Abstract
Conjugating certain types of lentiviral vectors with targeting ligands can redirect the vectors to specifically transduce desired cell types. However, extensive genetic and/or biochemical manipulations are required for conjugation, which hinders applications for targeting lentiviral vectors for broader research fields. We developed envelope proteins fused with biotin-binding molecules to conjugate the pseudotyped vectors with biotinylated targeting molecules by simply mixing them. The envelope proteins fused with the monomeric, but not tetrameric, biotin-binding molecules can pseudotype lentiviral vectors and be conjugated with biotinylated targeting ligands. The conjugation is stable enough to redirect lentiviral transduction in the presence of serum, indicating their potential in in vivo. When a signaling molecule is conjugated with the vector, the conjugation facilitates transduction and signaling in a receptor-specific manner. This simple method of ligand conjugation and ease of obtaining various types of biotinylated ligands will make targeted lentiviral transduction easily applicable to broad fields of research.
Keywords: lentiviral vectors, targeting, biotin, streptavidin, rhizavidin, signaling
Introduction
Lentiviral vectors are used as a gene transduction tool in both experimental and clinical settings that require long-term transgene expression (Naldini, Trono, and Verma, 2016). Their ability to integrate their transgenes into host chromosomes enables their transgenes to be expressed for long periods of time (Naldini et al., 1996a; Naldini et al., 1996b). Although integration of vectors into chromosomes enables long-term transgene expression, it can also cause insertional mutagenesis, which can affect expression of specific host genes (Fraietta et al., 2018) (Ranzani et al., 2013) (Cavazzana-Calvo et al., 2010). Therefore, it is still important to limit integration of the vectors only to the specific target cells, which can be achieved by ex vivo transduction methods, including isolation of target cells, transduction of the isolated cells in vitro, and infusion of the transduced cells back into the body. Because hematopoietic cells can be easily isolated from the body and subsequently infused back in, ex vivo transduction by lentiviral vectors is being successfully used for transduction of hematopoietic cells, especially for treatment of monogenic hematopoietic diseases and expression of chimeric antigen receptors in current clinical trials (Naldini, Trono, and Verma, 2016).
Long-term transgene expression is not only beneficial for treating diseases of hematopoietic cells, but also for other types of cells and tissues; for example, long-term expression of alpha-1 antitrypsin in the lungs and arylsulfatase A in the brain can be used for gene therapy of alpha-1-antitripsin deficiency and metachromatic leukodystrophy, respectively (Wilson et al., 2010) (Sessa et al., 2016). Because ex vivo transduction of cells from solid organs is difficult, there needs to be alternative transduction approaches if lentiviral vectors are to be used to treat diseases that require long-term transgene expression.
When systemically administered, lentiviral vectors pseudotyped with commonly used envelope proteins such as vesicular stomatitis virus glycoprotein (VSV-G) are trapped by the liver and/or spleen and transduce cells in these organs, which decreases the number of vector particles available to reach the target organs (Brown et al., 2006) (Morizono et al., 2005). To efficiently deliver transgenes to target cells and tissues and avoid unnecessary transduction of non-target cells in the liver and spleen, the vectors need to escape trapping and have the ability to specifically bind and transduce the desired cell types. Such vectors are called “targeting vectors” (Kasahara, Dozy, and Kan, 1994).
Because envelope proteins mediate binding of the pseudotypes to target cells, targeting lentiviral vectors are developed by changing the binding specificity of pseudotyping envelope proteins. This requires both eliminating their original tropisms and conferring binding affinities specific to the molecules expressed on target cells(Morizono and Chen, 2005).
The original tropisms of pseudotyping envelope proteins can usually be eliminated by mutating their receptor-binding regions and they are then used as backbones to conjugate the specific targeting ligands(Morizono and Chen, 2011; Nakamura et al., 2005). We previously succeeded in modifying the tropisms of lentiviral vectors by pseudotyping the vectors with modified Sindbis virus envelope proteins(Morizono et al., 2001; Morizono et al., 2010; Morizono et al., 2009a; Morizono et al., 2009b; Morizono et al., 2005). The Sindbis virus has two envelope proteins, E2, which mediates binding, and E1, which mediates fusion(Fields, Knipe, and Howley, 2013; Ohno et al., 1997). We mutated several receptor-binding regions of E2 to eliminate its original tropism(Dubuisson and Rice, 1993; Klimstra, Heidner, and Johnston, 1999; Morizono et al., 2005). This mutated Sindbis envelope protein that lacks its natural tropism provides an ideal basis to develop a targeting lentiviral vector by conjugation with targeting ligands(Ahani et al., 2016; Aires da Silva et al., 2005; Bergman et al., 2004; Kasaraneni et al., 2017; Kasaraneni et al., 2018; Yang et al., 2006).
Approaches for conjugating targeting ligands are largely categorized as either covalent or non-covalent conjugation. The first involves expression of targeting ligands on the viral envelope by making fusion proteins of envelope proteins or membrane-anchoring proteins with targeting ligands. While conjugation by this method is stable, conjugation of each targeting ligand requires DNA cloning and validation of structures and expression levels(Bender et al., 2016; Funke et al., 2008; Kasahara, Dozy, and Kan, 1994; Munch et al., 2011; Nakamura et al., 2005; Sandrin, Russell, and Cosset, 2003; Somia, Zoppe, and Verma, 1995). Additionally, the functions of fusion proteins must be retained for each targeting ligand, as fusion of targeting molecules sometimes affects the functions of envelope proteins and/or targeting ligands(Fielding et al., 1998). For example, fusion of murine leukemia virus envelope proteins with targeting ligands results in loss of the fusion activity of the envelope protein, which is indispensable for transduction(Zhao et al., 1999). The other method is to conjugate targeting molecules non-covalently to the vectors that have adaptor molecules on their surfaces. In this approach, once the function and expression levels of the adaptor molecule on the viral surface are validated, it is not necessary to clone expression plasmids for different types of target molecules and confirm those properties every time the targeting ligands are changed.
We previously used the ZZ peptide, IgG Fc-binding peptide derived from protein A, as an adaptor molecule fused with the mutated Sindbis virus envelope protein (2.2, Fig. 1A and B) to non-covalently conjugate targeting antibodies to lentiviral vectors. Lentiviral vectors pseudotyped with 2.2 can be easily conjugated with antibodies against various target molecules, including CD4, Transferrin receptor 1 (TfR1), PSCA, CD19, CD20, DC-SIGN, CD34, and P-glycoprotein, by simply mixing the vectors with antibodies(Liang et al., 2009a; Liang et al., 2009b; Morizono et al., 2001; Morizono et al., 2010; Morizono et al., 2006; Morizono et al., 2005; Pariente et al., 2007). The vectors specifically transduce cell types recognized by the conjugated antibodies. Due to the ease of conjugating antibodies, this targeting lentiviral vector system has been successfully used by our research group and those of others (Anderson et al., 2009; Bergman et al., 2004; Cao et al., 2016; Lafitte et al., 2012; Wu et al., 2012; Zhang et al., 2011; Zhang et al., 2009; Zhang and Roth, 2010). However, the conjugated antibodies are detached by competitive binding of serum antibodies to the ZZ domain when serum immunoglobulin is present(Morizono et al., 2010). Therefore, the current applications of this targeting lentiviral vector are limited to in vitro settings and an immunodeficient mouse model that does not have serum immunoglobulin(Liang et al., 2009a; Liang et al., 2009b; Morizono et al., 2005; Pariente et al., 2008; Pariente et al., 2007). More stable conjugation methods using adaptor molecules that have higher affinity for their binding molecules need to be developed to overcome this problem.
Fig. 1.
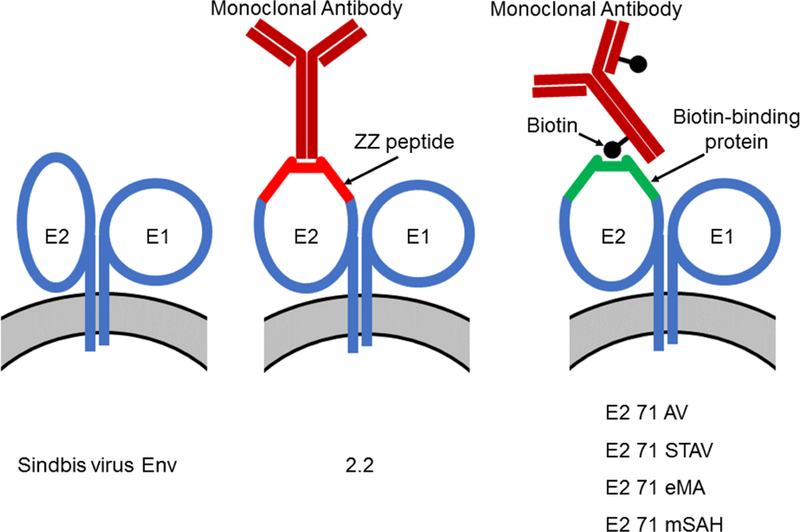
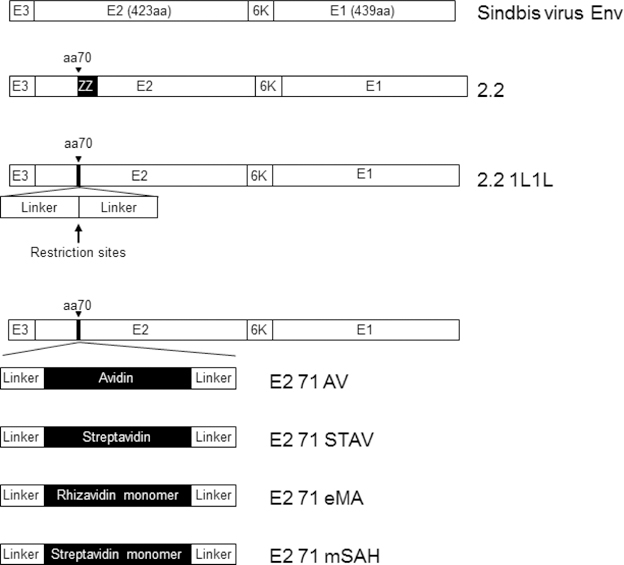
Strategies to conjugate targeting antibodies to lentiviral vectors. (A) Schematic representation of conjugating targeting antibodies to lentiviral vectors. Sindbis virus envelope proteins consist of two types of envelope proteins, E2, which mediates receptor binding, and E1, which mediates fusion between the cell membrane and viral envelope. The 2.2 pseudotype contains the ZZ peptide inserted into the E2 protein. The ZZ peptide binds the Fc region of antibodies. E2 71 AV, STAV, eMA, and mSAH have avidin, streptavidin, monomeric rhizavidin, and the monomeric streptavidin/rhizavidin hybrid, respectively, in E2. These molecules are known to bind biotin; therefore, the E2 71 AV, STAV, eMA, and mSAH pseudotypes are expected to be conjugated with biotinylated antibodies. (B) The two integral membrane glycoproteins, E1 and E2, form a heterodimer and function as a unit. E3 and 6K work as signal sequence peptides for E2 and E1, respectively. 2.2 contains the ZZ peptide at aa 71 of E2, and 2.2 1L1L replaces the ZZ peptide of 2.2 with flexible linkers encompassing restriction sites for cloning. E2 71 AV, STAV, eMA, and mSAH have core sequences of avidin, streptavidin, monomeric rhizavidin, and the monomeric rhizavidin/streptavidin hybrid between the flexible linkers at aa 71 of E2.
