Abstract
Adult tissue-specific stem cells are essential for homeostatic tissue maintenance and key to regeneration during injury repair or disease. Many critical stem cell functions rely on the presence of well-timed cues from the microenvironment or niche, which includes a diverse range of components, including neuronal, circulating and extracellular matrix inputs as well as an array of neighboring niche cells directly interacting with the stem cells. However, studies of stem cells and their niche have been challenging due to the complexity of adult stem cell functions, their intrinsic controls and the multiple regulatory niche components. Here, we review recent major advances in our understanding of the complex interplay between stem cells and their niche that were enabled by the tremendous technological leaps in single-cell transcriptome analyses, 3D in-vitro cultures and 4D in-vivo microscopy of stem cell niches.
Keywords: Stem cells, stem cell niche, hematopoiesis, hair follicle, intestine, single cell transcriptomics, single cell RNA sequencing, organoids, live imaging
GRAPHICAL ABSTARCT

Introduction
Each day, an adult human generates billions of new cells to replace those that are lost naturally or by damage. At the top of the production hierarchies are adult stem cells (SCs), defined by their abilities for long-term self-renewal and multipotent differentiation into several different lineage-restricted cell types. The first definitive proof of the existence of adult SCs came from work by Till, McCulloch and others in the 1960s demonstrating the existence of a hematopoietic SC (HSC) pool responsible for maintaining the entire blood-lineage throughout life [1–3]. Since then, multiple adult SCs have been discovered in several tissues and organs, such as the intestine [4,5], brain [6,7], mammary gland [8,9], and skin [10] including hair follicles [11–13].
While many of the special characteristics of SCs are intrinsic, no completely autonomous adult SC has been discovered. Rather, all known adult SCs rely to a large extent for proper function on external signals from its surroundings, termed the SC niche [14–16]. The niche communicates vital information regarding the regenerative needs of the tissue, the importance of which is exemplified by the detrimental effects of deviations from the crucial loss/production balance, as defects of SCs and their niches have been implicated in multiple human disorders and diseases [17].
The HSC sits atop a hierarchy of fate-committed multipotent progenitors (MPP) and terminally differentiated cells of the entire blood-lineage [18–20]. Self-renewal of HSCs and lineage-committed progenitors and their differentiation towards diverse blood lineages are regulated by multiple niche inputs from non-hematopoietic cell types, such as osteoblasts [21,22], peri-sinusoidal [23,24] and peri-arteriolar stromal cells [25], and endothelial cells [23,26], as well as from hematopoietic lineages, such as macrophages [27–29] and megakaryocytes [30].
Hair follicle SCs (HFSC) and downstream progenitors give rise to seven cell lineages that make up the hair shaft and its supporting channel during hair growth, a process that is interrupted by a naturally-occurring hair cycle of cyclical bouts of follicle destruction, a resting phase of relative quiescence, and re-growth [11–13,31]. Distinct niche signals controlling the balance of SC rest and activation are thought to emanate from essential niche components that include specialized mesenchymal dermal papilla cells [32–35], direct SC progeny of multipotent progenitors [36,37**] and neighboring nerves [38], as well as longer-range inputs from fibroblasts deep in the dermis [39], cells of the dermal adipocyte-lineage [40], and immune cells [41–43].
Intestinal SCs (ISC) residing in the intestinal crypt base constantly replenish the villus epithelium of rapidly turned-over enterocytes, goblet and enteroendocrine cells, and other SC progeny that are lost by conveyor belt-like upward displacement [4,5,44] The intestinal regulatory niche contains pericryptal mesenchymal fibroblasts [45,46], myofibroblasts and smooth muscle cells [47,48], as well as gut-lumen micro-biota [49,50]. Paneth cells, the only differentiated SC progeny that migrate to the crypt base, also provide regulatory niche signals to ISCs [51–53].
In this review we highlight recent major discoveries into the SC regulation by the niche that were made possible by groundbreaking new technological innovations of single cell-level profiling, complex cell cultures and organoids, and major advances in light microscopy (Figure 1). As we are not able to cover the entire extensive body of new knowledge gained in the SC field within the past few years, we refer to excellent recent reviews containing comprehensive updates in several SC niche systems [54–58].
Figure 1. Next-generation technologies, often used in combination or complementation, advance insights into SC niche complexities.
