Abstract
In March 2017, a novel highly pathogenic avian influenza A(H7N9) virus was detected at two commercial broiler breeder facilities in Tennessee, United States. In this study, a wild bird low pathogenic avian influenza A virus, A/blue-winged teal/Wyoming/AH0099021/2016(H7N9), was shown to be the probable precursor of the novel H7N9 virus; this low pathogenic virus has eight possible progenitor genes sharing > 99% sequence identity with the novel H7N9 virus. Phylogeographic analyses showed that viral gene constellations that formed and circulated among dabbling ducks contributed to the emergence of the novel H7N9 virus. This is in contrast to the virus that caused the 2016 H7N8 outbreak, which had more genetic contributions from viruses circulating among diving ducks. Study findings support the need for ongoing wild bird surveillance to monitor circulating viruses and to understand possible evolutionary pathways of virus emergence in poultry.
Keywords: avian influenza A virus, high pathogenic avian influenza, low pathogenic avian influenza, H7N9, wild birds, domestic poultry, dabbling duck, evolutionary pathway, phylogenetic analysis, phylogeographic analysis
Introduction
Influenza A viruses (IAVs) have been recovered from variety of bird species, including at least 105 species of 26 different families (1). Among wild birds, those of wetland and aquatic environments, such as birds in the order Anseriformes (particularly ducks, geese, and swans) and those in the order Charadriiformes (particularly gulls, terns, and waders) constitute the major natural IAV reservoir (2). These birds maintain a large IAV genetic pool, which contributes to the appearance of new IAVs in humans, lower mammals, and domestic poultry and other birds. Many Anseriformes and Charadriiformes birds perform regular long-distance migrations (3), and they are distributed globally, except in the most arid regions of the world (3). Wild bird migration facilitates virus transmission and genetic diversity through reassortments and adaptations among IAVs from different geographic locations.
It is well documented that wild birds have been the predominant source of IAVs detected in domestic poultry, and introductions of IAVs from wild birds to domestic poultry occasionally causes outbreaks among domestic poultry. Understanding the evolutionary pathway for emerging IAVs detected in domestic poultry, especially learning which wild bird species are involved in these introductions, will help us understand the natural history of IAVs and provide potential evidence to support the development of strategies for avian influenza surveillance among wild birds. Through retrospective analyses, Krauss and colleagues proposed shorebirds, gulls and dabbling ducks as the source of IAV introduction into domestic poultry in North America (4, 5). Through phylogeographic analyses, Xu et al. (6) suggested that viral gene constellations circulating among diving ducks can contribute toward the emergence of H7N8 IAVs that affect poultry, and Li et al. (7) indicated that, from 2001 to 2017, dabbling ducks and geese/swans could have facilitated emergences of the H5 IAVs associated with 18 individual introductions into domestic poultry in the United States. These studies suggested that certain species of wild bird may serve an important role in the maintenance, diversification, and transmission of IAVs in the wild bird reservoir.
After being introduced into domestic poultry, subtype H5 and H7 low pathogenic avian influenza (LPAI) viruses can mutate into highly pathogenic avian influenza (HPAI) viruses, often through acquisition of basic amino acids in the cleavage region of the HA protein by insertion or substitution (8) or through insertion of a long peptide sequence in the cleavage region of the HA protein from recombination with another gene segment(s) or host genome (9–11). However, whether and how a subtype H5 or H7 IAV would mutate from LPAI virus to HPAI virus cannot yet be predicted.
In March 2017, subtype H7N9 HPAI virus was detected in a commercial broiler breeder facility in Lincoln County, Tennessee, United States, with a single event of secondary spread within company. Subsequent surveillance detected H7N9 LPAI virus in Tennessee, as well as neighboring states of Alabama, Kentucky, and Georgia (12, 13). Nomenclature introduced in a previous study referred to the first identified HPAI virus as Commercial Poultry Farm (CPF)-TN-2017(H7N9) (7). Through genomic analyses, a prior report showed that it is probable that a H7N9 LPAI virus mutated to H7N9 HPAI virus after being introduced from wild birds to the commercial broilers in Tennessee (12, 14). However, the origin and evolution of wild bird virus precursors to CPF-TN-2017(H7N9), and which wild bird species contributed to such emergence is unclear. The aim of our study was to use phylogenetic and phylogeographic analyses to identify progenitor genes associated with CPF-TN-2017(H7N9), to illustrate the evolutionary pathway for the emergence of this virus, and to identify probable wild bird species that may contribute to its emergence.
Results
H7N9 HPAI viruses detected in Tennessee originated from IAVs in North American wild birds.
