 83.0, 80.7, 80.6, 80.5, 80.4, 80.3, 77.1, 76.7, 75.6, 74.3, 73.3, 72.0, 71.1, 68.1, 55.5, 53.0, 51.8, 50.2, 44.3, 44.0, 41.7, 35.2, 28.8–27.6 (Boc CH3 and aliphatic CH2).
1,3,2',6',2''',6'''-Hexaammonium-guanidinyl-5''-deoxy-5''-N-(hexadecafluorodecanoyl-neomycin hexakis(trifluoroacetate) (6). Yield = 91%; Rf 0.19 (NH4OH/MeOH/CH2Cl2 1:5:5); 1H-NMRR (300 MHz, CD3OD): δ 5.91 (d, 1H, J = 2.9 Hz), 5.11 (d, 1H, J = 3.7 Hz), 5.01 (br s, 1H), 4.25–4.20 (m, 1H), 4.14 (dd, 1H, J = 6.2, 3.8 Hz), 4.08 (dd, 1H, J = 10.2, 6.0 Hz), 4.02 (dd, 1H, J = 5.6, 2.5 Hz), 3.98 (dd, 1H, J = 8.6, 4.2 Hz), 3.81 (m, 5H), 3.66–3.49 (m, 9H), 3.38 (m, 3H), 2.73–2.40 (m, 4H), 2.13 (m, 1H), 1.74 (m, 1H); 13C-NMR (75 MHz, CD3OD): δ 173.77 (NH-CO-), 163.3 (TFA), 159.62, 159.51, 159.30, 159.25, 159.18, 158.56 (–NHC(NH)NH2), 129.85, 123.85, 123.44, 120.45, 120.09, 119.21, 116.22 (F- attached carbons), 111.63, 100.14, 97.19 (anomeric carbons), 87.67, 82.18, 80.07, 77.50, 76.65, 75.60, 74.22, 74.04, 71.97, 71.50, 70.89, 68.81, 57.14, 55.36, 53.63, 51.75, 43.46, 43.14, 42.34, 33.79, 28.10, 27.57; EIMS: calcd. for C40H63F17N19O13+ 1340.45. Found: 1340.9 [M+H]+.
3.7. Determination of the MIC Values for Aminoglycoside-Lipid Conjugates 1–6
Bacterial isolates were obtained from the American Type Culture Collection (ATCC). Isolates were kept frozen in skim milk at −80 °C until minimum inhibitory concentration (MIC) testing was carried out. Following two subcultures from frozen stock, the in vitro activities of peptides were determined by macrobroth dilution in accordance with the Clinical and Laboratory Standards Institute (CLSI) 2006 guidelines [19]. Stock solutions of peptides were prepared and dilutions made as described by CLSI. Test tubes contained doubling antimicrobial dilutions of cation adjusted Mueller-Hinton broth and inoculated to achieve a final concentration of approximately 5 × 105 CFU/mL then incubated in ambient air for 24 h prior to reading. Colony counts were performed periodically to confirm inocula. Quality control was performed using ATCC QC organisms.
3.8. Determination of the Hemolytic Activity for Aminoglycoside-Lipid Conjugates 5–6
Freshly isolated human erythrocytes isolated as recently described [20] were incubated for 1 h at 37 °C with 50, 100 µg/mL solution of peptide dissolved in PBS. The samples were centrifuged (4,000 rpm, 5 min) before the absorbance at 540 nm of the supernatant was measured by a microtiter negative and positive controls, respectively. Peptide concentrations corresponding to 50% hemolysis (EC50) were determined from the dose-response curves.
4. Conclusions
Six polycationic amphiphiles were synthesized, with the backbone of neomycin B providing cationic charge and either palmitic and arachidic bilipids or fluorinated undecanoic monolipid the hydrophobic region. The effect of basicity was evaluated by converting all six amine groups on neomycin B to guanidines, and all six compounds were tested against a panel of clinically relevant and ATCC Gram positive and Gram negative bacteria. Guanidinylation was found to increase activity 2–8 fold, with di-palmitoyl guandilyated neomycin B (3) displaying good activity against a number of Gram positive bacteria. When compared to previously synthesized monolipid PAs, compounds 1–6 displayed reduced activity. However, the PAs with fluorinated tails, 5 and 6, had significantly less toxicity towards red blood cells, suggesting that fluorination may increase the therapeutic window of membrane-active amphiphiles.
Acknowledgements
The authors thank the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Institutes for Health Research (CIHR) and the Manitoba Health Research Council (MHRC) for financial support.
Footnotes
Sample Availability: Samples of the compounds 1-6 are available from the authors.
