Abstract
Microglia are the resident immune cell of the central nervous system (CNS), and serve to protect and maintain the local brain environment. Microglia are critically dependent on signaling through the colony-stimulating factor 1 receptor (CSF1R); administration of CSF1R inhibitors that cross the blood brain barrier (BBB) lead to the elimination of up to 99% of microglia, depending on CNS exposure and treatment duration. Once microglia are depleted, withdrawal of inhibitor stimulates repopulation of the entire CNS with new cells, conceivably enabling a therapeutic strategy for beneficial renewal of the entire microglial tissue. We have explored the kinetics and limits of this repopulation event and show that the rate of microglial repopulation is proportional to the extent of microglial depletion – greater depletion of microglia results in more rapid repopulation. Using a CSF1R inhibitor formulation that eliminates ~99% of microglia within 7 days, we subjected mice to multiple rounds of elimination (7 days’ treatment) and repopulation (7 days’ recovery) and found that the brain only has the capacity for a single complete repopulation event; subsequent elimination and CSF1R inhibitor withdrawal fail to repopulate the brain. However, if the recovery time between, or after, cycles is extended sufficiently then the brain can ultimately repopulate. These kinetic studies define the opportunities and possible limits of the remarkable renewal capacities of microglia.
Keywords: proliferation, depletion, elimination, renewal, myeloid
Introduction
Microglia are the primary innate immune cells of the CNS, capable of sensing and responding to insults in the local environment (Nimmerjahn et al. 2005). During development, they also have a crucial role in synaptic sculpting (Paolicelli et al. 2011; Schafer et al. 2012), as well as regulation of neurogenesis (Antony et al. 2011); this function is maintained in the adult brain, where they continue to regulate neuronal structures and connections (Rice et al. 2017; Rice et al. 2015; Tremblay et al. 2010). Unique among myeloid cell types, microglia directly derive from yolk sac macrophage precursors, and colonize the CNS early in development (Ginhoux et al. 2010; Hoeffel et al. 2015). Due to the presence of the BBB, microglia are separated from myeloid cell populations in the periphery, and are long-lived and self-sustaining in their isolation (Lawson et al. 1992).
We previously discovered that microglia are dependent upon signaling through the colony-stimulating factor 1 receptor (CSF1R) for their survival (Elmore et al. 2014). Administration of BBB-permeant CSF1R inhibitors leads to the rapid elimination of up to 99% of microglia in a dose-dependent fashion (Dagher et al. 2015). Microglia can be depleted for the duration of CSF1R inhibitor administration, but withdrawal of inhibitors from the microglia-depleted brain stimulates rapid repopulation, which occurs from proliferation of cells contained within the CNS (Elmore et al. 2014; Huang et al. 2018). Overall, within ~14 days of inhibitor withdrawal, the CNS is fully repopulated with similar microglia densities, spacing, and morphologies to controls (Elmore et al. 2015). Similar repopulation kinetics have been reported in an alternative model of microglial depletion (Bruttger et al. 2015) utilizing an acute administration of diphtheria toxin, which also confirmed that repopulation occurred from within the CNS. These studies show that the adult brain has a remarkable capacity to repopulate the entire microglial tissue and gives insights into the regulation of microglial homeostasis and inflammatory responses.
Microglial elimination via CSF1R inhibitors has been shown to have wide ranging beneficial effects in various CNS disease models (Asai et al. 2015; Han et al. 2017; Janova et al. 2017; Klein et al. 2015; Schreiner et al. 2015; Spangenberg et al. 2016; Valdearcos et al. 2014), while depletion followed by repopulation in injured mice leads to resolution of chronic inflammatory responses, and promotes recovery (Rice et al. 2017). As such, CSF1R inhibitors are an attractive drug target for any disease characterized by neuroinflammation and microgliosis. This is further validated by the current clinical utility of CSF1R inhibitors (i.e. (Tap et al. 2015)) including a clinical trial demonstrating that CSF1R inhibitors also reduce microglia numbers in human brains (Butowski et al. 2016). Thus, it is imperative that we understand the consequences, kinetics, and capacity of the adult brain for microglial depletion and repopulation. Crucially, we find that the adult brain appears to only have the capacity for a single repopulation event, after which “normal” repopulation then fails to occur. However, if the recovery time between, or after, cycles is extended, then the brain can ultimately repopulate, suggesting an alteration in the mechanism or source of the cells that contribute to the new myeloid tissue. These findings help to define the complexities of myeloid repopulation and regulation in the adult brain.
Materials and Methods
Compounds
PLX3397 was provided by Plexxikon, Inc. and formulated in standard chow by Research Diets Inc. at 290- and 600 ppm. PLX3397 concentration in plasma and brain tissue were analyzed for pharmacokinetic (PK) data by Integrated Analytical Solutions, Inc. (Berkeley, CA, USA).
Animal Treatments
All rodent experiments were performed in accordance with animal protocols approved by the Institutional Animal Care and Use Committee at the University of California, Irvine. 2–9-month-old age-matched male C57b/6 wild-type or CX3CR1-GFP+/− mice were provided with control chow or PLX3397 chow for 7 days at a time, for 1, 2, or 3 cycles of treatment. Following each treatment cycle, PLX3397 was withdrawn and all mice maintained on control chow for 7 or 28 days. PDGFRa-CRE (C57BL/6-Tg(Pdgfrα-cre)1Clc/J) and Rosa26YFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J) reporter mice were obtained from The Jackson Laboratory. Crossing these mice yielded PDGFRα-CRE/Rosa26YFP (PGDFRα-YFP) progeny that express YFP in all cells that either transiently or constitutively express PGDFRα, which in the brain labels oligodendrocyte precursor cells and their progeny. Two to six-month-old PGDFRα-YFP mice were treated for seven days with PLX3397 (600 mg/kg in chow) to eliminate microglia, and then drug withdrawn for 7 days. At the conclusion of experiments, mice were sacrificed via CO2 inhalation and perfused transcardially with 1X phosphate-buffered saline (PBS). Brains were extracted and dissected down the midline, with one half flash-frozen on dry ice for subsequent RNA and protein analyses, and the other half drop-fixed in 4% paraformaldehyde (PFA) in 1X PBS. Fixed brains were cryopreserved in a 30% sucrose solution, frozen, and sectioned at 40 μm on a Leica SM2000 R sliding microtome for subsequent immunohistochemical analyses.