In this study, a total of eight H7N9 viruses were analyzed. Of these, three were LPAI isolates [A/chicken/Tennessee/17-007429-7/2017(H7N9), A/chicken/Tennessee/17-007429-3/2017(H7N9), and A/chicken/Tennessee/17-007431-4/2017(H7N9)], and five were H7N9 HPAI isolates representing the two affected premises: A/chicken/Tennessee/17-007147-3/2017(H7N9), A/chicken/Tennessee/17-007147-2/2017(H7N9), A/chicken/Tennessee/17-007147-5/2017(H7N9), A/chicken/Tennessee/17-007147-1/2017(H7N9) from the first premise, and A/chicken/Tennessee/17-008279-4/2017(H7N9) from the single event of secondary spread]. Sequence analyses showed that there are 98.13%–100.00% nucleotide sequence identity between HA genes of the H7N9 LPAI viruses and those of H7N9 HPAI viruses, 99.58%–100.00% nucleotide sequence identity between the NA genes of the H7N9 LPAI viruses and those of H7N9 HPAI viruses, and 99.72%–100.00% nucleotide sequence identity between six other genes of H7N9 LPAI viruses and those of H7N9 HPAI viruses. These viruses were collectively referred to as H7N9/TN.
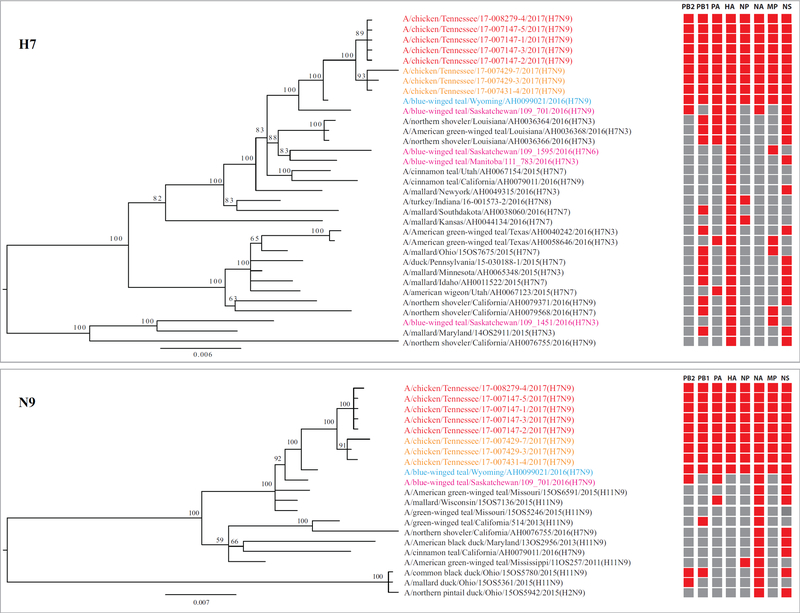
Consistent with findings in a prior study (12), our phylogenetic analyses of the HA, NA, and six internal genes suggested that these H7N9/TN viruses belong to the North American lineage and share high nucleotide sequence identity (>99%) with A/blue-winged teal/Wyoming/AH0099021/2016(H7N9) across all eight genes. Further analyses showed that the HA gene of H7N9/TN belongs to sub-lineage 3 (Figure S1), which also includes contemporary H7 viruses that are circulating among wild birds in North America and that have caused sporadic infections among US domestic poultry since 1996 (6). These H7N9/TN viruses are not genetically related to the enzootic H7N9 viruses in China nor to the enzootic H7N3 viruses in Mexico (Figure S1).
Molecular clock analyses suggest that the mean time to most recent common ancestor for H7N9/TN was during the 2016–17 fall–winter wild bird migration season (October 1, 2016, to January 14, 2017) (Table 1). The mean evolutionary rate of the H7 gene was 6.19 × 10−3 substitutions per site per year (subs/site/year) (95% highest posterior density = 5.62 × 10−3 – 6.80 × 10−3 subs/site/year), which is significantly higher than that for other gene segments (range, 2.58× 10−3 to 4.00 × 10−3 subs/site/year) (6).
Table 1.