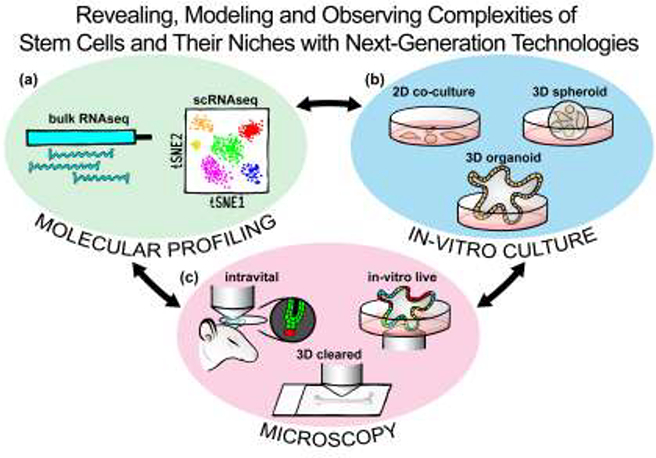
(a) Molecular profiling by population-based RNA sequencing and by single cell RNA sequencing reveal unprecedented level of gene expression complexities and cell heterogeneity in SCs and their niches. (b) In-vitro cultures with 2D engineered matrices and co-culture, 3D spheroid aggregates, and structured 3D organoids enable modeling of SC interactions with their niches. (c) 3D multicolor light microscopy of 3D cleared tissues and 4D intravital and in-vitro live imaging allow observation of SC niche complexities in vivo and in real-time.
Revealing complexity: single-cell profiling of organs
Precise regulation of gene expression in SCs and their niche is paramount for executing the molecular programs of SC quiescence, self-renewal and lineage differentiation. Specific sets of expressed genes and epigenetic configurations underlie functional distinctions between different cell types within complex tissues, including SCs and the cellular niche components. Since large-scale transcriptome analysis became technically feasible with the establishment of microarrays in Arabidopsis [59], it has been used with great impact as a window into SC and niche-specific properties and as a basis for discovering targets for functional studies in multiple SC niche systems [11,12,60–62]. Since then, technologies committed to monitoring the transcriptome of cells, as surrogate for protein expression, have flourished. RNA-sequencing (RNA-seq) was established in rapid succession in Arabidopsis [63], yeast [64] and mammalian cells [65] that surveys mRNA content in a manner that is relatively unbiased, when compared to microarrays, and with superior sensitivity [65,66], and was quickly utilized to analyze transcriptional patterns in mammalian cells, including SCs [67–69]. Purification of SCs and the diverse cell-types of their niche through cell sorting for transcriptomic analysis of bulk populations by RNA-seq enabled sensitive detection of gene expression, revealing their molecular identities with superior resolution and identifying the expression of ligand-receptor pairs between the SCs and their niche [70*,71]. While the sensitivity of this approach provides highly detailed molecular descriptions of the cell-types of interest, the data cannot resolve subtle heterogeneity within populations and detect the existence of rare sub-types. Immunofluorescence, flow cytometry, and mass cytometry [72] can enable deeper investigation of heterogeneity by interrogating single cells but are often limited by availability of detectors and/or antibodies.
With the advent of single-cell transcriptome analysis the transcriptional profiling power of RNA-seq can be combined with the ability to interrogate single cells [73]. Single-cell RNA-sequencing (scRNA-seq) and developed analysis algorithms that compress high-dimensional data into two or three dimensions, like t-stochastic neighbor embedding [74] or principle components analysis, allow for efficient identification of heterogenous cell subtypes. In addition, pseudotime [75] and FatelD [76] algorithms enable prediction of differentiation trajectories and reveal step-wise transcriptional changes as cell fates are determined. As a result, SCs, niche cells and other cell types previously considered as relatively homogenous have been shown to be remarkably complex and heterogenous, pointing to a diversity of unique cellular functions and processes. In the epidermis and hair follicle, pseudotime and pseudospace, a closely related algorithm predictive of spatial localization, were used in tandem to construct a map of HFSC differentiation and discover changes in expression of key signaling, extracellular matrix, and cell adhesion components [77]. A more recent study by Fuchs and colleagues identified heterogeneity amongst HF progenitors, and cognate heterogeneity in the dermal papilla, forming “micro-niches” along the epithelial-mesenchymal interface [37**]. Similar efforts have enabled the discovery of new, rare cell types in both intestinal organoids and the endogenous tissue, as well as heterogeneity amongst ISCs [78,79**]. Other studies have combined scRNA-seq with in-vivo and in-vitro observations of cells, to link transcriptome data with SC quiescence. Unique molecular profiles of isolated single HSCs and progenitors were associated with divisional history in culture, and identified niche components necessary for maintenance of dormancy [80*]. Additional scRNA-seq studies to identify cell-intrinsic factors regulating HSC quiescence revealed retinoic acid signaling as crucial for dormancy transcriptional programs [81]. Heterogeneity amongst niche components revealed the differential capacity to maintain adult SCs. Stromal osteolineage cells co-transplanted with HSCs/progenitors revealed distinct transcriptional signatures of these cells proximal and distal to engrafted HSCs/progenitors, and identified novel niche factors regulating HSC quiescence [82*]. Overall, scRNA-seq has revealed previously unknown complex spatial and temporal heterogeneity of both SCs and niche components and promises future discovery of key effectors of SC quiescence, maintenance and differentiation, as well as additions to the wide array of niche cell types and functions.