Confocal microscopy
Fluorescent immunolabeling followed a standard indirect technique (primary antibody followed by fluorescent secondary antibody), as previously described (Elmore et al., 2014). Primary antibodies used include Iba1 (1:1000; Wako), GFAP (1:10,000; Abcam), Cd11b (1:100; Bio-Rad). Colocalization scores and numbers of myeloid cells and astrocytes were determined using the colocalization and spots modules in Bitplane Imaris 7.5 software. 10X images were taken of hippocampal, cortical, and thalamic regions, and cell numbers were quantified throughout the image.
RNA Extraction and RNA-seq Processing
RNA was extracted and purified from frozen half brains using an RNA Plus Universal Mini Kit (Cat. #73404, Qiagen, Hilden, Germany) per the manufacturer’s instructions. Total RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip and Nanodrop absorbance ratios for 260/280nm and 260/230nm. Library construction was performed according to the Illumina TruSeq RNA v2 protocol. The input quantity for total RNA within the recommended range and mRNA was enriched using oligo dT magnetic beads. The enriched mRNA was chemically fragmented. First strand synthesis used random primers and reverse transcriptase to make cDNA. After second strand synthesis, the ds cDNA was cleaned using AMPure XP beads and the cDNA was end repaired and then the 3′ ends were adenylated. Illumina barcoded adapters were ligated on the ends and the adapter ligated fragments were enriched by nine cycles of PCR. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. Multiplexed libraries were sequenced using paired end 100 cycles chemistry for the HiSeq 2500. The version of HiSeq control software was HCS 2.2.58 with real time analysis software, RTA 1.18.64.
RNA-seq, Gene Ontology, and Pathway analysis
Single-end RNA-seq reads were first aligned using Bowtie v. 0.12.8 (Langmead et al. 2009) to the reference mouse transcriptome GENCODE,vM4. Gene expression level was quantified using RSEM v.1.2.12 (Li and Dewey 2011) with expression values normalized into Fragments Per Kilobase of transcript per Million mapped reads (FPKM). Samples displaying >20,000,000 uniquely mapped reads and >75% uniquely mapping efficiency were considered for downstream analyses. Differential expression analysis was performed using edgeR (Robinson et al. 2010) on protein-coding genes and lncRNAs. Differentially expressed genes were selected by using p-value <0.005 and false discovery rate (FDR) <0.05. Gene ontology analysis was performed with DAVID (Huang da et al. 2009a; Huang da et al. 2009b) Fisher’s exact test p-value lower than 0.05. KEGG pathways analysis was performed with PaintOmics v. 2.0 (Garcia-Alcalde et al. 2011).
Statistics
One-way ANOVAs with post-hoc Tukey’s Multiple Comparison tests were performed for all multiple comparisons (Graphpad Prism 6). For image quantification analyses, statistical significance was accepted at p<0.05; trends at p<0.10.
Results
The rate of microglial repopulation is proportional to the extent of microglial depletion
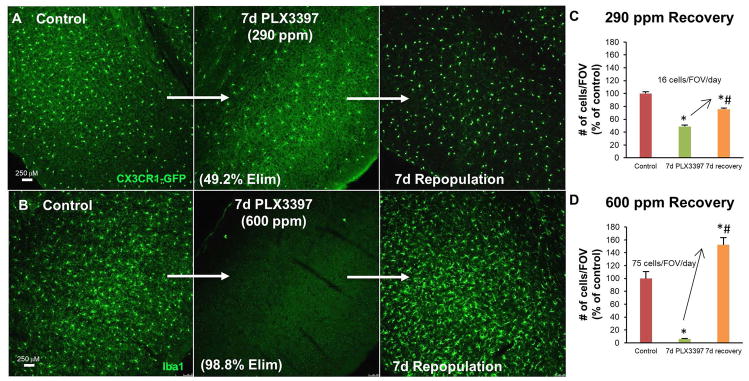
7 days’ treatment with the CSF1R/c-kit/Flt3 inhibitor PLX3397 at 290 ppm in chow reduced microglial numbers by 50%; upon inhibitor withdrawal only partial microglial repopulation was observed by 7 days (Figure 1A, C). To determine if a higher concentration of PLX3397 could more thoroughly ablate microglia in this time frame we reformulated chow at a concentration of 600 ppm in chow and found that 7 days’ treatment with this higher concentration led to 98.8% depletion of microglia (Figure 1B, D). Furthermore, upon inhibitor withdrawal, robust repopulation was observed, with brain microglial levels reaching 150% of controls within 7 days (Figure 1B, D). This overshoot is consistent with repopulation results from mice after being treated for 3 weeks with 290 ppm PLX3397, which led to a 95% depletion of microglia (Elmore et al. 2014). Comparing this rate of repopulation with that observed after 50% microglial depletion, it is apparent that repopulation is much more rapid and complete with higher levels of microglial depletion.
Figure 1.
Rate of microglial repopulation depends on extent of depletion. A, 10X representative images of cortical regions; mice treated with PLX3397 (290 ppm) for 7 days. B, 10X representative images of cortical regions; wild-type mice treated with higher dose of PLX3397 (600 ppm) for 7 days. C, Quantification of microglia cells/FOV with 290 ppm PLX3397, normalized to percent of Control, reveals an increase of an average of 16 cells/FOV/day between treatment and recovery groups; recovery group does not reach control levels. D, Quantification of Iba1+ cells/FOV with 600 ppm PLX3397, normalized to percent of Control, reveals an increase of an average of 75 cells/FOV/day between treatment and recovery groups; recovery group surpasses control levels. Error bars represent SEM, (n=4). p<0.05; significance symbols represent comparison groups: Control *, Elim 1 #.