Estimated time to most recent common ancestor (TMRCA) and nucleotide substitution rates for eight gene segments of H7N9 avian influenza viruses detected in chickens in Tennessee, United States, in 2017.
| Segment | Substitution rate (10−3 substitutions/site/year) | TMRCA | ||||
|---|---|---|---|---|---|---|
| Mean | 95% HPD low | 95% HPD high | Mean | 95% HPD low | 95% HPD high | |
| PB2 | 3.04 | 2.74 | 3.37 | Oct 10, 2016 | Jul 10, 2016 | Dec 28, 2016 |
| PB1 | 4.00 | 3.58 | 4.43 | Dec 5, 2016 | Oct 10, 2016 | Jan 28, 2017 |
| PA | 2.87 | 2.58 | 3.17 | Nov 30, 2016 | Sep 24, 2016 | Jan 27, 2017 |
| HA | 6.19 | 5.62 | 6.80 | Dec 15, 2016 | Oct 15, 2016 | Feb 5, 2017 |
| NP | 3.55 | 2.89 | 4.19 | Jan 14, 2017 | Nov 27, 2016 | Feb 24, 2017 |
| NA | 3.35 | 2.93 | 3.79 | Oct 11, 2016 | Jul 11, 2016 | Dec 31, 2016 |
| MP | 2.58 | 2.15 | 3.04 | Jan 11, 2017 | Nov 25, 2016 | Feb 22, 2017 |
| NS | 2.75 | 2.07 | 3.51 | Oct 1, 2016 | May 24, 2016 | Jan 19, 2017 |
HPD, highest posterior density.
Evolutionary pathways for emergence of H7N9/TN.
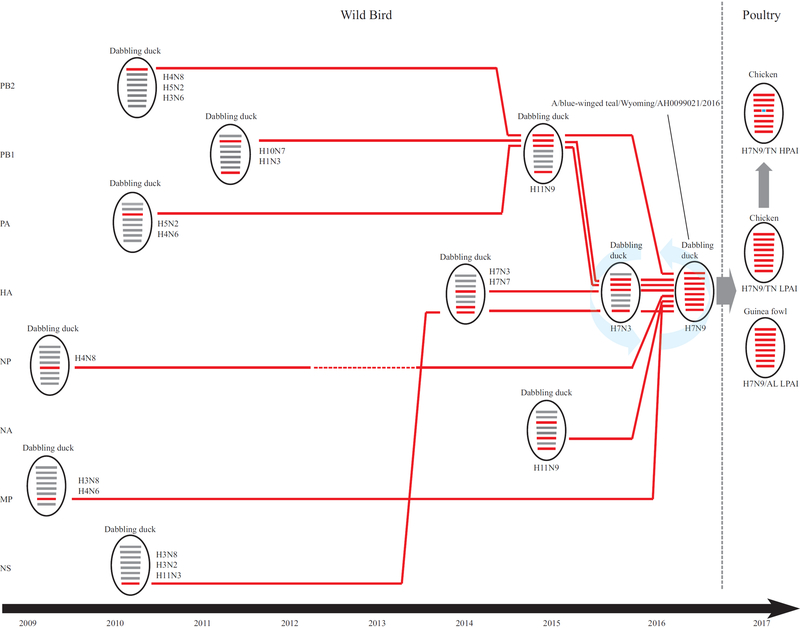
Our genotype analyses identified a single genotype for H7N9/TN HPAI viruses, and, as described above, A/blue-winged teal/Wyoming/AH0099021/2016(H7N9) had the same genotype; thus, this LPAI virus is a probable precursor virus for H7N9/TN. We further investigated possible genetic events that contributed to generation of this H7N9 precursor virus in wild birds. Examination of the eight gene segment–specific phylogenetic trees indicated that a group of H11N9 viruses isolated from dabbling ducks (referred to as H11N9–DD) in 2015 have three or four genetic segments, including the N9 gene, that are genetically close to those of the emerging A/blue-winged teal/Wyoming/AH0099021/2016(H7N9) virus. Furthermore, a group of H7N3 viruses isolated from dabbling ducks (referred to as H7N3–DD) in 2015 have three to four genes, including the HA gene, that are genetically close to genes of the emerging H7N9 virus. Thus, the genetic makeup of A/blue-winged teal/Wyoming/AH0099021/2016(H7N9) seems to have been derived from H7N3-DD and H11N9-DD (Figures 1 and 2).
Figure 1.
Genotypic analysis of H7 and N9 subtype viruses associated with novel H7N9 highly pathogenic avian influenza (HPAI) virus (H7N9-TN) responsible for a 2017 outbreak in Tennessee, United States. Genotypes were assigned by unique combinations of sub-lineages for each gene; sub-lineages were determined based on tree topology with at least a bootstrap value of 70 and a nucleotide sequence identity of 95%. Red squares indicate genotypes the same as that of the H7N9-TN virus; gray squares indicate other genotypes. Red font indicates H7N9-TN HPAI viruses; orange font indicates H7N9-TN low pathogenic (LPAI) viruses; blue font indicates the probable precursor H7N9 LPAI virus in wild birds; pink font indicates isolates from Canada. Scale bars indicate the scale of phylogenetic distance.