Modeling complexity: 2D co-cultures and 3D tissue-recapitulating organoids
Two-dimensional flat cell culture systems have long been used to model the cell relationships within tissues, while enabling relatively straightforward experimental manipulation. The simplicity of 2D cultures enables functional interrogation of identified targets - including those from scRNA-seq studies - in the SC niche for quiescence and differentiation programs. Critical niche components for maintaining murine HSCs have been identified in 2D culture using engineered hydrogel microwells that displayed scRNA-seq-identified adhesion receptors JamC and Esam [80*]. In similar assays, differentiation of T-lymphocytes from both murine and human HSCs was regulated by fibronectin-immobilized VCAM-1 and Delta-like-4 [83].
Whereas flat cultures lack the cellular and spatial complexity of tissues, three-dimensional cultures have made great strides to recapitulate elements of in-vivo spatial and structural organization by generating 3D structures out of biomaterials or by seeding cells within them. Seeded murine or human mesenchymal SCs onto 3D silk scaffolds generated a model for bone marrow adipose tissue (BMAT) and its bidirectional relationship with myeloma cell lines and for exploring BMAT-associated homeostatic and disease processes in the bone marrow [84*]. Another recent study showed that 3D epidermal and dermal clusters from young mouse back skin had the ability to self-organize and induce hair follicle formation. This capacity was lost in 3D clusters from old skin but could be rescued by addition of matrix metalloproteinases and other cytokines to the culture [85*].
While 3D cultures require artificial engineering, organoids, which can be generated from a single pluripotent or adult SC, have inherent self-organizing capabilities - reflective of in-vivo cell rearrangement - thus enabling simultaneous study of structure and function [86]. Organoids have great utility in screening genes and drug compounds but have also been used extensively in evaluating the “stemness” of adult SCs. Early work showed that mouse and human mammary epithelial SCs are capable of forming spheroids in culture, permitting efficient mammosphere production [87,88]. Many other organ systems have since established organoid protocols, including brain organoids derived from mouse and human induced pluripotent SCs [89,90]. A protocol for generating murine intestinal organoids was first described by the Clevers lab, in which a single ISC could differentiate into distinct crypt-villi structures of solely epithelial lineages [91]. Intestinal organoids have since been used to study many aspects of ISC and niche biology, utilizing both murine and human organoid systems, including coordinate metabolic requirements of ISCs and differentiated progeny [92*] and heterogeneity of differentiation programs [93]. Gene manipulation, using CRISPR, has also been effectively utilized in intestinal organoids; multigenic knock-outs of Wnt components in organoids demonstrated their requirement for organoid maintenance [94], and deletions of DNA-repair-associated genes were made in ISCs to identify which of these genes can recapitulate mutational signatures often found in human color cancers [95]. Supplementation of intestinal epithelial organoids with niche components mimicked elements of in-vivo SC interactions with the niche; addition of intestinal stromal cells to intestinal organoid cultures has elucidated key secreted factors from the gut mesenchyme for SC maintenance [96*]. Recently, the Koehler lab defined a protocol for generating murine skin organoids from pluripotent SCs, comprising of both epithelial and mesenchymal cell types, capable of making hair, sebaceous glands, and other accessory structures [97**]. In this system, HFSC precursors co-develop with dermal papillae niche precursors, reminiscent of in-vivo HF morphogenesis. While starting as a system to evaluate SC characteristics, organoid protocols have been adapted to tractably explore complex intercellular relationships, including SC interactions with their niche.
Observing complexity: advances in in-situ, intravital and ex-vivo imaging
New techniques in light microscopy have begun to reveal the 3D spatial complexities of SC niches to complement the heterogeneity and rare populations identified by single cell gene expression profiling. Through the use of multiplexed immunofluorescence with optical tissue clearing for extended imaging depths, Schroeder and colleagues were able to describe the spatial organization of the bone marrow HSC niche within an entire rodent femur [98]. With the advantages of more sensitive and quantitative measures of gene expression compared to immunofluorescence, single molecule fluorescence in situ hybridization (FISH) on intestinal crypts enabled expression analysis of several SC markers in spatially distinct ISCs and the verification of rare lineages at single-transcript resolution [78,99]. Multiplexed error-robust FISH made further improvements by enabling detection of up to 1000 distinct mRNA targets within single cells and tissues greatly expanding the power of this technique to visualize the spatial complexities of gene expression within tissues including SC niches [100,101].