Avidin and streptavidin are known to bind biotin at exceptionally high affinities. The dissociation constant (Kd) of binding between these two molecules is 10−15, which is 107−8 less than the Kd of the binding between the ZZ domain and the Fc region of antibodies (Laitinen et al., 2006). Therefore, molecules fused with them can be conjugated with biotinylated targeting ligands.
One group conjugated biotinylated antibodies on lentiviral vectors pseudotyped with both membrane-anchored avidin or streptavidin (for conjugation with biotinylated targeting ligands) and wild-type baculovirus envelope protein (for subsequent fusion) (Kaikkonen et al., 2009) (Huhtala et al., 2014) . Targeting antibodies were stably conjugated in both in vitro and in vivo settings, demonstrating the usefulness of avidin/biotin interactions for conjugation of virus with ligands. However, the targeting specificity of this vector is poor because this pseudotype still retains the broad tropism of pseudotyping baculovirus envelope protein. These studies indicated that the original receptor-binding regions of pseudotyping envelope proteins should be abolished for highly specific targeting lentiviral vectors, even when strong avidin-biotin interactions are used for ligand conjugation.
In the present study, we utilized the strong binding between biotin-binding molecules derived from streptavidin and rhizavidin with biotin and our mutated Sindbis virus envelope backbone to redirect the pseudotyped lentiviral vectors by stable conjugation with biotinylated targeting ligands.
Results
Designing biotin-binding envelope proteins.
The backbone construct, 2.2 1L1L, is derived from wild-type Sindbis virus with multiple mutations at the original receptor-binding regions (Fig. 1B) (Morizono et al., 2006; Morizono et al., 2005; Pariente et al., 2007). 2.2 1L1L contains restriction sites for inserting an adaptor molecule(s) between aa 71 and 74 of E2, which has flexible linkers before and after the insertion sites (Morizono et al., 2009a; Morizono et al., 2009b). At the insertion site, 2.2 contains the ZZ peptide (Fig. 1A and B). Since the conjugation of the ZZ domain with antibodies is unstable in serum, we attempted to develop strategies to stably conjugate targeting ligands to our targeting vectors. Because the binding of biotin and avidin/streptavidin is known to be stable in various in vivo and in vitro settings due to its extremely high affinity (Bayer and Wilchek, 1990), we inserted avidin or streptavidin into 2.2 1L1L, designated E2 71 AV and E2 71 STAV, respectively (Fig. 1A and B). Because the Sindbis virus envelope proteins form trimers (Li et al., 2010), whereas avidin and streptavidin form tetramers (Bayer and Wilchek, 1990), it is possible that fusion of the Sindbis envelope proteins with avidin/streptavidin impairs proper multimerization and structures on both sides. Thus, we inserted monomeric biotin-binding molecules eMA and mSAH instead of avidin and streptavidin into 2.21L1L, designated E2 71 eMA and E2 71 mSAH, respectively (Fig. 1A and B). eMA is a monomeric form mutant of rhizavidin, which is a dimeric biotin-binding molecule isolated from Rhizobium etli (Helppolainen et al., 2007; Lee et al., 2016). mSAH is a monomeric streptavidin developed by combining amino acid sequences of streptavidin with rhizavidin (Demonte et al., 2013). It was reported that the Kd of eMA and biotin was 3.1×10−11, and dissociation of eMA bound to biotin conjugate was not observed by surface plasmon resonance analysis (Lee et al., 2016). The Kd of mSAH and biotin was shown to be 7.3X10−10 (Demonte et al., 2013). Although the Kd of mSAH with biotin is higher than that of tetrameric streptavidin, binding of mSAH with biotinylated molecules was shown to be stable more than 1 hour in the presence of competing free biotin.
Pseudotyping lentiviral vectors with biotin-binding envelope proteins.
Because pseudotyping of envelope proteins occurs at the surface of vector producer cells, we first checked for expression of the envelope proteins on the surfaces of transfected 293T cells. Staining of transfected cells showed that E2 71 AV, E2 71 STAV, E2 71 eMA, and E2 71 mSAH are expressed on the cell surface (Fig. 2A). To investigate whether these envelope proteins can bind biotinylated molecules, we stained the transfected cells with biotinylated FITC and analyzed binding to transfected cells by flow cytometry (Fig. 2A). We observed no binding of biotinylated FITC to the cells expressing E2 71 AV or E2 71 STAV, demonstrating that avidin and streptavidin do not have the structures necessary for binding to biotin when fused to the Sindbis virus envelope protein. However, we observed that biotinylated FITC bound to cells transfected with E2 71 eMA or E2 71 mSAH.
Fig. 2.
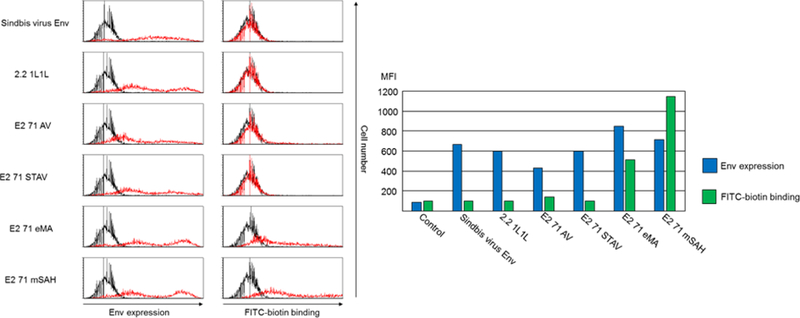
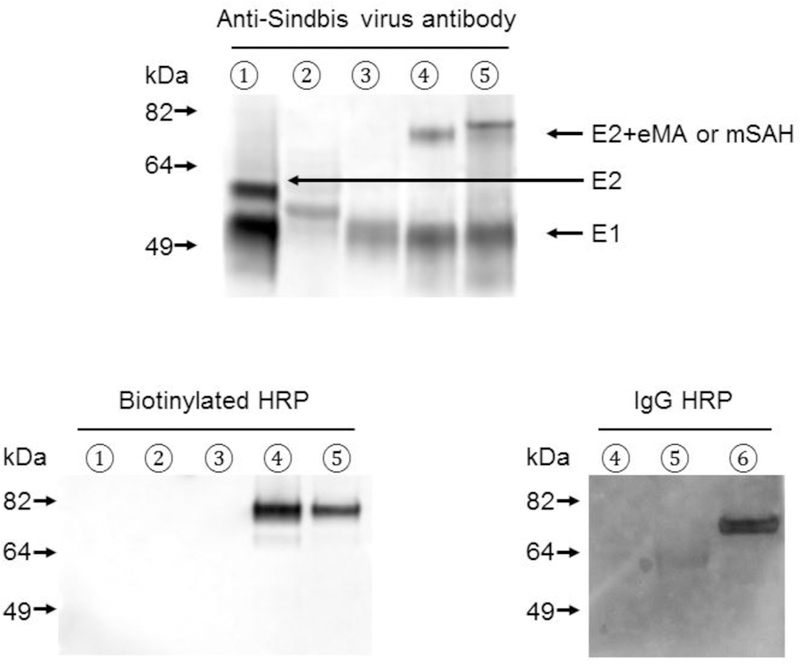
Expression of various envelope proteins on transfected cells and lentiviral vectors. (A) 293T cells transfected with control vector (black line) or envelope protein expression vectors (red line) were stained with anti-Sindbis virus antibody or biotinylated FITC and analyzed by flow cytometry. Flow cytometric profiles and mean fluorescence intensities of staining with anti-Sindbis virus envelope protein antibody and biotinylated FITC are shown. (B) Western blotting analysis of lentiviral vectors pseudotyped with: ① 2.2 1L1L, ② E2 71 AV, ③ E2 71 STAV, ④ E2 71 eMA, ⑤ E2 71 mSAH, or ⑥ 2.2. The same amounts of vectors (110 ng p24) were subjected to SDS-PAGE. After western blotting, the blotted membranes were stained with anti-Sindbis virus antibody, biotinylated HRP, or rabbit IgG-conjugated HRP.
We also investigated whether these envelope proteins can pseudotype lentiviral vectors, using western blotting analysis of the purified lentiviral vectors. The fusion envelope proteins were detected using anti-Sindbis virus antibodies and western blot analysis. The molecular weights of the E2 protein of E2 71 eMA were the same as that expected from fusion of the Sindbis virus E2 protein and eMA (~75 kDa) (Fig. 2B, top). The E2 protein of E2 71 mSAH showed a faint band of the expected size (~75 kDa) and a strong band at a higher than expected molecular weight (Fig. 2B, top). Because mSAH contains 5 N-glycosylation signal sequences (N-X-S/T), it is likely that the higher molecular weight represents N-glycosylation of mSAH. We did not detect immunoblot bands at the expected molecular weights (~75 kDa) of fusion proteins E2 71 AV and E2 71 STAV, demonstrating that Sindbis virus envelope proteins lose their ability to properly express and/or pseudotype lentiviral vectors when fused to avidin or streptavidin. The cryo-electron microscopic and crystal structure analyses of Sindbis virus envelope proteins previously demonstrated that a heterodimer of E1/E2 supports the structure of other heterodimers by forming the trimers of heterodimers (Li et al., 2010; Mukhopadhyay et al., 2006; Pletnev et al., 2001). It is possible that inhibition of the trimerization by avidin and/or streptavidin destabilizes E2 71 AV and E2 71 STAV, so these envelope proteins cannot survive viral purification procedures.
We next investigated whether the fusion proteins between E2 and eMA or mSAH can specifically bind biotinylated molecules. E2 71 AV and STAV did not show binding of biotinylated HRP by western blot analysis (Fig. 2B, bottom left), which is consistent with lack of binding activity to biotin shown by flow cytometric analysis (Fig 2A) and low expression on virions (Fig 2B, top). HRP conjugated with biotin specifically bound to the E2 protein of E2 71 eMA and the lower molecular weight form of E2 of E2 71 mSAH (Fig. 2B, bottom left). HRP conjugated with rabbit IgG specifically bound the E2 fusion protein with ZZ of the 2.2 pseudtotype via the interactions between the IgG Fc and ZZ domains, but did not bind E2 71 eMA or E2 71 mSAH (Fig. 2B, bottom right), demonstrating that binding of HRP-biotin to E2 71 eMA or E2 71 mSAH is biotin-dependent. These results demonstrate that biotinylated molecules can be conjugated to E2 71 eMA and E2 71 mSAH through the interactions between biotin and eMA or mSAH.