A restricted capacity for microglial repopulation with repeated short cycles of thorough depletion
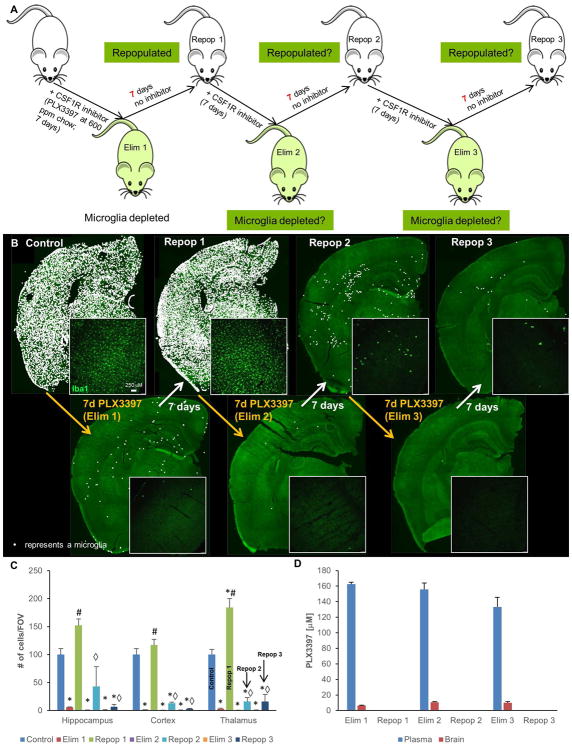
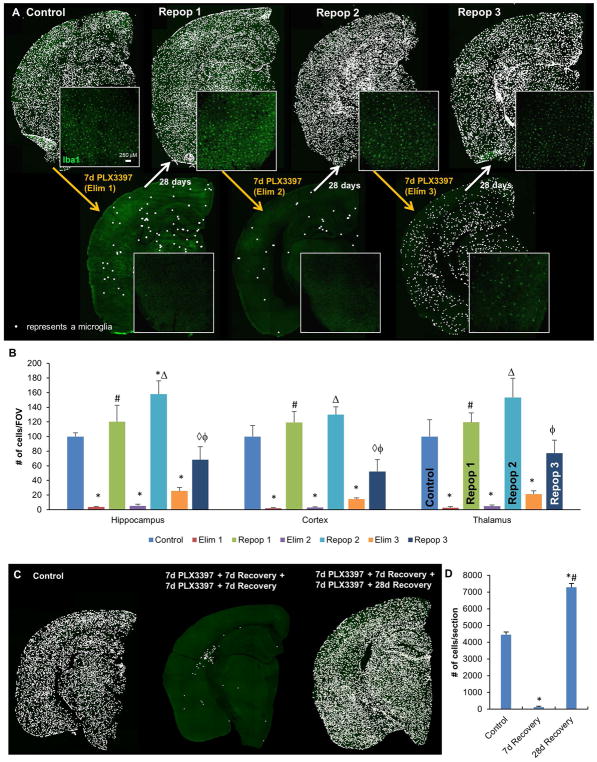
Having now identified a formulation of PLX3397 that potently eliminates ~99% of microglia within 7 days, we set out to determine whether there might be a finite capacity for repopulation with near-complete elimination cycles. To that end, we subjected 2-month old male wild-type mice to 3 cycles of elimination and repopulation with PLX3397 at 600 ppm in chow (7 days on drug, 7 days off drug per cycle; n=4/group; Figure 2A). Microglia were robustly eliminated for all 3 cycles, but repopulation was only observed in the first cycle (Figure 2B, C), with very few cells repopulating in cycles 2 and 3. Pharmacokinetic data shows that PLX3397 is rapidly washed out from both the plasma and brain, and that it does not accumulate with each progressive cycle (Figure 2D). Thus, the lack of repopulation in the second and third cycle is not due to accumulation of PLX3397. These results show that within these 7-day cycles, the adult brain has a restricted capacity for myeloid repopulation and appears to only have sufficient pools available for one complete repopulation event.
Figure 2.
Cyclic treatment of WT mice with 600 ppm PLX3397. A, Experimental paradigm: schematic depicting dose and duration of treatment and withdrawal cycles. B, Representative whole brain stitches of Iba1+ cells in each group. C, Quantification of number of Iba1+ cells/FOV for hippocampus, cortex, and thalamus. D, Pharmacokinetic data of PLX3397 levels in plasma and brain for each treatment cycle. Error bars represent SEM, (n=4). p<0.05; significance symbols represent comparison groups: Control *, Elim 1 #, Repop 1 ⋄, Elim 2 △, Repop 2 ϕ, Elim 3 θ, Repop 3 ●.