Figure 2.
Evolutionary pathway, from 2009 to 2017, leading to the generation of novel H7N9 highly pathogenic avian influenza (HPAI) viruses detected in chickens in Tennessee, United States, in 2017. Horizontal bars within ovals represent, from top to bottom, the eight gene segments shown on the right. Red horizontal bars indicate the gene constellations associated with novel H7N9 HPAI and LPAI viruses (shown on the right) detected in chickens in Tennessee and guinea fowl in Alabama (AL), United States, in 2017. Red horizontal bar with a blue mark indicates H7 gene of HPAI viruses that acquired additional amino acid insertions.
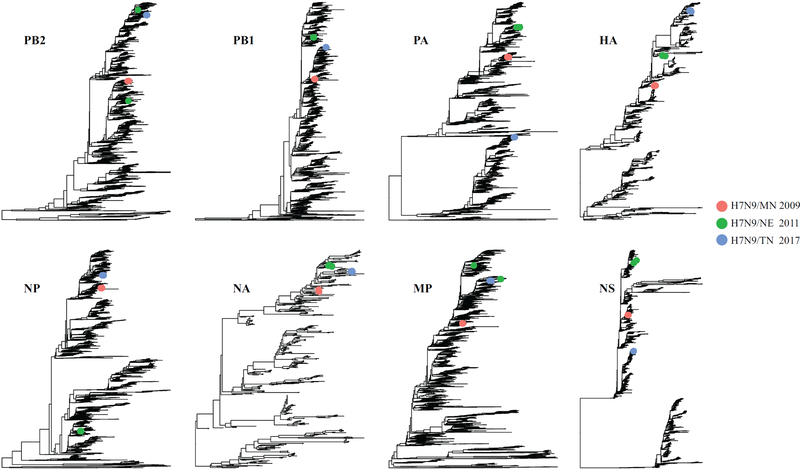
We performed phylogenetic analyses to identify possible genetic connections between the H7N9/TN viruses and two H7N9 low pathogenic avian influenza viruses, A/mallard/Minnesota/AI09–3770/2009 (H7N9) (abbreviated as H7N9/MN) and A/goose/Nebraska/17097–4/2011 (H7N9) (abbreviated as H7N9/NE), from two most recent cases [BYP-MN-2009(H7N9) and BYP-NE-2011(H7N9)] detected in domestic poultry of the United States. Results showed only four gene segments of H7N9/MN and that H7N9/NE HA are genetically associated with H7N9/TN. Specifically, HA, NA, and MP genes of both H7N9/MN and H7N9/NE, PB1 of H7N9/MN, and PB2 of H7N9/NE belong to the same genetic clade for the corresponding gene segment of H7N9/TN (Figure 3). Molecular characterization showed that the sequence identities between HA and NA genes of H7N9/TN and those of H7N9/MN were 95.54% and 97.10%, respectively and that the sequence identities between HA and NA genes of H7N9/TN and those of H7N9/NE were 96.25% and 97.96%.
Figure 3.
Phylogenetic relationship of eight gene segments between H7N9 outbreak in poultry in 2017 (H7N9/TN) and two historical H7N9 poultry detections in 2009 (H7N9/MN) and 2011 (H7N9/NE). As shown in the legend, H7N9/MN, H7N9/NE and H7N9/TN were indicated by red, greed and blue dots, respectively.
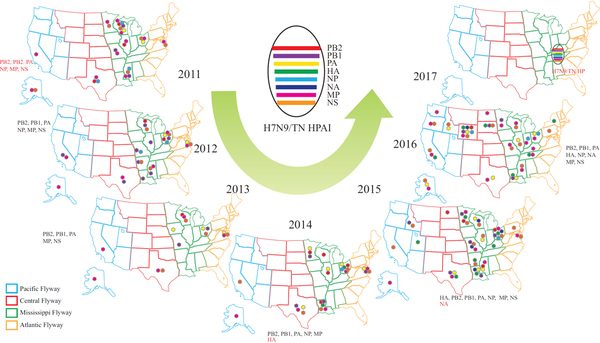
Spatial analyses from 2011 to 2016 of genetic clusters associated with H7N9 viruses suggested the progenitor gene segments were widely spread across all four North American migratory bird flyways (i.e., the Atlantic, Mississippi, Central, and Pacific Flyways (Figure 4), and the data are consistent with those of a previous publication that reported a lack of knowledge regarding the geographic patterns for different subtypes of IAV HA (except H5) and NA genes and for internal IAV genes (7).