Redirecting the E2 71 eMA and E2 71 mSAH pseudotypes by conjugation with biotinylated antibodies.
We next investigated whether the E2 71 eMA and mSAH pseudotypes can transduce cells when conjugated with biotinylated antibodies. We attempted to redirect the tropisms of the pseudotyped vectors by mixing them with biotinylated or non-biotinylated anti-human TfR1 (hTfR1) antibody and transducing the human T-cell line, Jurkat, which abundantly expresses hTfR1 (Fig. 3A). Conjugation of biotinylated and non-biotinylated anti-hTfR1 antibody with the 2.2 pseudotype drastically increased transduction efficiencies of Jurkat cells with the 2.2 pseudotype. Conjugation of the E2 71 eMA and mSAH pseudotypes with biotinylated anti-hTfR1 antibody but not the non-biotinylated antibody increased the transduction of Jurkat cells. Mixing the 2.2 1L1L pseudotype with biotinylated anti-hTfR1 antibodies did not mediate transduction of Jurkat cells. These results are consistent with the binding specificities of E2 71 eMA and mSAH shown by western blot analysis (Fig. 2B). Conjugation of these pseudotypes with biotinylated isotype control antibody did not mediate transduction of Jurkat cells, demonstrating that binding of conjugated antibodies to target cells is necessary for transduction by E2 71 eMA and mSAH pseudotypes. Although using higher concentrations of viral vectors yields higher percentages of transduction (as shown the in the results of subsequent experiments), we used the viral amounts that transduce less than 20% of those obtained for the experiments shown in Fig 3A to maintain the linear correlation between the percentages of EGFP+ cells and the titers of the vectors. These results demonstrated that conjugation of targeting antibodies with the E2 71 eMA and mSAH pseudotypes via the interactions between biotin and eMA and mSAH can redirect the tropisms of the vectors.
Fig. 3.
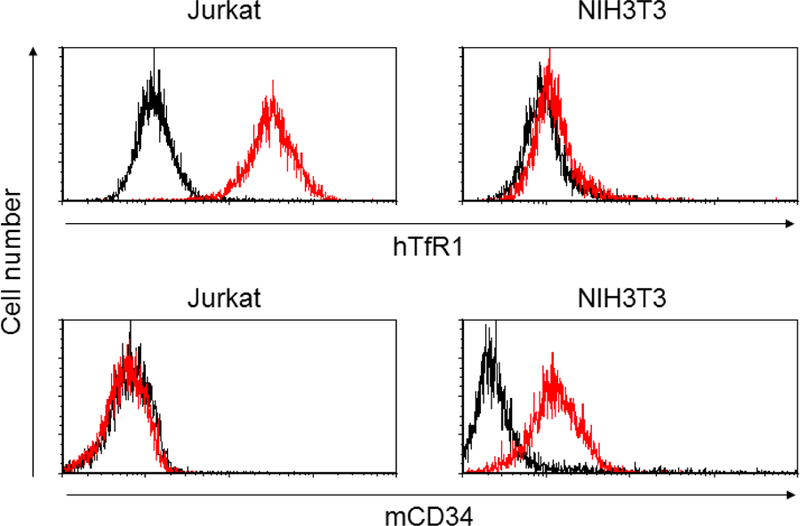
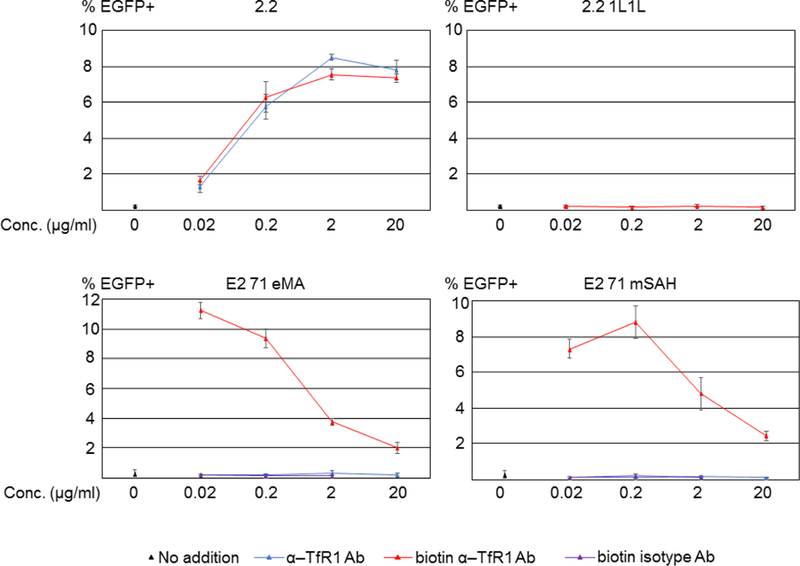
Targeted transduction with biotinylated antibodies. (A) Jurkat and NIH3T3 cells were stained with biotinylated anti-hTfR1, mCD34, or isotype control antibodies, followed by staining with Alexa 488-conjugated streptavidin. The staining was analyzed by flow cytometry. (B) Jurkat cells (1×105 cells) were infected with the same amount (1 ng p24/200 µl) of 2.2, 2.2 1L1L, E2 71 eMA, or E2 71 mSAH pseudotypes pre-incubated with or without targeting ligands (black dot, no ligand; blue line, anti-hTfR1 antibody; red line, biotinylated anti-hTfR1 antibody; purple line, biotinylated isotype control antibody) at different concentrations. Transgene (EGFP) expression was analyzed by flow cytometry 3 days post-transduction. The averages and standard derivations of triplicate experiments are shown.
Of note, while higher concentrations of antibodies do not inhibit antibody-directed transduction of the 2.2 pseudotype, the high concentrations of antibodies decrease transduction of E2 71 eMA and mSAH (Fig. 3B). A research group also reported that targeted lentiviral transduction was inhibited by the presence of excess amounts of targeting ligands when covalently conjugating the targeting ligands to the modified Sindbis virus envelope protein obtained from us (Kasaraneni et al., 2018). It is likely that: 1) excess amounts of the anti-hTFR1 biotinylated antibody competitively bind to hTfR1 expressed on Jurkat cells, thereby inhibiting transduction with the E2 71 eMA and mSAH pseudotypes stably conjugated with anti-hTfR1 antibody; and 2) due to the relatively low affinity of interactions between the ZZ and Fc domains, the 2.2 pseudotype prefers to bind hTfR1 antibodies bound on the surfaces of target cells rather than free excess amounts of the anti-hTFR1 antibody. We also conjugated E2 71 eMA with the lower concentration (0.002 μg/ml) of biotinylated anti-hTfR1 antibody, but the titer is then lower than when conjugated with the same antibody at 0.02 μg/ml (data not shown).
We previously showed that the antibodies conjugated on 2.2 are detached in the serum due to the competitive binding of serum immunoglobulin to the ZZ domain. To investigate the stability of antibody conjugation, we incubated the 2.2, E2 71 eMA, and E2 71 mSAH pseudotypes conjugated with biotinylated anti-hTfR1 antibodies with human serum before transduction. As we previously reported (Morizono et al., 2010), human serum IgG disrupts the conjugation of the antibodies from virus by competitive binding to the ZZ domain and, accordingly, the 2.2 pseudotype conjugated with anti-hTfR1 antibody lost its infectivity (Fig. 4A). On the other hand, E2 71 eMA and mSAH maintain their abilities to transduce Jurkat cells in the presence of human serum, demonstrating more stable conjugation with the antibodies.
Fig. 4.
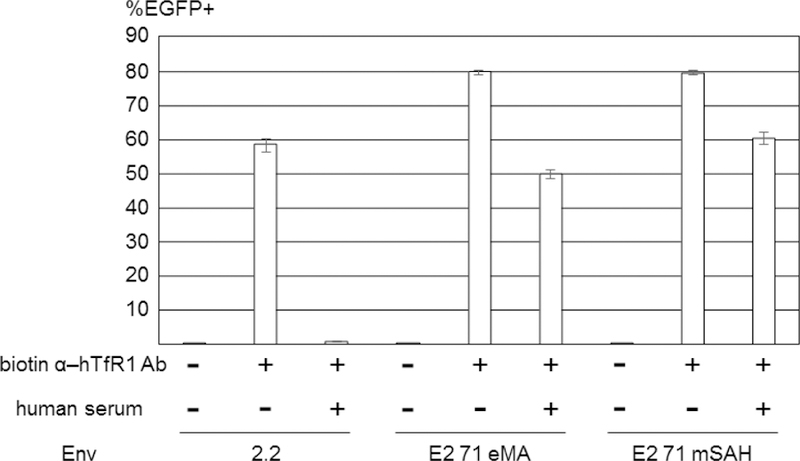
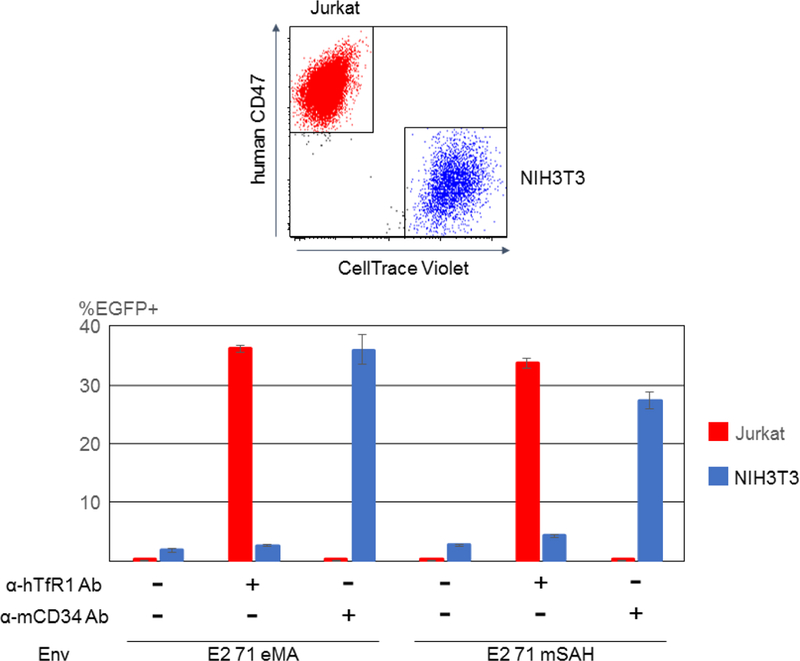
Properties of targeted transduction by the E2 71 eMA and E2 71 mSAH pseudotypes. (A) 2.2, E2 71 eMA, and E2 71 mSAH were conjugated with biotinylated anti-hTfR1 antibody. The conjugated vectors were incubated with or without human serum (50%) for 1 hour at 37ºC and used to transduce Jurkat cells. EGFP transgene expression was analyzed by flow cytometry. The averages and standard deviations of the triplicate experiments are shown. (B) E2 71 eMA and mSAH pseudotypes (100 ng p24/200 µl) were incubated with or without biotinylated anti-hTfR1 or mCD34 antibodies (200 pg). The vectors were then used to transduce mixtures of Jurkat (5×104) and CellTrace Violet-labeled NIH3T3 (2×104) cells. Three days post-transduction, the cells were stained with APC-conjugated anti-human CD47 antibody that specifically stains Jurkat cells. EGFP expression of Jurkat (APC+/CellTrace Violet-) and NIH3T3 (APC-/CellTrace Violet+) cells were analyzed by flow cytometry. The averages and standard derivations of triplicate experiments are shown.