Transcriptome analyses of microglia depleted and repopulated brains
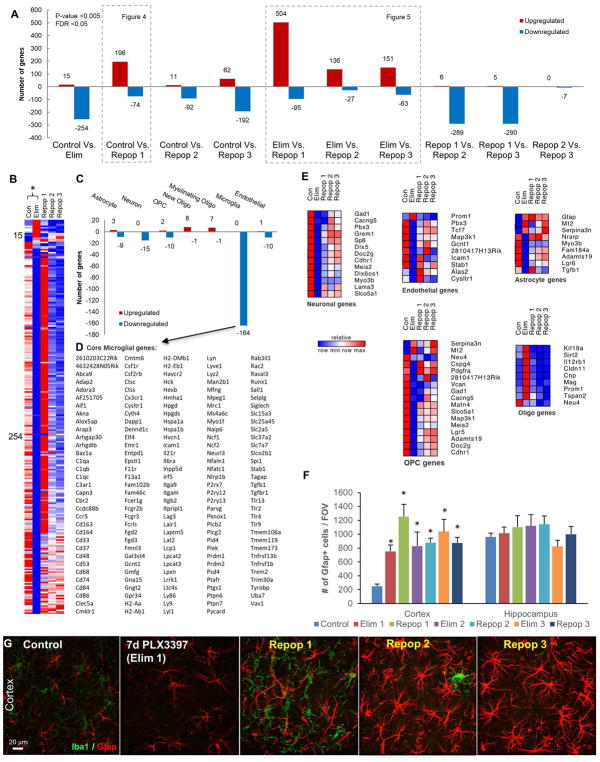
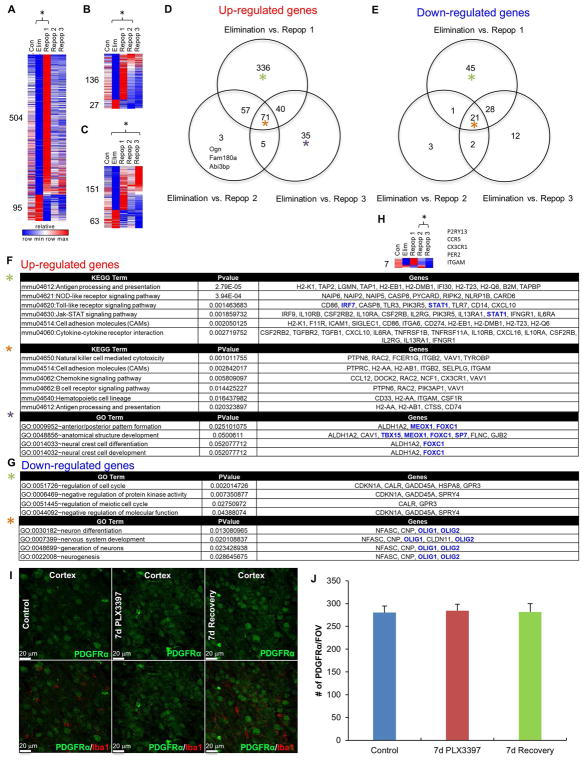
To explore the consequences and signaling pathways that were affected by microglial depletion, repopulation, and failed repopulation, we extracted whole brain RNA from Control, Elim (7 days 600 ppm PLX3397), Repop 1 (successful repopulation), and Repop 2 and 3 (unsuccessful repopulations), and performed RNA-seq. To aid in the exploration of gene expression changes we created a searchable website of expression values for each gene (http://rnaseq.mind.uci.edu/green/microglia_cycles/gene_search.php), where raw data points, mean, and standard error are plotted for all five groups. For more global changes and pathway analyses, FPKM values were obtained, and differential expression of significantly altered genes (p < 0.005 and FDR < 0.05) calculated (Figure 3). Analyses of Control vs. Elim brains revealed 15 upregulated genes, and reductions in 254 genes (Figure 3A, B). Using cell type-specific expression databases (Zhang et al. 2014), we found that the clear majority of the downregulated genes are expressed strongly, or solely, in microglia, allowing us to assemble a core set of microglia-expressed genes (Figure 3C, D), and validating several studies on isolated brain-extracted microglia (Bennett et al. 2016; Beutner et al. 2013; Butovsky et al. 2014; Chiu et al. 2013; Hickman et al. 2013; Wes et al. 2016). We divided other differentially expressed genes into those commonly expressed by astrocytes, neurons, oligodendrocyte precursors (OPCs), new oligodendrocytes, myelinating oligodendrocytes, or endothelial cells, and displayed relative expression of all groups in a heatmap (Figure 3E). Within the 15 upregulated genes between Control and Elim are the astrocyte-expressed genes Gfap, Mt2, and Serpina3n. Immunostaining for Gfap confirmed marked increases in Gfap expression with all cycles of elimination and repopulation (Figure 3F, G).
Figure 3.
Differentially expressed genes between treatment cycles with 600 ppm PLX3397, obtained via RNAseq analysis. A, Number of significant differentially expressed genes between each group analyzed. B–E, Differentially expressed genes between Con and Elim. B, Heatmap of significant differentially expressed genes. C, Differentially expressed genes stratified by those commonly expressed by CNS cell types reveals 164 downregulated microglial genes. D, List of 164 downregulated microglial genes termed “core” microglial genes. E, Heatmaps of differentially expressed genes stratified by CNS cell type reveals increase in common astrocyte-expressed genes, and decrease in common OPC-expressed genes. F–G, 63X representative images of cortex; immunostaining for Gfap and quantification of number of cells per FOV reveals the number of GFAP+ cells increases with Elim 1 and remains elevated for the remainder of cycles in the cortex, but remains unchanged in the hippocampus. All gene expression data can be accessed and searched at http://rnaseq.mind.uci.edu/green/microglia_cycles/gene_search.php. Error bars represent SEM, (n=4). * represents p<0.05 vs. Control.
In further exploring gene expression changes in non-microglial cells, we found that OPC genes were downregulated by microglial depletion, in line with recent reports that depletion lowers OPC numbers (Hagemeyer et al. 2017; Janova et al. 2017). However, mature oligodendrocyte-expressed genes were upregulated between Control and Elim (Figure 3E). The remaining downregulated genes are commonly expressed by neurons, astrocytes, and endothelial cells (Figure 3E), but could also be expressed in microglia, and hence their downregulation may be a direct result of microglial elimination, rather than an effect on a specific cell type.