Figure 4.
Temporal and geographic distribution, during 2011–2017, of gene constellations associated with the emergence of H7N9-TN highly pathogenic avian influenza (HPAI) virus in the United States in 2017. Dots indicate detections of specific gene constellations (see key) in viruses from US states within North American wild bird migration flyways. Four wild bird migratory flyways were indicated by different colors on maps. Red font indicates genotypes the same as that of the H7N9-TN virus of a specific gene segment firstly identified in that year.
The IAVs from dabbling ducks contributed to emergence of H7N9/TN.
To identify the specific wild bird species that contributed to the emergence of viruses associated with H7N9/TN viruses, we characterized the diffusion pathways of IAVs from different host groups. Results from phylogeographic analyses suggested that all gene segments except NP are significantly associated with one functional group of bird species: dabbling ducks (Table 2). Specifically, very strong support (Bayes factor = 189.32) was shown for the transmission of PA gene from dabbling ducks to the chickens in which H7N9/TN virus was detected; strong support was shown for transmission of PB2, PB1, HA, and NA genes (Bayes factor = 12.09, 36.53, 18.29, and 21.56, respectively) from dabbling ducks to the chickens; and support was shown for transmission of MP and NS genes (Bayes factor = 6.79 and 7.88, respectively) from dabbling ducks to the chickens. However, the Bayes factor was not significant to support that the NP gene was transmitted from dabbling ducks or any other wild bird species to the chickens in which H7N9/TN virus was detected.
Table 2.
Statistical support for the transmission of a novel H7N9 avian influenza virus from wild birds to chickens in Tennessee, United States, in 2017.
| Segment | Host group | Indicatora | Bayes factora | Level of support |
|---|---|---|---|---|
| PB2 | Dabbling duck | 0.74 | 12.09 | Strong support |
| PB1 | Dabbling duck | 0.89 | 36.53 | Strong support |
| PA | Dabbling duck | 0.97 | 189.32 | Very strong support |
| HA | Dabbling duck | 0.74 | 18.29 | Strong support |
| NP | - | |||
| NA | Dabbling duck | 0.77 | 21.56 | Strong support |
| MP | Dabbling duck | 0.52 | 6.79 | Support |
| NS | Dabbling duck | 0.60 | 7.88 | Support |
Only indicator > 0.5 and Bayes factor > 3 are considered as indicators of significant transmission.
-, No significant transmission was observed.
H7N9 LPAI virus evolved into an HPAI strain in chickens and has unique insertions from historical outbreaks.
With both the LPAI and HPAI H7N9 isolates detected in chicken flocks in Tennessee, we investigated how H7N9 LPAI virus evolved into an HPAI strain in chickens. Consistent with a prior study (12), we identified a 9–amino acid insertion at the cleavage site of HA, and BLAST analyses further demonstrated the 28-nucleotide insertion was identical to a fragment sequence of 28s rRNA in chicken genome. In chicken genome, there were a total of 75 copies of these 28-nucleotide sequence insertions across multiple chromosomes. Blast analyses also identified fragments (< 28 nucleotides) of these insertions in the 28s rRNA across a number of parasite and bacterial genomes.
Discussion
In this study, we characterized the origins of a subtype H7N9 LPAI virus that was introduced in 2017 from wild birds to domestic poultry in Tennessee and subsequently evolved into an HPAI virus, CPF-TN-2017(H7N9), among domestic poultry. Consistent with previous findings (7), our results identified a waterfowl virus, A/blue-winged teal/Wyoming/AH0099021/2016(H7N9), as the probable LPAI precursor virus; each of the eight gene segments in this virus shared > 99% sequence identity with those of the emerging H7N9 virus. Genetic analyses on the viruses isolated from CPF-TN-2017(H7N9) and those from two most recent introductions, BYP-MN-2009(H7N9) and BYP-NE-2011(H7N9), suggested CPF-TN-2017(H7N9) is a new and independent introduction directly from wild birds. Thus, the findings of this study support the premise that waterfowl are a key source for the emergence of economically costly IAVs among domestic poultry in North America.