The ability to observe living cells in real-time can provide a myriad of information of cellular dynamics often not easily captured by the snapshot methods that freeze cells and their molecular machinery in time, but imaging living tissue with traditional widefield and confocal microscopy has been challenging due to light scattering. Multiphoton (MP) microscopy with infrared light greatly expands tissue imaging depths and has enabled real-time observation of live SCs in their native environments. MP imaging of surgically exposed calvariae first provided short-term live glimpses of the cellular dynamics during HSC homing to the bone marrow niche [102,103]. Shortly after, Van Rheenen and colleagues visualized intestine with MP imaging through an implanted abdominal imaging window for lineage tracing of ISCs over multiple days. This uncovered a passive displacement of crypt base columnar SCs in the upper niche which were functionally replaced by ISC progeny from the lower crypt base indicating a positional competitive advantage and an uncoupling of SC fate decisions from division [104].
Recently Greco and colleagues have utilized MP imaging to great effect in studying the dynamics of epidermal and HFSCs in their natural state in living mice [33,105,106**]. Since the skin and hair are easily accessible for imaging and do not require surgical preparation, the same field and even individual hair follicles could be re-identified with the guidance of visible landmarks such as blood vessel patterns and tattooed dots and tracked over a span of many months in revisit experiments [107]. This enabled the discovery of a precise spatial organization of SC progeny divisions and coordinated cell movements that control the rapid expansion of growing hair follicles [105], and revealed the control of SC fate by the cell position within the niche [33]. In addition to deep tissue imaging, the infrared laser was repurposed to heat-ablate cells with surgical precision and its effects were observed immediately after or over a longer time-span. Laser ablation of HFSCs or niche components of individual follicles and later revisits using adjacent unperturbed follicles as a built-in control uncovered a plasticity between different HFSC domains and demonstrated an essential role of dermal papilla niche cells for control of hair regression and HFSC activation during the hair cycle [105,108].
The combined approach of utilizing live imaging on organoid cultures further alleviates the drawbacks of studying fixed tissues, which lack real-time visualization of intra- and intercellular dynamics, or of 2D cultures that do not necessarily recapitulate the complexities of in-vivo tissues, while adding the great advantage of scalability for high-throughput assays. For example, through live imaging of intestinal organoids and the use of Ca2+-activated fluorescent reporters Julius and co-workers recently identified enterochromaffin cells as responding to neural inputs and relaying information about luminal metabolites to the enteric nervous system via synapses [88]. Another recent study showed a distinct heterogeneity in oxygenation between intestinal organoids through live-imaging using a phosphorescent O2 probe [109]. Finally, the ability to generate organoids from human SCs for in vitro live imaging combined with the efficiency of generating fluorescence reporter lines with CRISPR-Cas9 genome editing greatly bolsters studies in human systems [110]. As a recent example, live imaging during the development of human brain organoids was used to model the biomechanics of cerebral folding, a characteristic of humans not shared with standard model rodent species [111*]. This study showcases the power of human organoid systems to pick up when there are limitations to animal models, static imaging and traditional cell culture in order to better recapitulate aspects of the complexity of SCs and the niche during development, homeostasis and disease.
Conclusions and outlook
Advances in single cell profiling, in-vitro culture and advanced light microscopy have further enabled SC researchers in the dissection of the complex intricacies of adult SC interactions with the niche on a larger scale and at a more refined level than ever before. These emerging technologies have begun to provide powerful new avenues to interrogate heterogeneity and complexity, diversity in function in SCs and niche components, and their essential crosstalk (Figure 2). However, as newly developed methods and technologies have to fully pass rigorous scrutiny and validation before ubiquitous acceptance and implementation to the modern biologist’s tool-kit, researchers should carefully consider their limitations when drawing conclusions and support findings with more traditional, well-established techniques. Regardless, these new technologies open the door to exploration of the complexities of SC functions and the regulation by the niche that was previously not thought possible and promise to continue the successful journey towards elucidating the key regulation behind SC identity and behavior that may allow for the development of regenerative therapies and treatment of disease.
Figure 2. Recent insights on hematopoietic, intestinal and skin/hair follicle SCs and their niches by new technologies.
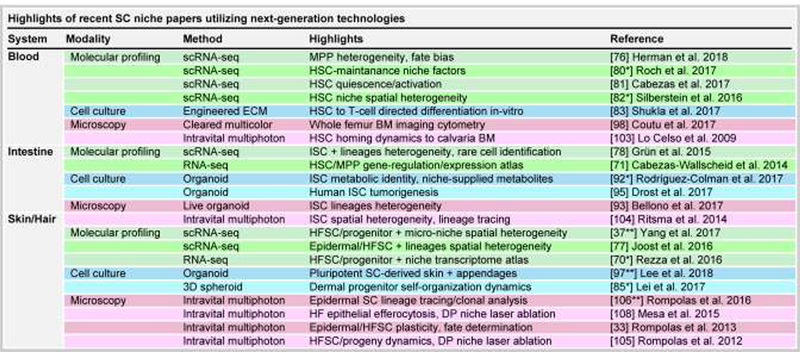
Major new advances from recent papers using new technologies are grouped by SC niche system and technology.