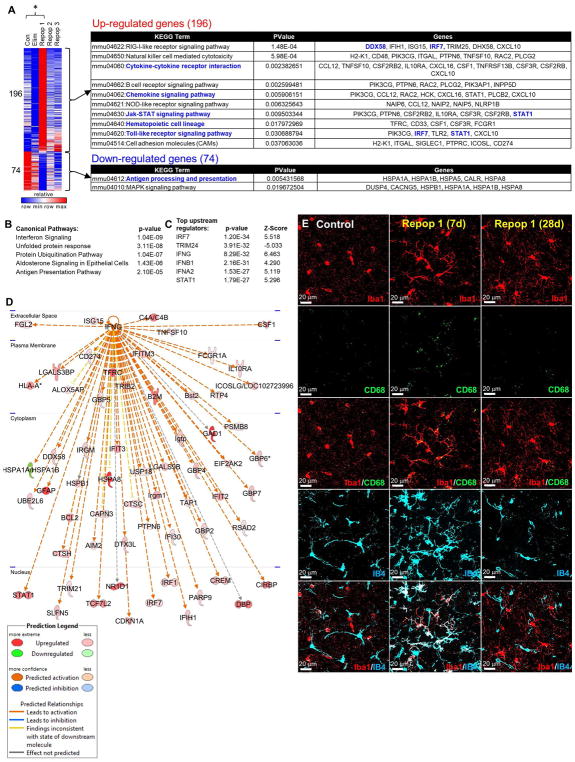
We next compared differentially expressed genes between Control and Repop 1 brains (the successful repopulation; Figure 4A). 196 genes were significantly upregulated, and the clear majority of these are commonly microglia-expressed (Figure 4A), in accordance with the overshoot in microglia numbers with 7-days repopulation (Figure 1B, D). Pathway analysis of altered genes revealed extensive activation of immune responses, including cytokine and chemokine signaling pathways, while downregulated genes were associated with antigen processing and presentation, as well as MAPK signaling (Figure 4A). Interferon signaling was identified as the top canonical pathway, and Ifng as a top upstream regulator (Figure 4B, C). Pathway mapping from Ifng was performed (Figure 4D), showing an IFNγ-activated gene signature, suggesting that repopulating microglia are in an activated state at this point. In agreement, we show that returning microglia at day 7 are less ramified with thicker processes, and stain positively for CD68 and the lectin IB4, markers of microglial activation, but that these markers are no longer found on microglia by day 28 of recovery, or in control brains (Figure 4E).
Figure 4.
Differentially expressed genes between Con and Repop 1 reveal an activated signature. A, Heatmap of significant differentially expressed genes; pathway analysis of differentially expressed genes between Con and Repop 1 reveals 196 upregulated genes, many of which are associated with cytokine and chemokine signaling, Jak-STAT signaling, and general immune cell identity and functions; downregulated genes involved in antigen processing and MAPK signaling. Transcription factors highlighted in blue. B, Top canonical pathways predicted to be activated in Repop 1 brains, and C, top predicted upstream regulators of those pathways. D, Ingenuity Pathway Analyses reveals upregulation of many downstream genes from IFNγ E, Immunostaining for activation markers CD68 and IB4 in Repop 1 (7d) and Repop 3 (28d), costained with Iba1.
Given the lack of repopulation in cycles 2 and 3, most gene expression changes relative to control brains were downregulated microglia-expressed genes, due to the cells’ absence. We therefore compared differentially expressed genes between Eliminated mice and Repop groups 1, 2, and 3 (Figure 5B–H). There were 336 unique upregulated and 45 downregulated genes between Elim vs. Repop 1, demonstrating that Repop 1 is unique from the other two Repop groups, as expected. Repop 2 and 3 are virtually identical to one another, with only 7 differentially expressed genes between them (Figure 5H). Pathway analysis was performed on the unique and shared gene expression changes between Elim vs. each Repop group and is summarized in Figure 5F–G. Notably, oligodendrocyte-expressed genes, such as Olig1, Olig2, and Cnp, were consistently elevated with elimination and downregulated with repopulation. As OPC-expressed genes are generally downregulated and oligodendrocyte-expressed genes are generally upregulated with elimination, we sought to determine if 1) repopulating microglia could be deriving from OPC’s/oligodendrocytes, and 2) if microglia elimination and repopulation was grossly altering the populations of these cells. To that end, we conducted a lineage tracing experiment whereby we crossed PDGFRα-CRE and Rosa26YFP reporter mice to yield PDGFRα-CRE/Rosa26YFP (PGDFRα-YFP) progeny that express YFP in all cells that either transiently or constitutively express PGDFRα, which in the brain labels OPC’s and their descendants (i.e. oligodendrocytes). Microglia were depleted with 7 days PLX3397 treatment, and repopulation stimulated with a 7-day recovery period (n=4 per group). Neither control or repopulated microglia expressed YFP, showing that neither derive from an OPC lineage (Figure 5I). Quantification of YFP+ cells in the cortex showed no changes in overall numbers of cells, suggesting that the collective number of OPC’s and oligodendrocytes does not change with treatments (Figure 5J), despite robust changes in gene expression.
Figure 5.
Pathway analysis and further comparison of differentially expressed genes, obtained via RNAseq. A–G, Differentially expressed genes between Elim vs. Repop 1, Elim vs. Repop 2, and Elim vs. Repop 3. D, E, Venn diagrams of common and unique differentially expressed genes between Elim vs. Repop 1, Elim vs. Repop 2, and Elim vs. Repop 3. F, Analysis of upregulated genes between the three comparisons, reveals 336 uniquely differentially expressed genes in Elim vs. Repop 1 comparison. G, Analysis of downregulated genes between the three comparisons. Transcription factors highlighted in blue. Colored asterisks for analyses match those in venn diagrams from D. H, Differentially expressed genes between Repop 2 and Repop 3. I, Representative images from PDGFRα-CRE/Rosa26YFP mice, which express YFP in OPC’s and their progeny (i.e. oligodendrocytes), from Control, 7d PLX3397 (600 ppm), and 7d recovery, showing YFP+ cells (green channel) and microglia (Iba1+; red channel). J, Quantification of (I). Error bars represent SEM, (n=4 per group).