The temporal and spatial analyses of progenitor genes associated with the H7N9 virus that emerged in Tennessee in 2017 suggest that emergence of a novel virus with risks to domestic poultry is complicated by multiple reassortment events. The analyses also suggest that progenitor genes associated with the emergence could be present in the wild bird population, especially dabbling ducks, across multiple migratory bird flyways and could have been present as early as 2009. Our findings support the premise that waterfowl maintain a large pool of genome-diverse IAVs and that gene flow has been very active among wild bird populations, both within and between migratory flyways. A group of H7N3 and H11N9 viruses identified from dabbling ducks from 2015 and 2016 likely contributed to the emergence of H7N9 virus, supporting the premise that dabbling ducks have the potential to maintain the virus pool associated with emergence in poultry. In an earlier study, diving ducks were shown to be associated with emergence of the H7N8 virus in Indiana, United States, in early 2016 (6); dabbing ducks and geese/swans were also suggested to be associated with emergence of 18 independent introductions of H5 LPAI viruses to domestic poultry in North America (7). In summary, while these studies repeatedly confirm waterfowl are primary sources of IAVs detected in domestic poultry, each introduction could be independent and be associated with one or more wild bird species sharing similar habitats and biologic properties.
Our findings suggest that the emerging H7N9 LPAIV in chickens acquired an insertion of a long nucleotide sequence from 28s rRNA, which is abundant in the host (i.e. chicken) genomes as well as in microbial genomes. Of interest, the size and sequences of the inserts for three individual cases were unique from each other (Figure 4-A). It is probable that the viruses acquired the insert through RNA non-homologous recombination, which has been proposed to occur rarely in negative-strand RNA viruses, including influenza viruses (15). Because 28s rRNA is one of the most abundant RNAs in host cells (16), it provides ample opportunities for RNA recombination events. On the other hand, it is unclear why the insert of a 28s rRNA gene fragment into H7 gene through RNA recombination was only identified in chickens but not in other bird species, although sequence analyses showed high nucleotide sequence identities between the 28s rRNA gene from chicken and those from other birds (e.g. mallard) in which the subtype H7 IAV was frequently identified.
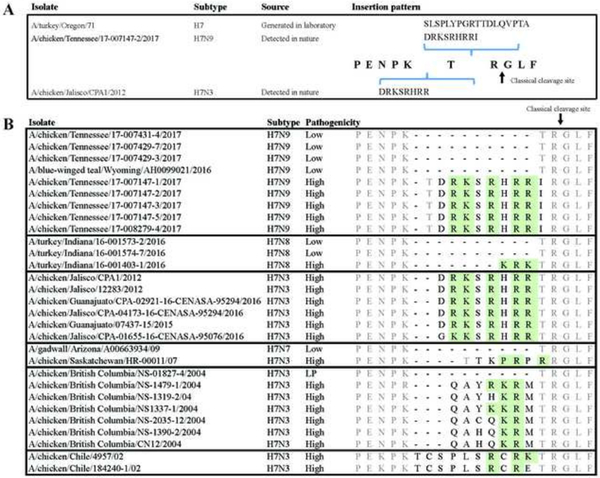
Insertion of a 28s rRNA fragment into the HA cleavage site was firstly reported in a laboratory strain of subtype H7N3 avian IAV (17) and a field strain of subtype H7N3 avian IAV (18). However, the inserts sequences were unique in the H7N9/TN virus and those H7N3 viruses (Figure 5-A). In addition, we compared the insertion sequences among the other subtype H7 HPAIVs detected in North American, including those from CPF-Chile-2002(H7N3), CPF-BC-2004(H7N3), CPF-SK-2007(H7N3), CPF-Mexico-2012(H7N3), CPF-IN-2016(H7N8) and CPF TN-2017(H7N9). Results showed that none of these insert sequences were identical, indicating a large diversity of the insert sequences at the HA cleavage site of subtype H7 HPAIVs (Figure 5-B).
Figure 5.
Insertion patterns at the cleavage site of HA proteins in H7 highly pathogenic avian influenza (HPAI) viruses from historic outbreaks in North America. A) Insertion patterns and positions among the three HPAI viruses that acquired insertions by recombination with host 28s rRNA. B) Insertion patterns among the H7 HPAI viruses. Green shadings indicate insertions of basic amino acids K and R of H7N9-TN outbreak and previous outbreaks, respectively. Inserted amino acids were colored in black and other amino acids were colored in gray.
Of note, non-homologous recombination has not been reported in subtype H5 avian IAV, which was frequently reported to mutate from a low pathogenic virus to a highly pathogenic virus. It is still unclear why subtype H7 avian IAV is more likely than subtype H5 avian IAV to have non-homologous recombination events.
A limitation of this study is that the genomic data from wild birds we used was acquired from public databases. The avian IAVs in wild birds are primarily detected in captured birds (e.g., birds caught by the United States Fish and Wildlife Service) or in birds killed by hunters; thus, the types and number of hosts associated with avian IAVs in the public databases may represent a biased population. To minimize potential biases from sampling, we used two data pre-processing steps: 1) we grouped the birds in species with similar habitats into the same functional groups; and 2) we balanced the number of samples from each functional group before conducting phylogeographic analyses. Despite the limitation of the genomic data, our findings suggest that the evolutionary pathway for emergence of H7N9/TN virus in 2017 was independent from that of H7N8 virus in Indiana in 2016.