Highlights.
New technologies overcome barriers from complexities of stem cells and their niches
Single-cell profiling reveals heterogeneity, rare cells and fate trajectories
3D organoid culture models stem cell and niche interactions in-vitro
3D multiplexed and 4D in-vitro and intravital live imaging resolves complexities
Acknowledgments
We thank all lab members for helpful comments on the manuscript and valuable discussions. N.H. was supported by grants F30AR070639 from NIH/NIAMS and T32GM007280 from NIH/NIGMS. N.S. was supported by NIH/NIAMS grant R01AR071047. M.R. was supported by grants from the NIH/NIAMS (R01AR071047; R01AR063151) and New York State Department of Health (NYSTEM-C029574), and a fellowship from the Irma T. Hirschl Trust. We apologize to all colleagues whose relevant work we could not discuss due to space limitations.
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Description for papers of special interest (*) or outstanding interest (**)
- 1.Till JE, McCulloch EA: A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res 1961,14:213–22. [PubMed] [Google Scholar]
- 2.Becker AJ, McCulloch EA, Till JE: Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 1963,197:452–4. [DOI] [PubMed] [Google Scholar]
- 3.Weissman IL: The road ended up at stem cells. Immunol Rev 2002,185:159–174. [DOI] [PubMed] [Google Scholar]
- 4.Bjerknes M, Cheng H: Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 1999, 116:7–14. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. : Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007,449:1003–7. [DOI] [PubMed] [Google Scholar]
- 6.Altman J, Das GD: Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol 1965,124:319–35. [DOI] [PubMed] [Google Scholar]
- 7.Temple S: Division and differentiation of isolated CNS blast cells in microculture. Nature 1989,340:471–3. [DOI] [PubMed] [Google Scholar]
- 8.Smith GH, Medina D: A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J Cell Sci 1988,90:173–83. [DOI] [PubMed] [Google Scholar]
- 9.Kordon EC, Smith GH: An entire functional mammary gland may comprise the progeny from a single cell. Development 1998,125:1921–30. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS, Morris RJ: Epithelial stem cells in vivo. J Cell Sci Suppl 1988,10:45–62. [DOI] [PubMed] [Google Scholar]
- 11.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E: Defining the epithelial stem cell niche in skin. Science 2004,303:359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G: Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 2004,22:411–7. [DOI] [PubMed] [Google Scholar]
- 13.Cotsarelis G, Sun TT, Lavker RM: Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 1990,61:1329–37. [DOI] [PubMed] [Google Scholar]
- 14.Moore KA, Lemischka IR: Stem cells and their niches. Science 2006,311:1880–5. [DOI] [PubMed] [Google Scholar]
- 15.Scadden DT: Nice neighborhood: emerging concepts of the stem cell niche. Cell 2014,157:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezza A, Sennett R, Rendl M: Adult stem cell niches: cellular and molecular components. Curr Top Dev Biol 2014,107:333–72. [DOI] [PubMed] [Google Scholar]
- 17.Hoggatt J, Kfoury Y, Scadden DT: Hematopoietic Stem Cell Niche in Health and Disease. Annu Rev Pathol 2016,11:555–81. [DOI] [PubMed] [Google Scholar]
- 18.Kondo M, Weissman IL, Akashi K: Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997,91:661–72. [DOI] [PubMed] [Google Scholar]
- 19.Akashi K, Traver D, Miyamoto T, Weissman IL: A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000,404:193–197. [DOI] [PubMed] [Google Scholar]
- 20.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E: New Evidence Supporting Megakaryocyte-Erythrocyte Potential of Flk2/Flt3+Multipotent Hematopoietic Progenitors. Cell 2006,126:415–426. [DOI] [PubMed] [Google Scholar]
- 21.Calvi L, Adams G, Weibrecht K, Weber J, Olson D, Knight M, Martin R, Schipani E, Divieti P, Bringhurst F, et al. : Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003,425:841–846. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Niu C, Ye L, Huang H, He X, Tong W-G, Ross J, Haug J, Johnson T, Feng JQ, et al. : Identification of the haematopoietic stem cell niche and control of the niche size. Nature 2003,425:836–41. [DOI] [PubMed] [Google Scholar]
- 23.Ding L, Saunders TL, Enikolopov G, Morrison SJ: Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012,481:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asada N, Kunisaki Y, Pierce H, Wang Z, Fernandez NF, Birbrair A, Ma’ayan A, Frenette PS: Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol 2017,19:214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. : Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 2013,502:637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, et al. : Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 2010,6:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler IG, Sims NA, Pettit AR, Barbier V, Nowlan B, Helwani F, Poulton IJ, van Rooijen N, Alexander KA, Raggatt LJ, et al. : Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 2010,116:4815–28. [DOI] [PubMed] [Google Scholar]
- 28.Chow A, Lucas D, Hidalgo A, Mendez-Ferrer S, Hashimoto D, Scheiermann C, Battista M, Leboeuf M, Prophete C, van Rooijen N, et al. : Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med 2011,208:261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christopher MJ, Rao M, Liu F, Woloszynek JR, Link DC: Expression of the G-CSF receptor in monocytic cells is sufficient to mediate hematopoietic progenitor mobilization by G-CSF in mice. J Exp Med 2011,208:251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura-lshizu A, Takubo K, Fujioka M, Suda T: Megakaryocytes are essential for HSC quiescence through the production of thrombopoietin. Biochem Biophys Res Commun 2014,454:353–7. [DOI] [PubMed] [Google Scholar]