To confirm that the E2 71 eMA and mSAH pseudotypes can be redirected according to the specificity of the conjugated antibodies, we transduced mixed cultures of two cell types, using these pseudotypes conjugated with antibodies specific to one cell type in the culture. Anti-mouse CD34 (mCD34) monoclonal antibody binds NIH3T3 but not Jurkat cells (Fig. 3A), whereas anti-hTfR-1 antibody binds Jurkat but not NIH3T3 cells (Fig. 3A). We transduced co-cultures of Jurkat and NIH3T3 cells with E2 71 eMA or mSAH conjugated with either anti-hTfR1 or mCD34 antibody, and 3 days post-transduction, we analyzed transgene expression (EGFP) of Jurkat and NIH3T3 cells. To distinguish between these two cells types, NIH3T3 was labeled with CellTrace Violet before co-culture, and Jurkat cells were stained with APC-conjugated anti-human CD47 antibody that specifically stains Jurkat cells. EGFP expression of Jurkat (APC+, CellTrace Violet-) and NIH3T3 (APC-, CellTrace Violet+) cells was analyzed individually (Fig. 4B). When the pseudotypes were not conjugated with any antibodies, both cell types were minimally transduced. Conjugation of the pseudotypes with anti-hTfR1 antibody specifically increased transduction of Jurkat cells, and conjugation with anti-mCD34 specifically increased transduction of NIH3T3 cells. These results confirm that the tropisms of the E2 71 eMA and mSAH pseudotypes are determined by the specificities of the conjugated antibodies.
The effects of biotinylation sites of targeting ligands on the titers of E2 71 eMA and E2 71 mSAH pseudotypes.
The antibodies against hTfR1 and mCD34 used in the previous experiments were purchased from biotech companies as regular catalog items. Commercial biotinylated antibodies are usually biotinylated by random biotinylation of exposed lysine, using NHS-biotin. Because biotinylation can occur on any exposed lysine, topology of biotinylated antibodies could be in any direction when conjugated with the E2 71 eMA or mSAH pseudotypes (Fig. 5A). If antigen-binding regions of antibodies are not directed toward targeting antigens, the antibody will not be able to efficiently bind to target cells and mediate transduction. Therefore, it is ideal for biotinylation to occur specifically at the C-terminus of antibodies for efficient transduction of target cells with the E2 71 eMA and mSAH pseudotypes (Fig. 5B). For this purpose, we attempted to perform specific biotinylation of the C-terminus of the anti-hTfR1 antibody. We first reduced the S-S binding between heavy chains of anti-hTfR1 antibodies, using β-mercaptoethylamine, followed by biotinylation of the reduced cysteines with Maleimide-PEG2-Biotin (Fig. 5B). To expose conjugated biotin at the C-terminus for better binding to eMA and mSAH, we eliminated the Fc domain of the antibodies by digestion with pepsin (Fig. 5B) and designated this antibody as biotin anti-hTfR1 Fab. SyproRuby staining showed that the majority of heavy chains were eliminated with biotin anti-hTfR1 Fab (Fig. 5C). Western blotting analysis with streptavidin-HRP showed that biotinylation specifically occurs at heavy chains of biotin anti-hTfR1 Fab, while non-specific biotinylation of lysines biotinylate both light and heavy chains of commercial anti-hTfR 1 antibody. These results demonstrated that biotin anti-hTfR1 Fab possesses biotin at the C-terminus of its heavy chain. We next investigated whether biotin anti-hTfR1 Fab mediates transduction of the E2 71 eMA and mSAH pseudotypes more efficiently than biotinylated anti-hTfR1 antibody (Fig. 5D) and found that biotin anti-hTfR1 Fab mediates transduction two-fold more efficiently than biotinylated anti-hTfR1 antibody. These results indicate that controlled biotinylation of the C-terminus of antibodies is ideal for efficient targeting of conjugated vectors.
Fig. 5.

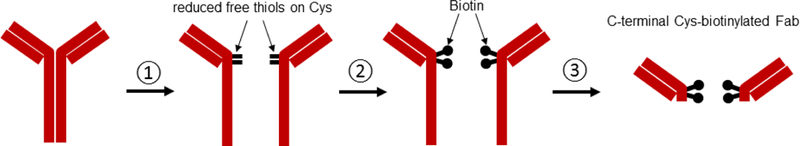
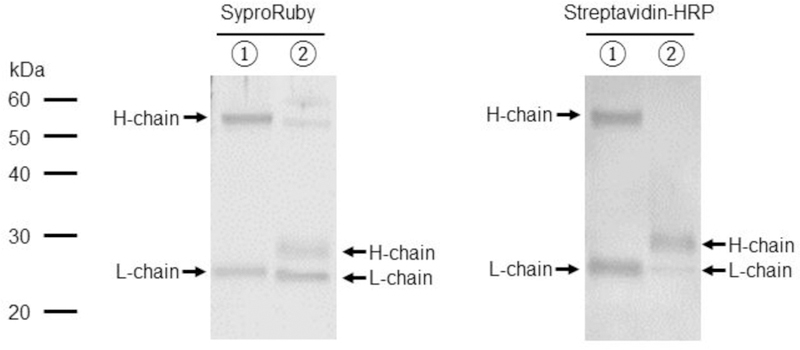
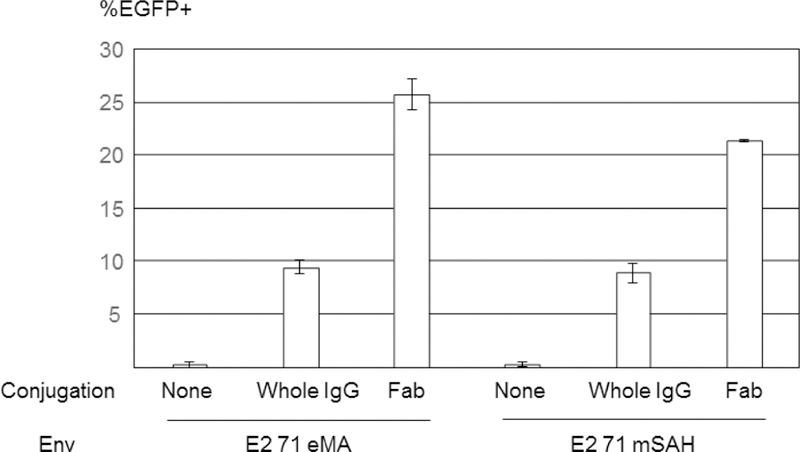
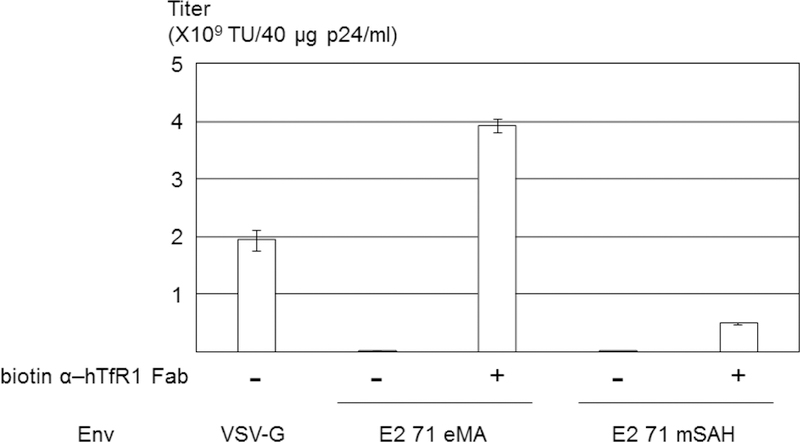
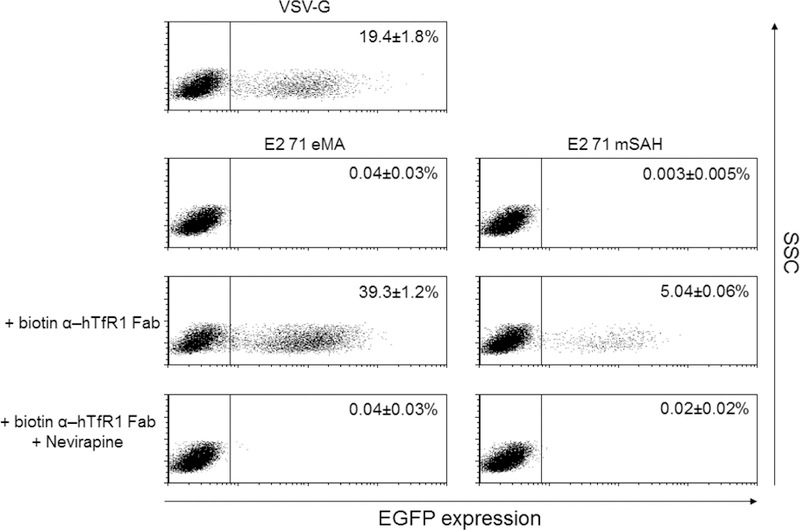
Site-specific biotinylation of the anti-hTfR1 antibody. (A) Schematic representation of conjugation of antibodies randomly biotinylated at lysine residues, the Fab fragment specifically biotinylated at C-terminal cysteines, and the biotinylated EGF. (B) ① disulfide bonds between heavy chains of anti-hTfR1 antibody are reduced by 2-mercaptethanolamine; ② the reduced cysteines were biotinylated with maleimide biotin; and ③ Fc regions of the biotinylated antibody are digested with pepsin. (C) ① Biotinylated anti-hTfR1 antibody; and ② the Fab fragment of cysteine-biotinylated anti-hTfR1 antibody subjected to SDS-PAGE, followed by SyproRuby staining and western blotting, using HRP-conjugated streptavidin. (D) Jurkat cells (1×105 cells) were infected with the same amount (1 ng p24/200 µl) of E2 71 eMA or E2 71 mSAH pseudotypes pre-incubated with or without biotinylated anti-hTfR1 antibody or biotinylated Fab fragment of anti-hTfR1 antibody at 200 ng/ml. Transgene (EGFP) expression was analyzed by flow cytometry 3 days post-transduction. The averages and standard derivations of triplicate experiments are shown. (E) The titers (TU/40 µg p24/ml) of VSV-G pseudotype and the E2 71 eMA and mSAH pseudotypes with or without biotin α–hTfR1 Fab conjugation. The titers were calculated by triplicated transduction of Jurkat cells with EGFP-expressing vectors. The averages and standard derivations are shown. (F) Flow cytometric profiles of Jurkat cells (1×105) transduced with VSV-G, E2 71 eMA or mSAH pseudotypes (1 ng p24/200 µl) conjugated with the Fab fragment of cysteine-biotinylated anti-hTfR1 antibody (10 ng) in the presence or absence of nevirapine (1 µM).