Expanded capacity for myeloid cell repopulation with increased recovery time
Having shown that when challenged with short cycles of thorough microglial depletion the adult brain is limited to one complete round of microglial repopulation, we wanted to determine if the myeloid cell repopulation capacity in the brain could be increased by allowing more recovery time between cycles. To that end, groups of male mice (n=4/group) were treated with cycles of 7 days PLX3397 (600 ppm), followed by 28 days of recovery (Figure 6A). This time, all three rounds of recovery resulted in repopulation of myeloid cells, showing that the adult brain does have the ability to reconstitute itself with Iba1+ myeloid cells given sufficient recovery time between cycles for repopulation (Figure 6A, B).
Figure 6.
Cyclic treatment of WT mice with 600 ppm PLX3397, and 28d withdrawal periods. A, Representative whole brain stitches of Iba1+ cells in each group. B, Quantification of number of Iba1+ cells/FOV in hippocampus, cortex, and thalamus. C, Mice treated for two 7d elimination cycles (Elim 2 with 7d recovery), then allowed to recover for 28 days; representative half-brain stitches of Iba1+ cells and quantification demonstrate full repopulation after 28 days recovery. D, Quantification of (C). Error bars represent SEM, (n=4). p<0.05; significance symbols represent comparison groups: Control *, Elim 1 #, Repop 1 ⋄, Elim 2 △, Repop 2 ϕ, Elim 3 θ, Repop 3 ●.
Finally, we investigated whether 28 days of recovery would be sufficient to replenish the myeloid cell compartment in a brain that has been through 2 short cycles of 7d treatment/7d recovery. We found that although the brain fails to repopulate in 7 days after 2 short cycles of depletion (7d treatment/7d recovery), it fully repopulates with Iba1+ cells within 28 days of recovery after the second depletion cycle (Figure 6C, D), indicating that complete repopulation with myeloid cells, although delayed, is still possible.
Discussion
We previously demonstrated that microglial elimination via CSF1R inhibition and the subsequent removal of these antagonists stimulates repopulation of the brain (Elmore et al. 2014). In a “normal” repopulation event, the brain rapidly repopulates without peripheral contributions, and fully fills with new microglia within just 7 days. These new microglia initially appear reactive but assume homeostatic phenotypes within another 7–14 days (Elmore et al. 2015). In this study, we sought to determine the limits and kinetics of microglia repopulation events by challenging the adult brain to sequential cycles of microglia depletion and repopulation. We found that the adult brain appears to only have the capacity for a single repopulation event, after which “normal” repopulation then fails to occur, despite the presence of surviving microglia in the brain. However, if the recovery time between, or after, cycles is extended, then the brain can ultimately repopulate with myeloid cells, suggesting that the brain may have multiple mechanisms to repopulate outside of the “normal” repopulation described thus far. For example, other models of microglia depletion using genetic approaches have described a slower mechanism of repopulation that derives from peripheral cells (Varvel et al. 2012). Furthermore, continuous tamoxifen-induced genetic CSF1R deletion in microglia leads to chronic partial microglial depletion, which over time causes the brain to partly (~50%) fill with peripherally derived macrophages that take up residence within the brain parenchyma (Cronk et al. 2018). Although our previous studies have shown that with a single “normal” repopulation event following CSF1R inhibition (Elmore et al. 2014), peripherally derived cells do not infiltrate into the brain, it is yet to be determined whether peripherally derived cells contribute to the myeloid cell repopulation in either of the extended repopulation events detailed here.
Interestingly, the limited capacity of microglia to repopulate with repeated rapid depletion cycles mirrors findings from oligodendrocyte precursors (OPC’s). Administration of cuprizone leads to demyelination of axons through the selective death of oligodendrocytes, and once demyelinated, OPCs then proliferate and differentiate into new oligodendrocytes, effectively remyelinating the axons (Mason et al. 2000; Sachs et al. 2014). Using a model of chronic cuprizone treatment, it has been shown that OPCs become progressively depleted resulting in failed remyelination (Mason et al. 2004). Paralleling these observations, we found that with rapid cycles of microglial elimination and recovery (7 days’ treatment, 7 days’ withdrawal), microglia failed to repopulate a second or third time, suggesting the source of repopulating microglia becomes partially exhausted. Further similarities were also found between oligodendrocyte depletion and subsequent remyelination. Varying the dose and/or time of CSF1R inhibitor treatment affects the extent/speed of microglial depletion. For example, administration of PLX3397 at 290 ppm in chow leads to 50% depletion of microglia within 7 days. Repopulation following 50% microglial depletion is slow, and does not recover microglial numbers to control levels within 7 days following inhibitor withdrawal. On the other hand, inhibitor withdrawal following >95% microglial elimination, either through 21 days’ treatment with PLX3397 at the same dose (290 ppm in chow; (Elmore et al. 2014), or with 7 days’ treatment with PLX3397 at 600 ppm in chow, is rapid, with microglial densities exceeded within 7 days. Thus, the brain reacts differently to differing extents of microglial depletion, suggesting a sensing mechanism to mount an appropriate repopulation response, with greater depletion requiring more powerful and rapid repopulation. Similar observations have been made with the remyelination that occurs following cuprizone-mediated demyelination; remyelination only occurs with extensive demyelination rather than incomplete demyelination (Doan et al. 2013). These similarities to oligodendrocyte repopulation experiments suggest there may be common renewal properties between source cells for microglial repopulation and progenitor cells from the oligodendrocyte lineage. However, it is important to highlight that lineage tracing from OPC’s shows that repopulating microglia do not derive from this lineage.