- 31.Sennett R, Rendl M: Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin Cell Dev Biol 2012,23:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clavel C, Grisanti L, Zemla R, Rezza A, Barros R, Sennett R, Mazloom AR, Chung C-Y, Cai X, Cai C-L, et al. : Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev Cell 2012,23:981–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rompolas P, Mesa KR, Greco V: Spatial organization within a niche as a determinant of stem-cell fate. Nature 2013,502:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshimori N, Fuchs E: Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 2012,10:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E: A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 2009,4:155–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu Y-C, Li L, Fuchs E: Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 2014,157:935–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Yang H, Adam RC, Ge Y, Hua ZL, Fuchs E: Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 2017,169:483–496. In this seminal article, the authors reveal with single cell RNA-seq a heterogeneity amongst HFSCs and transit-amplifying cells in the matrix and spatial “microniches” made by columnar DP, which act to regulate and govern epithelial progenitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL: Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 2011,8:552–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plikus ΜV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong C-M: Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 2008,451:340–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Festa E, Fretz J, Berry R, Schmidt B, Rodeheffer M, Horowitz M, Horsley V: Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell 2011,146:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellana D, Paus R, Perez-Moreno M: Macrophages contribute to the cyclic activation of adult hair follicle stem cells. PLoS Biol 2014,12:e1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C-C, Wang L, Plikus ΜV., Jiang TX, Murray PJ, Ramos R, Guerrero-Juarez CF, Hughes MW, Lee OK, Shi S, et al. : Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 2015,161:277–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong H-A, Lai K, Ahn R, Corbin K, Lowe MM, Scharschmidt TC, et al. : Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 2017,169:1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt GH, Wilkinson MM, Ponder BAJ: Cell migration pathway in the intestinal epithelium: an in situ marker system using mouse aggregation chimeras. Cell 1985,40:425–9. [DOI] [PubMed] [Google Scholar]
- 45.Aoki R, Shoshkes-Carmel M, Gao N, Shin S, May CL, Golson ML, Zahm AM, Ray M, Wiser CL, Wright CVE, et al. : Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol 2016,2:175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stzepourginski I, Nigro G, Jacob J-M, Dulauroy S, Sansonetti PJ, Eberl G, Peduto L: CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A 2017,114:E506–E513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He XC, Zhang J, Tong W-G, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. : BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat Genet 2004,36:1117–1121. [DOI] [PubMed] [Google Scholar]
- 48.Kosinski C, Li VSW, Chan ASY, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, et al. : Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A 2007,104:15418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam M, Midtvedt T, Uribe A: Differential cell kinetics in the ileum and colon of germfree rats. Scand J Gastroenterol 1994,29:445–451. [DOI] [PubMed] [Google Scholar]
- 50.Kaiko GE, Ryu SH, Koues Ol, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, Stappenbeck TS: The Colonic Crypt Protects Stem Cells from Microbiota- Derived Metabolites. Cell 2016,165:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, Clevers H: Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 2005,129:626–38. [DOI] [PubMed] [Google Scholar]
- 52.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H: Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011,469:415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durand A, Donahue B, Peignon G, Letourneur F, Cagnard N, Slomianny C, Perret C, Shroyer NF, Romagnolo B: Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A 2012,109:8965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge Y, Fuchs E: Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet 2018,19:311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao X, Xu C, Asada N, Frenette PS: The hematopoietic stem cell niche: from embryo to adult. Development 2018,145:dev139691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trentesaux C, Romagnolo B: Intestinal Stem Cells and Their Defining Niche In Advances in Stem Cells and their Niches. . Elsevier Inc.; 2018:1–40. [Google Scholar]
- 57.Gonzales KAU, Fuchs E: Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev Cell 2017,43:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crane GM, Jeffery E, Morrison SJ: Adult haematopoietic stem cell niches. Nat Rev Immunol 2017,17:573–590. [DOI] [PubMed] [Google Scholar]
- 59.Schena M, Shalon D, Davis RW, Brown PO: Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995,270:467–70. [DOI] [PubMed] [Google Scholar]
- 60.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR: A stem cell molecular signature. Science 2002,298:601–4. [DOI] [PubMed] [Google Scholar]
- 61.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA: “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 2002,298:597–600. [DOI] [PubMed] [Google Scholar]
- 62.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. : The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002,111:241–50. [DOI] [PubMed] [Google Scholar]