Transduction of Jurkat cells with the anti-hTfR1 Fab-conjugated E2 71 eMA pseudotype was very efficient, and the titers for the same amount of the viral protein (p24) were higher than with the titers of VSV-G pseudotype (Fig. 5E) [VSV-G: 1.9×109 transduction unit (TU)/40 µg p24/ml, E2 71 eMA + anti-hTfR1 Fab: 3.9×109 TU/40 µg p24/ml]. Since the titers are exceptionally high, it is possible that the EGFP expression of the cells transduced by the E2 71 eMA and SAH pseudotypes is mediated by pseudo-transduction, which is the binding of the EGFP protein associated with the vectors (Kim et al., 2017). The flow cytometric profile of EGFP expression from the transduced Jurkat cells showed distinct EGFP-positive populations (Fig. 5F), which indicates endogenous EGFP expression. We further investigated whether this expression is mediated by the transduced EGFP gene, using the reverse transcriptase inhibitor, nevirapine (Morizono et al., 2010). Nevirapine eliminated more than 99% of EGFP expression of Jurkat cells transduced with anti-hTfR1 Fab conjugated with the E2 71 eMA and mSAH pseudotypes, demonstrating that these vectors induce EGFP expression by transduction.
Redirecting E2 71 eMA and mSAH pseudotypes by biotinylated EGF.
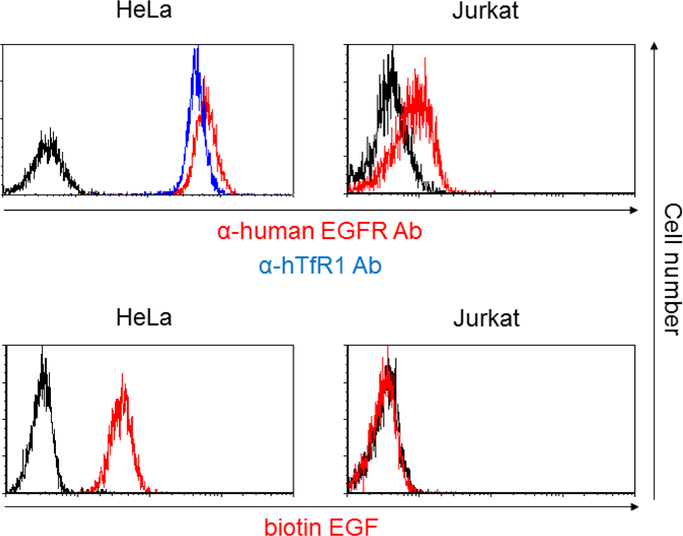
Because conjugation of antibodies to E2 71 eMA and mSAH occurs via the interactions between biotin and eMA and mSAH, the targeting ligand is not limited to antibodies, but occurs with any biotinylated molecule. To test this hypothesis, we attempted to target EGFR by conjugating biotinylated EGF with the E2 71 eMA and mSAH pseudotypes. HeLa cells abundantly express EGFR, but Jurkat cells do not (Fig. 3A and 6A). Both cell types expressed hTfR1 (Fig. 3A and 6A). HeLa cells can be efficiently transduced by the wild-type Sindbis virus pseudotype. Mutations introduced into its receptor-binding regions decreased the titers of the 2.2, E2 71 eMA and mSAH pseudotypes by more than 30-fold (Fig. 6B). Addition of biotinylated EGF increased transduction of HeLa cells with the E2 71 eMA and mSAH pseudotypes, but not 2.2 (Fig. 6B). Transduction of Hela cells with EGF-conjugated E2 71 eMA and mSAH pseudotypes was blocked by antibodies against EGFR, while transduction of anti-hTfR1 antibody-conjugated E2 71 eMA and mSAH pseudotypes was not (Fig. 6C). Transduction of EGFR-negative Jurkat cells with the E2 71 eMA and mSAH pseudotypes was not enhanced by addition of biotinylated EGFR (Fig. 6B). These results demonstrated that the E2 71 eMA and mSAH pseudotypes can be redirected by any biotinylated molecules.
Fig. 6.
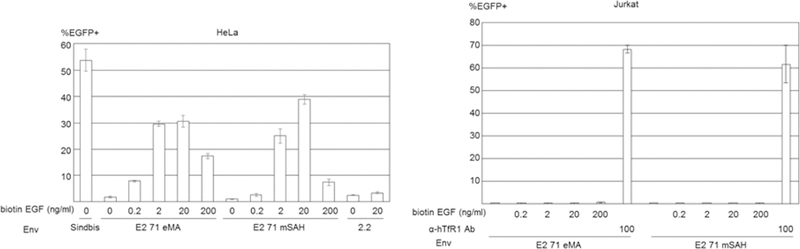
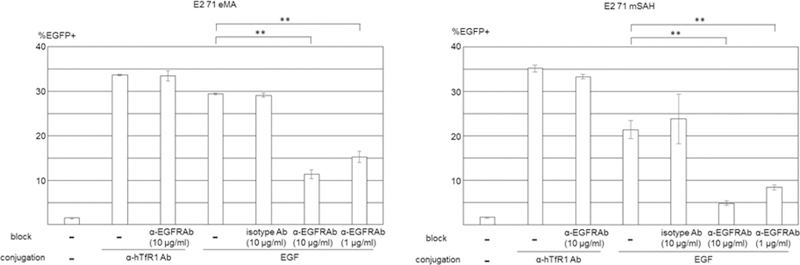
Targeted transduction by conjugation with biotinylated EGF. (A) HeLa and Jurkat cells were stained with APC-conjugated anti-human EGFR (red line), hTfR1 (blue line), and its isotype control antibodies (black line). Hela and Jurkat cells were incubated with biotinylated EGF (red line) or buffer only (black line), followed by incubation with APC-conjugated streptavidin. (B) HeLa cells (5×104) were transduced with the same amount of wild-type Sindbis virus envelope protein and the E2 71 eMA and mSAH, or 2.2 pseudotype (100 ng p24/200 µl) with or without conjugation with various concentrations of biotinylated EGF. Jurkat cells (1×105) were transduced with the E2 71 eMA and mSAH pseudotypes (100 ng p24/200 µl) with or without conjugation with biotinylated EGF (0.2–200 ng/ml) or anti-hTfR1 antibody (100 ng/ml). EGFP transgene expression was analyzed 3 days post-transduction. The averages and standard derivations of the triplicate experiments are shown. (C) HeLa cells (5×104) were incubated with anti-EGFR antibody (1 or 10 µg/ml) or its isotype control antibody (10 µg/ml), followed by transduction with the E2 71 eMA or mSAH pseudotypes (100 ng p24/200 µl) with or without conjugation with biotinylated EGF (20 ng/ml) or anti-hTfR1 antibody (200 ng/ml). EGFP transgene expression was analyzed 3 days post-transduction. The averages and standard derivations of the triplicate experiments are shown. Significance was calculated by comparing anti-hTfR1 antibody and EGF-mediated transduction without blocking antibody to those with blocking antibodies, using a two-sample two-sided unpaired student t-test (**, p<0.01).
EGF induces signaling via a receptor tyrosine kinase, EGFR, by phosphorylation of the cytoplasmic tyrosine residues of EGFR. It was recently shown that Gas6, a ligand of the TAM family of tyrosine kinase receptors, elicits signaling more efficiently when conjugated on the surface of virions than on free protein (Bhattacharyya et al., 2013). We investigated whether EGFR conjugated on virions can induce phosphorylation of EGFR more efficiently than unconjugated (free) EGFR. We added EGF conjugated with E2 71 eMA pseudotype to HeLa cells and analyzed phosphorylation of EGFR cytoplasmic tyrosine at various time points by Luminex assay. As a control of phosphorylation by free EGF with unconjugated lentiviral vector, we added the same amount of EGF and 2.2 pseudotype. Increases in phosphorylation were observed from 30 sec and peaked at 10 min after addition of EGF (Fig. 7). The EGF mixed with E2 71 eMA induced two-fold more phosphorylation of EGFR than free EGF, which is observed from 30 sec to 2 hours after addition of virus and EGF. These results demonstrated that EGF conjugated on the surface of virus can induce intracellular signaling more efficiently than free EGF.
Fig. 7.
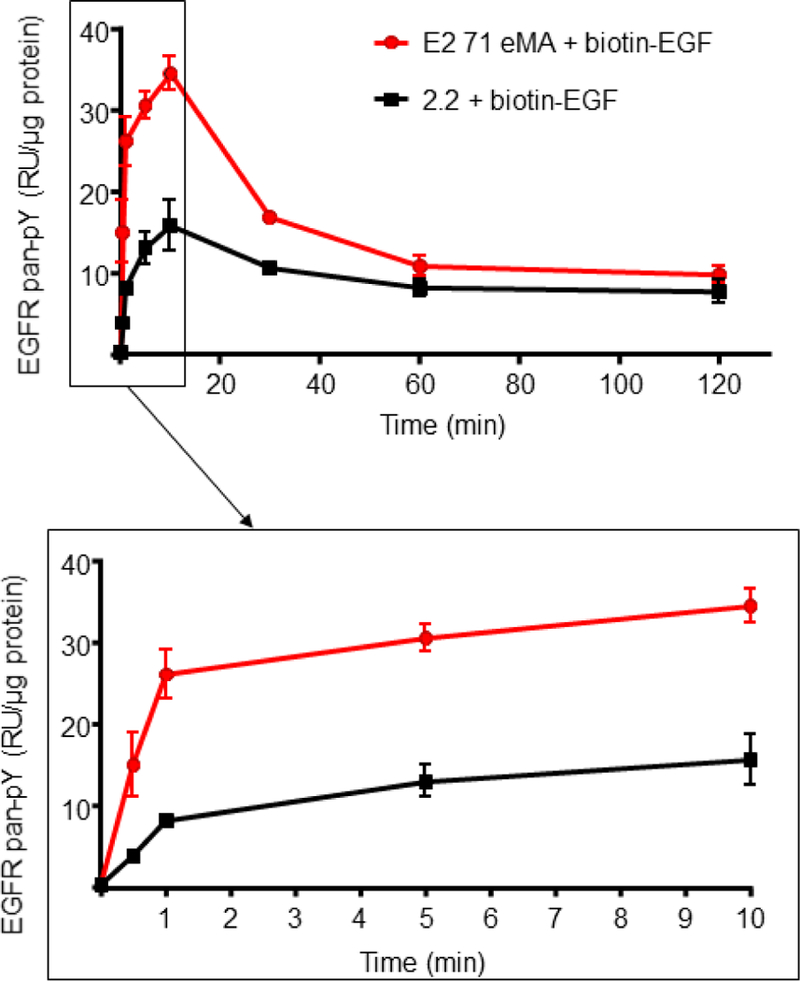
Signaling elicited by EGF conjugated with the E2 71 eMA pseudotype. Phosphorylation of EGFR of Hela cells after 30 sec, 1 min, 5 min, 10 min, 30 min, 1 hour, or 2 hours of incubation with 200 µl of the E2 71 eMA or 2.2 pseudotype (100 ng p24/100 µl) and biotinylated EGF (2 ng/ml). The averages and standard derivations of the triplicate experiment are shown.