We sought to better understand the signals coordinating successful vs. unsuccessful repopulation cycles through whole-brain gene expression profiling, but not surprisingly, the great majority of genes that were differentially expressed between groups with and without microglia, are typically expressed by microglia. As expected, profiling of the microglia-depleted brain revealed a core set of genes that were markedly downregulated with CSF1R inhibitor treatment known to be expressed solely or highly in microglia, and confirm several studies detailing gene expression in microglia extracted from brains (Bennett et al. 2016; Beutner et al. 2013; Butovsky et al. 2014; Chiu et al. 2013; Hickman et al. 2013). These genes include P2ry12, Tmem119, Fcrls, and Tgfβr1. By profiling total brain RNA rather than isolated microglia, we can further show the relative quantities of these genes in non-microglial cells (i.e. their specificity to microglia), as well as confirm that their expression has not been switched on/upregulated by the microglial extraction processes used by the prior studies. Interestingly, we found that OPC-expressed genes are decreased with microglial elimination, supporting the findings of recent studies highlighting the role of adult microglia in OPC population maintenance (Hagemeyer et al. 2017; Janova et al. 2017). However, we also find that oligodendrocyte expressed genes are upregulated at the same time, and lineage tracing from OPC’s (via PDGFRα-cre) showed that overall levels of OPC’s and their descendants (i.e. oligodendrocytes) were not altered with microglial elimination or repopulation. Further gene expression profiling of repopulated brains showed that the returning microglia are clearly in an activated state, with a characteristic IFNγ-induced profile, and an upregulation of many pro-inflammatory cytokines and chemokines. Indeed, our prior results show that although they initially appear primed to protect against an attack, in the absence of actual pathogens or damage, microglia revert to a resting/surveillant state within 14 days (Elmore et al. 2015).
The ability to eliminate microglia and then repopulate has potential therapeutic implications. For example, after traumatic brain injuries, microglia become chronically activated, and impede functional recovery (Cherry et al. 2016; Loane et al. 2014; Nagamoto-Combs et al. 2007; Ramlackhansingh et al. 2011; Smith et al. 1997). The ability to remove these reactive microglia and then repopulate with fresh microglial cells offers a strategy to resolve neuroinflammation and promote recovery (Rice et al. 2017), however, there appear to be limits on the frequency and timing of “normal” repopulation events. In conclusion, we have more fully defined the extent of the adult brain’s capacity for renewal of microglial tissue, helping to set the potential opportunities and limits of microglial depletion/repopulation as a therapeutic strategy.
Acknowledgments
This work was supported by the National Institutes of Health under awards R01NS083801 (NINDS), P50 AG016573 (NIA) to KNG, and AG00096 to ARN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Dan Hoang, Ken Nguyen, and Michael Phelan Ph.D., for the setting up of gene expression data at http://rnaseq.mind.uci.edu/green/microglia_cycles/gene_search.php.
Footnotes
Financial Disclosures:
Dr. Najafi reports no financial interests or conflicts of interest; Mr. Crapser reports no financial interests or conflicts of interest; Ms. Jiang reports no financial interests or conflicts of interest; Ms. Ng reports no financial interests or conflicts of interest; Dr. Mortazavi reports no financial interests or conflicts of interest; Dr. West is an employee of Plexxikon Inc.; Dr. Green reports no financial interests or conflicts of interest.
References
- Antony JM, Paquin A, Nutt SL, Kaplan DR, Miller FD. Endogenous microglia regulate development of embryonic cortical precursor cells. J Neurosci Res. 2011;89:286–98. doi: 10.1002/jnr.22533. [DOI] [PubMed] [Google Scholar]
- Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, Wolozin B, Butovsky O, Kugler S, Ikezu T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–93. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016;113:E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner C, Linnartz-Gerlach B, Schmidt SV, Beyer M, Mallmann MR, Staratschek-Jox A, Schultze JL, Neumann H. Unique transcriptome signature of mouse microglia. Glia. 2013;61:1429–42. doi: 10.1002/glia.22524. [DOI] [PubMed] [Google Scholar]
- Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, et al. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity. 2015;43:92–106. doi: 10.1016/j.immuni.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–43. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butowski N, Colman H, De Groot JF, Omuro AM, Nayak L, Wen PY, Cloughesy TF, Marimuthu A, Haidar S, Perry A, et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol. 2016;18:557–64. doi: 10.1093/neuonc/nov245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JD, Tripodis Y, Alvarez VE, Huber B, Kiernan PT, Daneshvar DH, Mez J, Montenigro PH, Solomon TM, Alosco ML, et al. Microglial neuroinflammation contributes to tau accumulation in chronic traumatic encephalopathy. Acta Neuropathol Commun. 2016;4:112. doi: 10.1186/s40478-016-0382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O’Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk JC, Filiano AJ, Louveau A, Marin I, Marsh R, Ji E, Goldman DH, Smirnov I, Geraci N, Acton S, et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J Exp Med. 2018 doi: 10.1084/jem.20180247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher NN, Najafi AR, Kayala KM, Elmore MR, White TE, Medeiros R, West BL, Green KN. Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J Neuroinflammation. 2015;12:139. doi: 10.1186/s12974-015-0366-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan V, Kleindienst AM, McMahon EJ, Long BR, Matsushima GK, Taylor LC. Abbreviated exposure to cuprizone is sufficient to induce demyelination and oligodendrocyte loss. J Neurosci Res. 2013;91:363–73. doi: 10.1002/jnr.23174. [DOI] [PubMed] [Google Scholar]