- 63.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR: Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008,133:523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M: The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008,320:1344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B: Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008, 5:621–8. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, Gerstein M, Snyder M: RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009, 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lien W-H, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E: Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 2011, 9:219–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugimura R, He XC, Venkatraman A, Arai F, Box A, Semerad C, Haug JS, Peng L, Zhong X-B, Suda T, et al. : Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell 2012, 150:351–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. : The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 2012, 149:146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Rezza A, Wang Z, Sennett R, Qiao W, Wang D, Heitman N, Mok KW, Clavel C, Yi R, Zandstra P, et al. : Signaling Networks among Stem Cell Precursors, Transit- Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Rep 2016, 14:3001–18. The authors used cell sorting with fluorescent reporters and antibody labeling to purify HFSC, HF progenitors, and key niche components for bulk RNA-seq to generate a transcriptome atlas of key HF populations with superior sensitivity and resolution of mRNA transcript levels, identifying a multitude of newly discovered signature genes and potential signaling partners. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, et al. : Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 2014, 15:507–522. [DOI] [PubMed] [Google Scholar]
- 72.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, Bruggner R V, Melamed R, Trejo A, Ornatsky Ol, et al. : Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. : mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009, 6:377–82. [DOI] [PubMed] [Google Scholar]
- 74.van der Maaten L, Hinton G: Visualizing data using t-SNE. J Mach Learn Res 2008, 9:2579–2605. [Google Scholar]
- 75.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL: The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014, 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herman JS, Sagar Grün D : FatelD infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods 2018, 15:379–386. [DOI] [PubMed] [Google Scholar]
- 77.Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, Kasper M: Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 2016, 3:221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A: Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015, 525:251–5. [DOI] [PubMed] [Google Scholar]
- 79**.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. : A single-cell survey of the small intestinal epithelium. Nature 2017, 551:333–339. In this seminal article, the authors utilize single cell RNA-seq of 53,193 intestinal epithelial cells to identify novel intestinal epithelial cell subtypes, and heterogeneity amongst enteroendocrine cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80*.Roch A, Giger S, Girotra M, Campos V, Vannini N, Naveiras O, Gobaa S, Lutolf MP: Single-cell analyses identify bioengineered niches for enhanced maintenance of hematopoietic stem cells. Nat Commun 2017, 8:221 This article links single cell RNA-seq of isolated cultured HSCs with their in-vitro division history to identify adhesion receptors, JamC and Esam, in promoting quiescence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cabezas-Wallscheid N, Buettner F, Sommerkamp P, Klimmeck D, Ladel L, Thalheimer FB, Pastor-Flores D, Roma LP, Renders S, Zeisberger P, et al. : Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 2017, 169:807–823. [DOI] [PubMed] [Google Scholar]
- 82*.Silberstein L, Goncalves KA, Kharchenko P V., Turcotte R, Kfoury Y, Mercier F, Baryawno N, Severe N, Bachand J, Spencer JA, et al. : Proximity-Based Differential Single-Cell Analysis of the Niche to Identify Stem/Progenitor Cell Regulators. Cell Stem Cell 2016, 19:530–543. In this paper, the authors transplanted RFP-labeled HSCs/progenitors into stromal osteolineage reporter mice, identified distinct engraftment locations relative to HSCs and discovered with scRNA-seq novel niche factors regulating HSC quiescence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shukla S, Langley MA, Singh J, Edgar JM, Mohtashami M, Zúñiga-Pflücker JC, Zandstra PW: Progenitor T-cell differentiation from hematopoietic stem cells using Delta-like-4 and VCAM-1. Nat Methods 2017, 14:531–538. [DOI] [PubMed] [Google Scholar]
- 84*.Fairfield H, Falank C, Farrell M, Vary C, Boucher JM, Driscoll H, Liaw L, Rosen CJ, Reagan MR: Development of a 3D bone marrow adipose tissue model. Bone 2018, doi:10.1016/j.bone.2018.01.023. This article demonstrates modeling of bone marrow adipose tissue (BMAT) with murine and human MSCs seeded onto 3D silk scaffolds. Co-culture with myeloma cell lines then revealed a bidirectional relationship between BMAT and myeloma cells resulting in delipidation of BMAT adipocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85*.Lei M, Schumacher LJ, Lai Y-C, Juan W-T, Yeh C-Y, Wu P, Jiang T-X, Baker RE, Widelitz RB, Yang L, et al. : Self-organization process in newborn skin organoid formation inspires strategy to restore hair regeneration of adult cells. Proc Natl Acad Sci U S A 2017, 114:E7101–E7110. In this paper, epidermal and dermal cells were aggregated from neonatal or adult back skin to demonstrate the self-organization hair follicle induction capacity of aggregates from young skins. Adult back skins were rescued to re-acquire this ability by activating signaling pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lancaster MA, Knoblich JA: Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014, 345:1247125. [DOI] [PubMed] [Google Scholar]