In summary, our findings support the premise that wild bird H7 IAV introductions present an ongoing threat to domestic poultry, and the findings highlight the importance of avian IAV surveillance at the wild bird–domestic poultry interface. The integration of genomic approaches with surveillance activities will increase our understanding of the evolutionary pathways of IAVs and could help assess IAV-associated risks for domestic poultry.
Materials and Methods
Data.
To identify the genesis of novel H7N9 HPAI virus in Tennessee, we retrieved the genomic sequences for all avian-origin IAVs from the Influenza Virus Resource (https://www.ncbi.nlm.nih.gov/genomes/FLU) (19), the Influenza Research Database (https://www.fludb.org) (20), and GISAID (https://www.gisaid.org/) (21). In total, 29, 210 IAVs were collected worldwide, of which 9,395 (32%) were from North America (Table S1).
Sequence alignment and phylogenetic analysis.
Multiple sequence alignments were generated using MAFFT v7.273 (22). The preliminary phylogenic analyses were performed using an approximate maximum-likelihood method with a generalized time–reversible substitution model and the “CAT” approximation rate model in FastTree 2.1 (23). The refined phylogenetic tree for each gene segment was then inferred using a maximum-likelihood method by running RAxML v8.2.9 and using a GAMMA model of rate heterogeneity and a generalized time–reversible substitution model (24). Phylogenetic trees were visualized by ggtree v1.6.11 (25) and FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Genotype analyses and assignment of possible progenitor gene and potential precursor viruses.
The genotype for a specific influenza virus was determined by using a unique combination of genetic clusters for each segment of the virus, as described elsewhere (6). A minimum bootstrap value of 70 for tree topology and a nucleotide sequence identity of ≥ 95% were used to define genetic clusters. The possible progenitor gene and potential precursor viruses for a specific influenza virus were also determined based on tree topology and sequence identities for all viruses detected the same year as or in a year prior to detection of the testing virus. A possible progenitor gene was assigned only when the following criteria were met: 1) the candidate gene and the testing gene were located in the same genetic cluster with a minimum bootstrap value of 70; 2) the candidate gene and the testing gene shared ≥ 98% nucleotide sequence identity; 3) the candidate gene had the maximum nucleotide sequence identity in its genetic cluster; and 4) the candidate gene was not detected in a year later than the year the testing virus was detected. A virus was assigned as a potential precursor virus when it had a minimum of three possible progenitor genes for a testing virus.
Molecular clock analyses.
The Bayesian Markov Chain Monte Carlo method implemented in BEAST v1.8.4 (26) was used to estimate the time-scale phylogenies, substitution rate, and time to most recent common ancestor for each gene segment. Molecular clock analyses were performed as previously described (6). SRD06 partitioned substitution model, uncorrelated lognormal relaxed clock model, and Bayesian skyline coalescent tree prior were applied in the analyses. Two independent runs with a chain length of 100 million (sample frequency = 10,000) were performed for each gene segment. Results from two independent runs were combined by LogCombiner v1.8.4 (http://beast.community/logcombiner) and then analyzed by Tracer v1.6 using a 10% burn-in rate. The maximum clade credibility trees were summarized using TreeAnnotator v1.8.4 (http://beast.community/treeannotator) and edited by FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Phylogeographic analyses to infer the potential wild bird host group(s) contributing to emergence of the influenza virus causing a specific outbreak in domestic poultry.
Phylogeographic analyses were performed, as described elsewhere (6, 27), to infer the host group associated with the novel CPF-TN-2017(H7N9) virus. In brief, based on ecologic attributes, the avian species were categorized into 9 functional groups: dabbling duck, diving/sea duck, goose/swan, gull/tern/seabird, poultry, raptor, shorebird, other avian, and unknown. To avoid sampling bias, we used a resampling strategy, limiting samples to 10 for each group in each year. Phylogeographic analyses were performed using Beast v1.84 (26). An asymmetric substitution model with Bayesian stochastic search variable selection and a strict clock model were applied in the analyses. The Markov Chain Monte Carlo chain length was set to 100 million, with sampling every 10,000 states. The convergence of each run was checked by Tracer v1.6 (http://beast.community/tracer) before continuing to the next step. All poorly configured states were removed according to a 10% burn-in rate. Maximum clade credibility phylogenetic trees were generated using TreeAnnotator v1.8.4 (http://beast.community/treeannotator) and visualized by using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). Bayes factor was calculated to indicate the statistical support level for transmission of H7N9 virus from wild birds to domestic poultry. Significant statistical support was indicated by a combination of Bayes factor ≥ 3 and mean indicator ≥ 0.5. Different statistical support levels were defined as follows: 3 ≤ Bayes factor ≤ 10 indicates support; 10 ≤ Bayes factor ≤ 100 indicates strong support; 100 ≤ Bayes factor ≤ 1,000 indicates very strong support; and Bayes factor ≥ 1,000 indicates decisive support.