Discussion
Intensive and time-consuming molecular and/or biochemical manipulations have been required for redirecting lentiviral vectors to individual target molecules (Ahani et al., 2016; Kasaraneni et al., 2017; Kasaraneni et al., 2018; Morizono et al., 2009a) (Morizono et al., 2009b), which has hindered wide applications of targeted lentiviral transduction. Here we report development of lentiviral vectors that can be stably conjugated with biotinylated targeting molecules and redirected, requiring only simple mixing. Wide varieties of biotinylated antibodies and ligands are commercially available from many manufacturers. For example, the three targeting molecules used in this study, biotinylated anti-hTfR1, mCD34 antibodies, and biotinylated EGF, are regular catalog products of globally accessible manufacturers (Biolegend and ThermoFisher Scientific). In addition, conjugating biotin to lysine residues is a relatively simple procedure that does not require special devices or techniques; thus, researchers can biotinylate targeting ligands of interest if they are not commercially available.
The highly stable binding of this targeting technology will be important for various in vitro and in vivo settings. The conjugation method involves simply mixing virus and biotinylated targeting ligands, and the availability of wide varieties of biotinylated molecules from various companies enables many researchers to utilize this technology to target cells of interest.
Fusion of the E2 protein with wild-type avidin and streptavidin interfered with the ability of the envelope to pesudotype lentiviral vectors and of avidin and streptavidin to bind biotin. It is likely that tetramerization induced by wild-type avidin and streptavidin interferes with trimerization of Sindbis virus envelope proteins, which is required for proper folding and expression on the envelope. Development of monomeric rhizavidin and streptavidin, which bind biotin at high affinities, enabled us to express biotin-binding envelope proteins on the envelope. Viral envelope proteins of various viruses are known to form trimers (Fields, Knipe, and Howley, 2013) (Gibbons et al., 2000; Wilson, Skehel, and Wiley, 1981; Zhu et al., 2006). eMA and mSAH may be able to fuse with such envelope proteins without interfering with the expression and functions of the envelope proteins. In addition, receptor-binding proteins of non-enveloped viruses such as adenovirus also form trimers (Xia et al., 1994). Fusion of these proteins with eMA and/or mSAH may facilitate redirection of the tropisms of non-envelope viral vectors by conjugation with biotinylated ligands.
Our results comparing non-specific lysine and site-specific biotinylation indicate that the topology of conjugated antibodies is important for efficient transduction of target cells. The placement of targeting ligands on the viral surface can adversely affect the antigen-binding regions of these ligands. In the case of redirecting measles virus envelope proteins fused with targeting ligands, linkers between targeting ligands, and envelope proteins resulted in reduced titers of the pseudotyped lentiviral vectors (Rasbach et al., 2013).
The importance of the topology of conjugated targeting ligands will be dependent on how the ligands bind to target antigens. A previous study by another research group showed that the targeting ligand conjugated on the virus cannot efficiently bind the membrane proximal site of the HER2/nre receptor, while the conjugated ligand targeting the membrane distal sites with the same receptor can do so efficiently (Kasaraneni et al., 2018) . If the binding sites of the ligands are localized to the exposed uppermost surface of targeted molecules, the topology of the targeting ligands conjugated on the envelope may not affect binding and transduction efficiencies of vectors. When the binding site is located at the plasma membrane-proximal sites, it is likely that the targeting ligands must be directed towards the binding site to optimally access and bind targeted cell surface molecules.
Our results showed that intracellular EGFR signaling was triggered by the binding of our conjugated vectors to the surface receptor. We previously reported that Gas6, which binds the viral envelope via binding to the envelope lipid, phosphatidylserine (PtdSer), drastically enhanced lentiviral transduction and infection of replication-competent vaccinia virus by 10–50-fold (Morizono and Chen, 2014; Morizono et al., 2011). Subsequent studies of other groups showed that 80% of this enhancement is mediated by signaling via the binding of Gas6 to its receptors (Bhattacharyya et al., 2013). While binding of Gas6 to virus is specifically mediated by the PtdSer-binding region of Gas6 and PtdSer of the viral envelope, E2 71 eMA and mSAH will enable the display of any biotinylated signaling molecule.
Antibodies against CD3 fused with GPI-anchors were previously displayed on the viral membrane to induce signaling in T-cells (Derdak et al., 2006). Signaling via CD3 is known to facilitate a post-binding step(s) of lentiviral transduction and HIV infection (Korin and Zack, 1998; Zack et al., 1990). While these studies used displayed ligands on vector particles for the purpose of cell activation and not for targeting, our system allows for targeted transduction that is facilitated by both specific binding and induction of supportive signaling for lentiviral transduction. Because any type of biotinylated molecule can be used as a conjugating ligand, this technology will enable researchers in various research fields to transduce target cells and/or elicit signaling, using a wide variety of targeting and/or signaling ligands.
We and other research groups have successfully used our previous targeting lentiviral vector system with the ZZ domain for specific transduction of desired cell types in immunodeficient mice (Lafitte et al., 2012; Morizono et al., 2005; Pariente et al., 2007; Zhang et al., 2011; Zhang et al., 2009), demonstrating the proof of principle of the targeted lentiviral transduction. High stability of binding between E2 71 eMA and mSAH and biotinylated ligands will enable this targeting lentiviral transduction system to be applicable to immunocompetent animal experimental settings, facilitating application of targeted lentiviral transduction to broad fields of research.
Materials and Methods
Plasmids, antibodies, proteins and chemicals.
Expression vectors of wild-type Sindbis virus envelope protein 2.2 and 2.2 1L1L were described previously(Morizono et al., 2001; Morizono et al., 2009a; Pariente et al., 2007) . Expression vectors of E2 71 AV, STAV, eMA, and mSAH were constructed by inserting the core sequences of Avidin, streptavidin, monomeric rhizavidin, and monomeric streptavidin/rhizavidin hybrids, respectively, between flexible linkers at amino acid (aa) 71 of E2. Anti-hTfR1 and EGFR antibodies were purchased from Bio X Cell (West Lebanon, NH). Biotin and APC-conjugated anti-hTfR1 antibody, Alexa 488 and HRP-conjugated streptavidin, goat Alexa 488 and HRP-conjugated anti-mouse IgG, Alexa 488-conjugated goat anti-rabbit IgG, and biotin-conjugated EGF were purchased from ThermoFisher Scientific (Canoga Park, CA). Biotin-conjugated anti-mCD34 antibody, APC-conjugated anti-human EGFR antibody, and APC-conjugated anti-human CD47 antibody were purchased from Biolegend (San Diego, CA). Biotinylated FITC was purchased from Sigma-Aldrich (St. Louis, MO).
Cells and viruses.
293T cells were cultured in DMEM (ThermoFisher Scientific) containing 10% dialyzed FCS (Sigma-Aldrich) and antibiotics. Jurkat, NIH3T3, and HeLa cells were cultured in IMDM (ThermoFisher Scientific) containing 10% BaCl2-precipitated FCS. All lentiviral vectors were produced in 293T cells, using TransIT LT1 (Mirus Bio, Madison, WI). Briefly, 293T cells (1.4×107) were transfected with one type of protein expression vector (6–7 µg), packaging plasmid ps PAX2, (6–7 µg), and either lentiviral vector or cppt2e (6–7 µg). Two days post-transfection, the supernatant was subjected to ultracentrifugation (20,000 rpm, 4°C, 2 hours) by SW32 rotor (Beckman-Coulter, Brea, CA), using PBS containing 25% sucrose and 1 mM EGTA. The pellet containing the virus was resuspended in Hanks buffered saline (100-fold concentration).
Western blotting.
The amounts of viral vectors were normalized to the amount of HIV p24 (1 mg/ml) and mixed with LDS sample buffer (ThermoFisher Scientific) with 2-mercaptoethanol. Each sample (20 µl) was subjected to electrophoresis through an SDS 12% polyacrylamide gel (ThermoFisher Scientific). Immunoblot analyses of envelope proteins were performed with: 1) rabbit anti-Sindbis virus polyclonal antibody and horseradish peroxidase (HRP)-conjugated goat anti-rabbit polyclonal antibody (ThermoFisher Scientific); 2) HRP-conjugated rabbit goat anti-goat immunoglobulin antibody (ThermoFisher Scientific); and 3) biotinylated HRP (ThermoFisher Scientific). The protein bands were visualized by ECL plus substrate (ThermoFisher Scientific) and ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA).
Site-specific biotinylation of OKT9.
The disulfide bonds of the hinge region of OKT9 were reduced by incubation with 50 mM β-mercaptoethylamine (ThermoFisher Scientific) for 90 min at 37˚C, and the reduced OKT9 was biotinylated with EZ-Link Maleimide-PEG2-Biotin (ThermoFisher Scientific) according to the manufacturer’s protocol. The Fab fragment of biotinylated OKT9 was generated with the F(ab’)2 preparation kit (ThermoFisher Scientific) according to the manufacturer’s protocol. Fragmentation of biotinylated OKT9 and biotinylation of the heavy and light chains were analyzed by SyproRuby staining (ThermoFisher Scientific) and western blotting, using HRP-conjugated streptavidin, respectively, following SDS-PAGE, using a 12% polyacrylamide gel in a reduced condition.
Flow cytometric analysis of the modified Sindbis virus envelope proteins hTfR1, mCD34, and human EGFR.
293T cells were transfected with expression vectors of wild-type Sindbis virus envelope protein, 2.2 1L1L, E2 71 AV, E2 71 STAV, E2 71 eMA, or E2 71 mSAH, using TransIT LT1. Two days post-transfection, expression of the envelope proteins was analyzed by staining the transfected cells with rabbit anti-Sindbis virus antibody, followed by staining with Alexa 488-conjugated goat anti-rabbit IgG antibody (ThermoFisher Scientific). The binding of biotin was analyzed by staining the transfected cells with biotinylated FITC. Expression of hTfR1 and mCD34 on Jurkat and NIH3T3 cells was analyzed by staining cells with either anti-hTfR1, mCD34, or isotype control antibodies, followed by staining with Alexa 488-conjugated streptavidin (ThermoFisher Scientific). Expression of hTfR1 and EGFR on HeLa and Jurkat cells was analyzed by staining the cells with APC-conjugated anti-hTfR1, EGFR, or its isotype control antibody. Binding of EGFR to HeLa and Jurkat cells was analyzed by staining the cells with biotinylated EGF, followed by staining with APC-conjugated streptavidin (Biolegend). Flow cytometric data were acquired by FACScan (BD) upgraded with a red laser (Cytek, Fremont, CA) and analyzed by FCSExpress 5 (De Novo Software, Los Angeles, CA).