- 87.Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ: Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci U S A 1987, 84:136–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS: In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003, 17:1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y: Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3:519–32. [DOI] [PubMed] [Google Scholar]
- 90.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA: Cerebral organoids model human brain development and microcephaly. Nature 2013, 501:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. : Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459:262–5. [DOI] [PubMed] [Google Scholar]
- 92*.Rodríguez-Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras-Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ, et al. : Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543:424–427. In this article, the authors identify differential metabolic dynamics of ISCs and Paneth cells. Whereas ISCs have high mitochondrial activity, Paneth cells preferentially undergo glycolysis for regulating ISC functions by supplying lactate to support oxidative phosphorylation in ISCs. [DOI] [PubMed] [Google Scholar]
- 93.Bellono NW, Bayrer JR, Leitch DB, Castro J, Zhang C, O’Donnell TA, Brierley SM, Ingraham HA, Julius D: Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell 2017, 170:185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Merenda A, Andersson-Rolf A, Mustata RC, Li T, Kim Η, Koo B-K: A Protocol for Multiple Gene Knockout in Mouse Small Intestinal Organoids Using a CRISPR-concatemer. J Vis Exp 2017, doi: 10.3791/55916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, et al. : Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96*.Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, Virshup DM: PDGFRα + pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A 2018, 115:E3173–E3181. This study demonstrates an important function of pericryptal myofibroblast-derived RSPO3 to stimulate Wnt/β-catenin signaling in the intestinal crypt and contribute to Paneth cell differentiation, both essential for intestinal organoid growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97**.Lee J, Böscke R, Tang P-C, Hartman BH, Heller S, Koehler KR: Hair Follicle Development in Mouse Pluripotent Stem Cell-Derived Skin Organoids. Cell Rep 2018, 22:242–254. In this seminal study, the authors successfully generated differentiated skin and hair in organoids derived from mouse PSCs that essentially recapitulate elements of embryonic skin and hair follicle morphogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coutu DL, Kokkaliaris KD, Kunz L, Schroeder T: Multicolor quantitative confocal imaging cytometry. Nat Methods 2018, 15:39–46. [DOI] [PubMed] [Google Scholar]
- 99.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A: Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol 2011, 14:106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X: RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X: High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci U S A 2016, 113:14456–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sipkins DA, Wei X, Wu JW, Runnels JM, Côté D, Means TK, Luster AD, Scadden DT, Lin CP: In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature 2005, 435:969–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Côté D, Rowe DW, Lin CP, Scadden DT: Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 2009, 457:92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ritsma L, Ellenbroek SIJ, Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H, van Rheenen J: Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 2014, 507:362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, Greco V: Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 2012, 487:496–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106**.Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V: Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 2016, 352:1471–4. In this groundbreaking study, the authors used intravital multiphoton imaging for longitudinal clonal analyses of individual basal epidermal SCs to determine the spatiotemporal coordination of SC behaviors and support a model of equipotency amongst epidermal SCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pineda CM, Park S, Mesa KR, Wolfel M, Gonzalez DG, Haberman AM, Rompolas P, Greco V: Intravital imaging of hair follicle regeneration in the mouse. Nat Protoc 2015, 10:1116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mesa KR, Rompolas P, Zito G, Myung P, Sun TY, Brown S, Gonzalez DG, Blagoev KB, Haberman AM, Greco V: Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 2015, doi:10.1038/nature14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Okkelman IA, Foley T, Papkovsky DB, Dmitriev Rl: Live cell imaging of mouse intestinal organoids reveals heterogeneity in their oxygenation. Biomaterials 2017, 146:86–96. [DOI] [PubMed] [Google Scholar]
- 110.Rios AC, Clevers H: Imaging organoids: a bright future ahead. Nat Methods 2018, 15:24–26. [DOI] [PubMed] [Google Scholar]
- 111*.Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O: Human Brain Organoids on a Chip Reveal the Physics of Folding. Nat Phys 2018, 14:515–522. Engineered human brain organoids were used to model the intercellular and cytoskeletal mechanisms of cerebral folding, a feature of humans not present in rodent models, highlighting the power of human organoids to model processes that are limited in animal models and 2D culture. [DOI] [PMC free article] [PubMed] [Google Scholar]