References
- 1.Tadesse D.A., Zhao S., Tong E., Ayers S., Singh A., Bartholomew M.J., McDermott P.F. Antimicrobial Drug Resistance in Escherichia Coli from Humans and Food Animals, United States, 1950-2002. Emerg. Infect. Dis. 2012;18:741–749. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontes L.C., Neves P.R., Oliveira S., Silva K.C., Hachich E.M., Sato M.I., Lincopan N. Isolation of Pseudomonas Aeruginosa Coproducing Metallo-Beta-Lactamase SPM-1 and 16S rRNA Methylase RmtD1 in an Urban River. Antimicrob. Agents Chemother. 2011;55:3063–3064. doi: 10.1128/AAC.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonnell G., Russell A.D. Antiseptics and Disinfectants: Activity, Action, and Resistance. Clin. Microbiol. Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipsky B.A., Holroyd K.J., Zasloff M. Topical Versus Systemic Antimicrobial Therapy for Treating Mildly Infected Diabetic Foot Ulcers: A Randomized, Controlled, Double-Blinded, Multicenter Trial of Pexiganan Cream. Clin. Infect. Dis. 2008;47:1537–1545. doi: 10.1086/593185. [DOI] [PubMed] [Google Scholar]
- 5.Findlay B., Zhanel G.G., Schweizer F. Cationic Amphiphiles, a New Generation of Antimicrobials Inspired by the Natural Antimicrobial Peptide Scaffold. Antimicrob. Agents Chemother. 2010;54:4049–4058. doi: 10.1128/AAC.00530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brogden K.A. Antimicrobial Peptides: Pore Formers Or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 7.Yeung A.T., Gellatly S.L., Hancock R.E. Multifunctional Cationic Host Defence Peptides and their Clinical Applications. Cell Mol. Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bera S., Zhanel G.G., Schweizer F. Synthesis and Antibacterial Activity of Amphiphilic Lysine-Ligated Neomycin B Conjugates. Carbohydr. Res. 2011;346:560–568. doi: 10.1016/j.carres.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Bera S., Zhanel G.G., Schweizer F. Antibacterial Activity of Guanidinylated Neomycin B- and Kanamycin A-Derived Amphiphilic Lipid Conjugates. J. Antimicrob. Chemother. 2010;65:1224–1227. doi: 10.1093/jac/dkq083. [DOI] [PubMed] [Google Scholar]
- 10.Bera S., Zhanel G.G., Schweizer F. Antibacterial Activities of Aminoglycoside Antibiotics-Derived Cationic Amphiphiles. Polyol-Modified Neomycin B-, Kanamycin A-, Amikacin-, and Neamine-Based Amphiphiles with Potent Broad Spectrum Antibacterial Activity. J. Med. Chem. 2010;53:3626–3631. doi: 10.1021/jm1000437. [DOI] [PubMed] [Google Scholar]
- 11.Bera S., Zhanel G.G., Schweizer F. Evaluation of Amphiphilic Aminoglycoside-Peptide Triazole Conjugates as Antibacterial Agents. Bioorg. Med. Chem. Lett. 2010;20:3031–3035. doi: 10.1016/j.bmcl.2010.03.116. [DOI] [PubMed] [Google Scholar]
- 12.Bera S., Zhanel G.G., Schweizer F. Design, Synthesis, and Antibacterial Activities of Neomycin-Lipid Conjugates: Polycationic Lipids with Potent Gram-Positive Activity. J. Med. Chem. 2008;51:6160–6164. doi: 10.1021/jm800345u. [DOI] [PubMed] [Google Scholar]
- 13.Francois B., Russell R.J., Murray J.B., Aboul-ela F., Masquida B., Vicens Q., Westhof E. Crystal Structures of Complexes between Aminoglycosides and Decoding A Site Oligonucleotides: Role of the Number of Rings and Positive Charges in the Specific Binding Leading to Miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael K., Wang H., Tor Y. Enhanced RNA Binding of Dimerized Aminoglycosides. Bioorg. Med. Chem. 1999;7:1361–1371. doi: 10.1016/S0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 15.Findlay B., Zhanel G.G., Schweizer F. Investigating the Antimicrobial Peptide “Window of Activity” using Cationic Lipopeptides with Hydrocarbon and Fluorinated Tails. Int. J. Antimicrob. Agents. 2012;40:36–42. doi: 10.1016/j.ijantimicag.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Baker T.J., Luedtke N.W., Tor Y., Goodman M. Synthesis and Anti-HIV Activity of Guanidinoglycosides. J. Org. Chem. 2000;65:9054–9058. doi: 10.1021/jo001142e. [DOI] [PubMed] [Google Scholar]
- 17.Zhanel G.G., DeCorby M., Laing N., Weshnoweski B., Vashisht R., Tailor F., Nichol K.A., Wierzbowski A., Baudry P.J., Karlowsky J.A., et al. Antimicrobial-Resistant Pathogens in Intensive Care Units in Canada: Results of the Canadian National Intensive Care Unit (CAN-ICU) Study, 2005-2006. Antimicrob. Agents Chemother. 2008;52:1430–1437. doi: 10.1128/AAC.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Chiang F.I., Wu L., Czyryca P.G., Li D., Chang C.W. Surprising Alteration of Antibacterial Activity of 5"-Modified Neomycin Against Resistant Bacteria. J. Med. Chem. 2008;51:7563–7573. doi: 10.1021/jm800997s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standard Institute. Clinical and Laboratory Standard Institute; Wayne, PA, USA: 2006. Document: M100-S16 CLSI.NCCLS. [Google Scholar]
- 20.Dathe M., Schumann M., Wieprecht T., Winkler A., Beyermann M., Krause E., Matsuzaki K., Murase O., Bienert M. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry. 1996;35:12612–12622. doi: 10.1021/bi960835f. [DOI] [PubMed] [Google Scholar]