Transduction of Jurkat and NIH3T3 cells, 2.2, and 2.2 1L1L.
E2 71 eMA and E2 71 mSAH (2 ng p24/ml) were conjugated with different concentrations of anti-hTfR1, biotinylated anti-hTfR1, biotinylated isotype control antibodies, or the biotinylated Fab fragment of anti-hTfR1 at room temperature for 30 min. Cells (1×105) were transduced with 200 µl of virus at 37ºC for 2 hours. After transduction, cells were cultured for 3 days. EGFP expression was analyzed by flow cytometry. NIH3T3 cells were labeled with CellTrace Violet (ThermoFisher Scientific). The labeled NIH3T3 (2×104) and Jurkat cells (5×104) were infected with 200 µL of E2 71 eMA or mSAH (100 ng p24/ml) with or without conjugation of 500 ng/ml biotinylated anti-hTFR1 or mCD34 antibodies. After incubation with the vectors for 2 hours, cells were cultured in medium for 3 days. The cells were then harvested and stained with APC-conjugated anti-human CD47 antibody. Flow cytometric data were acquired by BDFortessa. EGFP expression of Jurkat cell populations (APC-positive/CellTraceViolet-negative) and NIH3T3 cell populations (APC-negative/CellTraceViolet-positive) were analyzed by FCSExpress 5.
Inhibition of Jurkat transduction by human serum and Nevirapine.
The E2 71 eMA and mSAH pseudotypes (1 µg p24/ml) were conjugated with biotinylated anti-hTfR1 antibody (1 µg/ml) and incubated with or without 50% human AB serum (Sigma-Aldrich) for 1 hour at 37ºC. The pseudotypes were diluted with PBS to 100 ng/ml p24/ml. Jurkat cells were incubated with 200 µL of the vectors, and EGFP expression was analyzed by flow cytometry 3 days post-transduction. For inhibition of reverse transcription, Jurkat cells were incubated with 20 nM Nevirapine (NIH AIDS Reagent Program) for 1 hour prior to transduction. The cells were then transduced with VSV-G, E2 71 eMA or mSAH pseudotype with or without conjugation of the biotinylated Fab fragment of anti-hTfR1 in the presence or absence of 20 nM Nevirapine. The cells were cultured for 3 days post-transduction in the absence or presence of 20 nM Nevirapine. EGFP expression was analyzed by flow cytometry 3 days post- transduction.
Transduction by EGF conjugation.
The E2 71 eMA, mSAH, and 2.2 pseudotypes (100 ng p24/ml) were incubated with different concentrations of biotinylated EGF for 30 min at room temperature. HeLa and Jurkat cells (1×105) were incubated with 200 µl of wild-type Sindbis virus envelope, 2.2, E2 71 eMA, or mSAH pseudotype with or without biotinylated EGF conjugation. Three days post-transduction, EGFP expression was analyzed by flow cytometry. To confirm the roles of EGFR in EGF-mediated transduction, HeLa cells were incubated with anti-EGFR (Bio X cell) or its isotype control (Biolegend) antibodies at 1 or 10 µg/ml, followed by transduction with E2 71 eMA or mSAH pseudotype (100 ng p24/ml) conjugated with biotinylated EGF (20 ng/ml) or anti-hTfR1 antibody (200 ng/ml) in the absence or presence of the blocking antibodies. EGFP expression was analyzed by flow cytometry 3 days post- transduction.
Quantification of EGFR phosphorylation.
HeLa cells were incubated with 2.2 or E2 71 eMA pseudotype (100 ng p24)/ml, then incubated with EGF-biotin (2 ng/ml) for 30 sec to 2 hours. Cells were promptly lysed after indicated incubation times in 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 1% Triton-X 100, 0.1% Na deoxycholate, 0.1% SDS, and 140 mM NaCl, with protease and phosphatase inhibitor supplemented before use (Boston Bioproducts, Ashland, MA). Protein concentrations were quantitated with a bicinchoninic acid assay. A magnetic bead-based ELISA assay was used for phosphorylation measurement. Lysates were incubated with EGFR antibody-coupled magnetic beads (Bio-Rad Laboratories) overnight at 4°C, then the beads were washed with 0.1% (v/v) Tween-20 in TBS. Phospho-tyrosine biotinylated antibody (R&D Systems) and streptavidin– phycoerythrin (Bio-Rad Laboratories) were incubated for 60 and 15 min, respectively, at room temperature. Signaling was quantified using a MagPix Luminex reader (Bio-Rad Laboratories), then normalized to protein concentration.
Highlights:
We have developed lentiviral vectors whose tropisms can be redirected by conjugation with biotinylated targeting molecules.
The binding of biotinylated targeting molecules and lentiviral vectors is stable so that the vectors can specifically transduce desired cell types in the presence of human serum.
The simple conjugation method of this targeting system is ideal for many researchers to utilize this technology to specifically transduce cell types of their interest.
Acknowledgements
We thank Dr. Nazzarian Ramin for providing NIH3T3 cells. We thank Drs. Irvin Chen and Jocelyn Kim for discussion, and Ms. Wendy Aft for editing of the manuscript. This work was supported by the UCLA AIDS Institute and the UCLA Center for AIDS Research, the James B. Pendleton Trust and McCarthy Foundation, and U.S. National Institute of Health grants R21AI095004 (K.M), R01AI108400 (K. M.) and DP5-OD019815 (A. M. and S. Y. B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We have no financial conflicts of interest to declare.
Declarations of interest: none
References:
- Ahani R, Roohvand F, Cohan RA, Etemadzadeh MH, Mohajel N, Behdani M, Shahosseini Z, Madani N, and Azadmanesh K (2016). Sindbis Virus-Pseudotyped Lentiviral Vectors Carrying VEGFR2-Specific Nanobody for Potential Transductional Targeting of Tumor Vasculature. Molecular biotechnology 58(11), 738–747. [DOI] [PubMed] [Google Scholar]
- Aires da Silva F, Costa MJ, Corte-Real S, and Goncalves J (2005). Cell type-specific targeting with sindbis pseudotyped lentiviral vectors displaying anti-CCR5 single-chain antibodies. Hum Gene Ther 16(2), 223–34. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Walker J, Nolta JA, and Bauer G (2009). Specific transduction of HIV-susceptible cells for CCR5 knockdown and resistance to HIV infection: a novel method for targeted gene therapy and intracellular immunization. Journal of acquired immune deficiency syndromes 52(2), 152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EA, and Wilchek M (1990). Biotin-binding proteins: overview and prospects. Methods in enzymology 184, 49–51. [DOI] [PubMed] [Google Scholar]
- Bender RR, Muth A, Schneider IC, Friedel T, Hartmann J, Pluckthun A, Maisner A, and Buchholz CJ (2016). Receptor-Targeted Nipah Virus Glycoproteins Improve Cell-Type Selective Gene Delivery and Reveal a Preference for Membrane-Proximal Cell Attachment. PLoS pathogens 12(6), e1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman I, Whitaker-Dowling P, Gao Y, and Griffin JA (2004). Preferential targeting of vesicular stomatitis virus to breast cancer cells. Virology 330(1), 24–33. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Zagorska A, Lew ED, Shrestha B, Rothlin CV, Naughton J, Diamond MS, Lemke G, and Young JA (2013). Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell host & microbe 14(2), 136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L, and Naldini L (2006). Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med 12(5), 585–91. [DOI] [PubMed] [Google Scholar]
- Cao Z, Lis R, Ginsberg M, Chavez D, Shido K, Rabbany SY, Fong GH, Sakmar TP, Rafii S, and Ding BS (2016). Targeting of the pulmonary capillary vascular niche promotes lung alveolar repair and ameliorates fibrosis. Nat Med 22(2), 154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chretien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, Bank A, Soulier J, Larghero J, Kabbara N, Dalle B, Gourmel B, Socie G, Chretien S, Cartier N, Aubourg P, Fischer A, Cornetta K, Galacteros F, Beuzard Y, Gluckman E, Bushman F, Hacein-Bey-Abina S, and Leboulch P (2010). Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 467(7313), 318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonte D, Drake EJ, Lim KH, Gulick AM, and Park S (2013). Structure-based engineering of streptavidin monomer with a reduced biotin dissociation rate. Proteins 81(9), 1621–33. [DOI] [PubMed] [Google Scholar]
- Derdak SV, Kueng HJ, Leb VM, Neunkirchner A, Schmetterer KG, Bielek E, Majdic O, Knapp W, Seed B, and Pickl WF (2006). Direct stimulation of T lymphocytes by immunosomes: virus-like particles decorated with T cell receptor/CD3 ligands plus costimulatory molecules. Proc Natl Acad Sci U S A 103(35), 13144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson J, and Rice CM (1993). Sindbis virus attachment: isolation and characterization of mutants with impaired binding to vertebrate cells. J Virol 67(6), 3363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AK, Maurice M, Morling FJ, Cosset FL, and Russell SJ (1998). Inverse targeting of retroviral vectors: selective gene transfer in a mixed population of hematopoietic and nonhematopoietic cells. Blood 91(5), 1802–9. [PubMed] [Google Scholar]
- Fields BN, Knipe DM, and Howley PM (2013). “Fields virology” 6th ed. 2 vols. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- Fraietta JA, Nobles CL, Sammons MA, Lundh S, Carty SA, Reich TJ, Cogdill AP, Morrissette JJD, DeNizio JE, Reddy S, Hwang Y, Gohil M, Kulikovskaya I, Nazimuddin F, Gupta M, Chen F, Everett JK, Alexander KA, Lin-Shiao E, Gee MH, Liu X, Young RM, Ambrose D, Wang Y, Xu J, Jordan MS, Marcucci KT, Levine BL, Garcia KC, Zhao Y, Kalos M, Porter DL, Kohli RM, Lacey SF, Berger SL, Bushman FD, June CH, and Melenhorst JJ (2018). Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558(7709), 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke S, Maisner A, Muhlebach MD, Koehl U, Grez M, Cattaneo R, Cichutek K, and Buchholz CJ (2008). Targeted cell entry of lentiviral vectors. Mol Ther 16(8), 1427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Ahn A, Chatterjee PK, and Kielian M (2000). Formation and characterization of the trimeric form of the fusion protein of Semliki Forest Virus. Journal of virology 74(17), 7772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helppolainen SH, Nurminen KP, Maatta JA, Halling KK, Slotte JP, Huhtala T, Liimatainen T, Yla-Herttuala S, Airenne KJ, Narvanen A, Janis J, Vainiotalo P, Valjakka J, Kulomaa MS, and Nordlund HR (2007). Rhizavidin from Rhizobium etli: the first natural dimer in the avidin protein family. The Biochemical journal 405(3), 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhtala T, Kaikkonen MU, Lesch HP, Viitala S, Yla-Herttuala S, and Narvanen A (2014). Biodistribution and antitumor effect of Cetuximab-targeted lentivirus. Nucl Med Biol 41(1), 77–83. [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Lesch HP, Pikkarainen J, Raty JK, Vuorio T, Huhtala T, Taavitsainen M, Laitinen T, Tuunanen P, Grohn O, Narvanen A, Airenne KJ, and Yla-Herttuala S (2009). (Strept)avidin-displaying lentiviruses as versatile tools for targeting and dual imaging of gene delivery. Gene Ther 16(7), 894–904. [DOI] [PubMed] [Google Scholar]