- Elmore MR, Lee RJ, West BL, Green KN. Characterizing newly repopulated microglia in the adult mouse: impacts on animal behavior, cell morphology, and neuroinflammation. PLoS One. 2015;10:e0122912. doi: 10.1371/journal.pone.0122912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–97. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alcalde F, Garcia-Lopez F, Dopazo J, Conesa A. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27:137–9. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–5. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer N, Hanft KM, Akriditou MA, Unger N, Park ES, Stanley ER, Staszewski O, Dimou L, Prinz M. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017 doi: 10.1007/s00401-017-1747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Harris RA, Zhang XM. An updated assessment of microglia depletion: current concepts and future directions. Mol Brain. 2017;10:25. doi: 10.1186/s13041-017-0307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–78. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, Wang J, Zhao L, Liang YX, Wu T, et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci. 2018;21:530–540. doi: 10.1038/s41593-018-0090-8. [DOI] [PubMed] [Google Scholar]
- Janova H, Arinrad S, Balmuth E, Mitjans M, Hertel J, Habes M, Bittner RA, Pan H, Goebbels S, Begemann M, et al. Microglia ablation alleviates myelin-associated catatonic signs in mice. J Clin Invest. 2017 doi: 10.1172/JCI97032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D, Patzko A, Schreiber D, van Hauwermeiren A, Baier M, Groh J, West BL, Martini R. Targeting the colony stimulating factor 1 receptor alleviates two forms of Charcot-Marie-Tooth disease in mice. Brain. 2015;138:3193–205. doi: 10.1093/brain/awv240. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Gordon S. Turnover of resident microglia in the normal adult mouse brain. Neuroscience. 1992;48:405–15. doi: 10.1016/0306-4522(92)90500-2. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loane DJ, Kumar A, Stoica BA, Cabatbat R, Faden AI. Progressive neurodegeneration after experimental brain trauma: association with chronic microglial activation. J Neuropathol Exp Neurol. 2014;73:14–29. doi: 10.1097/NEN.0000000000000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Jones JJ, Taniike M, Morell P, Suzuki K, Matsushima GK. Mature oligodendrocyte apoptosis precedes IGF-1 production and oligodendrocyte progenitor accumulation and differentiation during demyelination/remyelination. J Neurosci Res. 2000;61:251–62. doi: 10.1002/1097-4547(20000801)61:3<251::AID-JNR3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Mason JL, Toews A, Hostettler JD, Morell P, Suzuki K, Goldman JE, Matsushima GK. Oligodendrocytes and progenitors become progressively depleted within chronically demyelinated lesions. Am J Pathol. 2004;164:1673–82. doi: 10.1016/S0002-9440(10)63726-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamoto-Combs K, McNeal DW, Morecraft RJ, Combs CK. Prolonged microgliosis in the rhesus monkey central nervous system after traumatic brain injury. J Neurotrauma. 2007;24:1719–42. doi: 10.1089/neu.2007.0377. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–8. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–8. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Ramlackhansingh AF, Brooks DJ, Greenwood RJ, Bose SK, Turkheimer FE, Kinnunen KM, Gentleman S, Heckemann RA, Gunanayagam K, Gelosa G, et al. Inflammation after trauma: microglial activation and traumatic brain injury. Ann Neurol. 2011;70:374–83. doi: 10.1002/ana.22455. [DOI] [PubMed] [Google Scholar]
- Rice RA, Pham J, Lee RJ, Najafi AR, West BL, Green KN. Microglial repopulation resolves inflammation and promotes brain recovery after injury. Glia. 2017;65:931–944. doi: 10.1002/glia.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice RA, Spangenberg EE, Yamate-Morgan H, Lee RJ, Arora RP, Hernandez MX, Tenner AJ, West BL, Green KN. Elimination of Microglia Improves Functional Outcomes Following Extensive Neuronal Loss in the Hippocampus. J Neurosci. 2015;35:9977–89. doi: 10.1523/JNEUROSCI.0336-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs HH, Bercury KK, Popescu DC, Narayanan SP, Macklin WB. A new model of cuprizone-mediated demyelination/remyelination. ASN Neuro. 2014:6. doi: 10.1177/1759091414551955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner B, Romanelli E, Liberski P, Ingold-Heppner B, Sobottka-Brillout B, Hartwig T, Chandrasekar V, Johannssen H, Zeilhofer HU, Aguzzi A, et al. Astrocyte Depletion Impairs Redox Homeostasis and Triggers Neuronal Loss in the Adult CNS. Cell Rep. 2015;12:1377–84. doi: 10.1016/j.celrep.2015.07.051. [DOI] [PubMed] [Google Scholar]
- Smith DH, Chen XH, Pierce JE, Wolf JA, Trojanowski JQ, Graham DI, McIntosh TK. Progressive atrophy and neuron death for one year following brain trauma in the rat. J Neurotrauma. 1997;14:715–27. doi: 10.1089/neu.1997.14.715. [DOI] [PubMed] [Google Scholar]
- Spangenberg EE, Lee RJ, Najafi AR, Rice RA, Elmore MR, Blurton-Jones M, West BL, Green KN. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-beta pathology. Brain. 2016;139:1265–81. doi: 10.1093/brain/aww016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap WD, Wainberg ZA, Anthony SP, Ibrahim PN, Zhang C, Healey JH, Chmielowski B, Staddon AP, Cohn AL, Shapiro GI, et al. Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor. N Engl J Med. 2015;373:428–37. doi: 10.1056/NEJMoa1411366. [DOI] [PubMed] [Google Scholar]
- Tremblay ME, Lowery RL, Majewska AK. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 2010;8:e1000527. doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdearcos M, Robblee MM, Benjamin DI, Nomura DK, Xu AW, Koliwad SK. Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep. 2014;9:2124–38. doi: 10.1016/j.celrep.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varvel NH, Grathwohl SA, Baumann F, Liebig C, Bosch A, Brawek B, Thal DR, Charo IF, Heppner FL, Aguzzi A, et al. Microglial repopulation model reveals a robust homeostatic process for replacing CNS myeloid cells. Proc Natl Acad Sci U S A. 2012;109:18150–5. doi: 10.1073/pnas.1210150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Holtman IR, Boddeke EW, Moller T, Eggen BJ. Next generation transcriptomics and genomics elucidate biological complexity of microglia in health and disease. Glia. 2016;64:197–213. doi: 10.1002/glia.22866. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]