Supplementary Material
Acknowledgement
This project was supported by the United States Department of Agriculture and the National Institutes of Health (NIH) [grant number R21AI135820].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. 2006. Global patterns of influenza a virus in wild birds. Science 312:384–388. [DOI] [PubMed] [Google Scholar]
- 2.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous 1996. Handbook of the Birds of the World vol 3 Lynx Edicions, Barcelona,. [Google Scholar]
- 4.Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector-Borne & Zoonotic Diseases 4:177–189. [DOI] [PubMed] [Google Scholar]
- 5.Krauss S, Stucker KM, Schobel SA, Danner A, Friedman K, Knowles JP, Kayali G, Niles LJ, Dey AD, Raven G. 2015. Long-term surveillance of H7 influenza viruses in American wild aquatic birds: are the H7N3 influenza viruses in wild birds the precursors of highly pathogenic strains in domestic poultry? Emerging microbes & infections 4:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Ramey AM, Bowman AS, DeLiberto TJ, Killian ML, Krauss S, Nolting JM, Torchetti MK, Reeves AB, Webby RJ. 2017. Low-pathogenic influenza A viruses in North American diving ducks contribute to the emergence of a novel highly pathogenic influenza A (H7N8) virus. Journal of Virology 91:e02208–02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Bowman AS, DeLiberto TJ, Killian ML, Krauss S, Nolting JM, Torchetti MK, Ramey AM, Reeves AB, Stallknecht DE, Webby RJ, Wan X-F. 2018. Genetic evidence supports sporadic and independent introductions of low pathogenic H5 avian influenza A viruses from wild birds to domestic poultry in North America. Journal of Virology In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia M, Crawford JM, Latimer JW, Rivera-Cruz E, Perdue ML. 1996. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. Journal of General Virology 77:1493–1504. [DOI] [PubMed] [Google Scholar]
- 9.Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee C-W, Manvell RJ, Mathieu-Benson C, Moreno V, Pedersen JC. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg Infect Dis 10:693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y. 2005. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. Journal of General Virology 86:727–731. [DOI] [PubMed] [Google Scholar]
- 11.Maurer-Stroh S, Lee RT, Gunalan V, Eisenhaber F. 2013. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virology journal 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee DH, Torchetti MK, Killian ML, Berhane Y, Swayne DE. 2017. Highly Pathogenic Avian Influenza A(H7N9) Virus, Tennessee, USA, March 2017. Emerg Infect Dis 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.USDA-APHIS. 2017. Epidemiologic and other analyses of HPAI/LPAI affected poultry flocks: June 26, 2017 Report. USDA:APHIS:VS:STAS:Center for Epidemiology and Animal Health Fort Collins, CO: doi:Doc #391.0317 V2. 39 pgs. [Google Scholar]
- 14.USDA-APHIS. 2017. Epidemiologic and Other Analyses of HPAI/LPAI Affected Poultry Flocks June 26, 2017 Report. https://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/downloads/epi-ai.pdf. Accessed July 28.
- 15.Han GZ, Worobey M. 2011. Homologous recombination in negative sense RNA viruses. Viruses 3:1358–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ide S, Miyazaki T, Maki H, Kobayashi T. 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327:693–696. [DOI] [PubMed] [Google Scholar]
- 17.Khatchikian D, Orlich M, Rott R. 1989. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature 340:156–157. [DOI] [PubMed] [Google Scholar]
- 18.Maurer-Stroh S, Lee RT, Gunalan V, Eisenhaber F. 2013. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virology journal 10:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. Journal of virology 82:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squires RB, Noronha J, Hunt V, García - Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN. 2012. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza and other respiratory viruses 6:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogner P, Capua I, Lipman DJ, Cox NJ. 2006. A global initiative on sharing avian flu data. Nature 442:981–981. [Google Scholar]
- 22.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS one 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatakis A 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8:28–36. [Google Scholar]
- 26.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS computational biology 5:e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.