- Kasahara N, Dozy AM, and Kan YW (1994). Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science 266(5189), 1373–6. [DOI] [PubMed] [Google Scholar]
- Kasaraneni N, Chamoun-Emanuelli AM, Wright G, and Chen Z (2017). Retargeting Lentiviruses via SpyCatcher-SpyTag Chemistry for Gene Delivery into Specific Cell Types. MBio 8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaraneni N, Chamoun-Emanuelli AM, Wright GA, and Chen Z (2018). A simple strategy for retargeting lentiviral vectors to desired cell types via a disulfide-bond-forming protein-peptide pair. Sci Rep 8(1), 10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JT, Liu Y, Kulkarni RP, Lee KK, Dai B, Lovely G, Ouyang Y, Wang P, Yang L, and Baltimore D (2017). Dendritic cell-targeted lentiviral vector immunization uses pseudotransduction and DNA-mediated STING and cGAS activation. Sci Immunol 2(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra WB, Heidner HW, and Johnston RE (1999). The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate. J Virol 73(8), 6299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin YD, and Zack JA (1998). Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol 72(4), 3161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafitte M, Rousseau B, Moranvillier I, Taillepierre M, Peuchant E, Guyonnet-Duperat V, Bedel A, Dubus P, de Verneuil H, Moreau-Gaudry F, and Dabernat S (2012). In vivo gene transfer targeting in pancreatic adenocarcinoma with cell surface antigens. Mol Cancer 11, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen OH, Hytonen VP, Nordlund HR, and Kulomaa MS (2006). Genetically engineered avidins and streptavidins. Cell Mol Life Sci 63(24), 2992–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Kim JA, Yen TC, Lee IH, Ahn B, Lee Y, Hsieh CL, Kim HM, and Jung Y, (2016). A Rhizavidin Monomer with Nearly Multimeric Avidin-Like Binding Stability Against Biotin Conjugates. Angewandte Chemie 55(10), 3393–7. [DOI] [PubMed] [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, and Rossmann MG (2010). Structural changes of envelope proteins during alphavirus fusion. Nature 468(7324), 705–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Morizono K, Pariente N, Kamata M, Lee B, and Chen IS (2009a). Targeted transduction via CD4 by a lentiviral vector uses a clathrin-mediated entry pathway. J Virol 83(24), 13026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Pariente N, Morizono K, and Chen IS (2009b). Targeted transduction of CD34+ hematopoietic progenitor cells in nonpurified human mobilized peripheral blood mononuclear cells. J Gene Med 11(3), 185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Bristol G, Xie YM, Kung SK, and Chen IS (2001). Antibody-directed targeting of retroviral vectors via cell surface antigens. J Virol 75(17), 8016–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, and Chen IS (2005). Targeted gene delivery by intravenous injection of retroviral vectors. Cell Cycle 4(7), 854–6. [DOI] [PubMed] [Google Scholar]
- Morizono K, and Chen IS (2011). Receptors and tropisms of envelope viruses. Current opinion in virology 1(1), 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, and Chen IS (2014). Role of phosphatidylserine receptors in enveloped virus infection. Journal of virology 88(8), 4275–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Ku A, Xie Y, Harui A, Kung SK, Roth MD, Lee B, and Chen IS (2010). Redirecting lentiviral vectors pseudotyped with Sindbis virus-derived envelope proteins to DC-SIGN by modification of N-linked glycans of envelope proteins. J Virol 84(14), 6923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Pariente N, Xie Y, and Chen IS (2009a). Redirecting lentiviral vectors by insertion of integrin-tageting peptides into envelope proteins. J Gene Med 11(7), 549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Ringpis GE, Pariente N, Xie Y, and Chen IS (2006). Transient low pH treatment enhances infection of lentiviral vector pseudotypes with a targeting Sindbis envelope. Virology 355(1), 71–81. [DOI] [PubMed] [Google Scholar]
- Morizono K, Xie Y, Helguera G, Daniels TR, Lane TF, Penichet ML, and Chen IS (2009b). A versatile targeting system with lentiviral vectors bearing the biotin-adaptor peptide. J Gene Med 11(8), 655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, and Chen IS (2011). The Soluble Serum Protein Gas6 Bridges Virion Envelope Phosphatidylserine to the TAM Receptor Tyrosine Kinase Axl to Mediate Viral Entry. Cell Host Microbe 9(4), 286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Xie Y, Ringpis GE, Johnson M, Nassanian H, Lee B, Wu L, and Chen I(2005). Lentiviral vector retargeting to P-glycoprotein on metastatic melanoma through intravenous injection. Nat Med 11(3), 346–52. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, and Rossmann MG (2006). Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14(1), 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch RC, Muhlebach MD, Schaser T, Kneissl S, Jost C, Pluckthun A, Cichutek K, and Buchholz CJ (2011). DARPins: An Efficient Targeting Domain for Lentiviral Vectors. Mol Ther [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Peng KW, Harvey M, Greiner S, Lorimer IA, James CD, and Russell SJ (2005). Rescue and propagation of fully retargeted oncolytic measles viruses. Nat Biotechnol 23(2), 209–14. [DOI] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gage FH, Trono D, and Verma IM (1996a). Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A 93(21), 11382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, and Trono D (1996b). In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272(5259), 263–7. [DOI] [PubMed] [Google Scholar]
- Naldini L, Trono D, and Verma IM (2016). Lentiviral vectors, two decades later. Science 353(6304), 1101–2. [DOI] [PubMed] [Google Scholar]
- Ohno K, Sawai K, Iijima Y, Levin B, and Meruelo D (1997). Cell-specific targeting of Sindbis virus vectors displaying IgG-binding domains of protein A. Nat Biotechnol 15(8), 763–7. [DOI] [PubMed] [Google Scholar]
- Pariente N, Mao SH, Morizono K, and Chen IS (2008). Efficient targeted transduction of primary human endothelial cells with dual-targeted lentiviral vectors. J Gene Med 10(3), 242–8. [DOI] [PubMed] [Google Scholar]
- Pariente N, Morizono K, Virk MS, Petrigliano FA, Reiter RE, Lieberman JR, and Chen IS (2007). A novel dual-targeted lentiviral vector leads to specific transduction of prostate cancer bone metastases in vivo after systemic administration. Mol Ther 15(11), 1973–81. [DOI] [PubMed] [Google Scholar]
- Pletnev SV, Zhang W, Mukhopadhyay S, Fisher BR, Hernandez R, Brown DT, Baker TS, Rossmann MG, and Kuhn RJ (2001). Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105(1), 127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani M, Annunziato S, Adams DJ, and Montini E (2013). Cancer gene discovery: exploiting insertional mutagenesis. Mol Cancer Res 11(10), 1141–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasbach A, Abel T, Munch RC, Boller K, Schneider-Schaulies J, and Buchholz CJ (2013). The receptor attachment function of measles virus hemagglutinin can be replaced with an autonomous protein that binds Her2/neu while maintaining its fusion-helper function. J Virol 87(11), 6246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrin V, Russell SJ, and Cosset FL (2003). Targeting retroviral and lentiviral vectors. Curr Top Microbiol Immunol 281, 137–78. [DOI] [PubMed] [Google Scholar]
- Sessa M, Lorioli L, Fumagalli F, Acquati S, Redaelli D, Baldoli C, Canale S, Lopez ID, Morena F, Calabria A, Fiori R, Silvani P, Rancoita PM, Gabaldo M, Benedicenti F, Antonioli G, Assanelli A, Cicalese MP, Del Carro U, Sora MG, Martino S, Quattrini A, Montini E, Di Serio C, Ciceri F, Roncarolo MG, Aiuti A, Naldini L, and Biffi A (2016). Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 388(10043), 476–87. [DOI] [PubMed] [Google Scholar]
- Somia NV, Zoppe M, and Verma IM (1995). Generation of targeted retroviral vectors by using single-chain variable fragment: an approach to in vivo gene delivery. Proc Natl Acad Sci U S A 92(16), 7570–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AA, Murphy GJ, Hamakawa H, Kwok LW, Srinivasan S, Hovav AH, Mulligan RC, Amar S, Suki B, and Kotton DN (2010). Amelioration of emphysema in mice through lentiviral transduction of long-lived pulmonary alveolar macrophages. J Clin Invest 120(1), 379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, and Wiley DC (1981). Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289(5796), 366–73. [DOI] [PubMed] [Google Scholar]
- Wu DT, Seita Y, Zhang X, Lu CW, and Roth MJ (2012). Antibody-directed lentiviral gene transduction for live-cell monitoring and selection of human iPS and hES cells. PloS one 7(4), e34778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Henry LJ, Gerard RD, and Deisenhofer J (1994). Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure 2(12), 1259–70. [DOI] [PubMed] [Google Scholar]
- Yang L, Bailey L, Baltimore D, and Wang P (2006). Targeting lentiviral vectors to specific cell types in vivo. Proc Natl Acad Sci U S A 103(31), 11479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, and Chen IS (1990). HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61(2), 213–22. [DOI] [PubMed] [Google Scholar]
- Zhang KX, Kim C, Chow E, Chen IS, Jia W, and Rennie PS (2011). Targeting trastuzumab-resistant breast cancer cells with a lentivirus engineered to bind antibodies that recognize HER-2. Breast Cancer Res Treat 125(1), 89–97. [DOI] [PubMed] [Google Scholar]
- Zhang KX, Moussavi M, Kim C, Chow E, Chen IS, Fazli L, Jia W, and Rennie PS (2009). Lentiviruses with trastuzumab bound to their envelopes can target and kill prostate cancer cells. Cancer gene therapy 16(11), 820–31. [DOI] [PubMed] [Google Scholar]
- Zhang X, and Roth MJ (2010). Antibody-directed lentiviral gene transduction in early immature hematopoietic progenitor cells. The journal of gene medicine 12(12), 945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhu L, Lee S, Li L, Chang E, Soong NW, Douer D, and Anderson WF (1999). Identification of the block in targeted retroviral-mediated gene transfer. Proc Natl Acad Sci U S A 96(7), 4005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J Jr., Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, and Roux KH (2006). Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441(7095), 847–52. [DOI] [PubMed] [Google Scholar]


