Abstract
Toxoplasma gondii, an obligate intracellular apicomplexan, causes latent infection in about one-third of the human population. During latent infection T. gondii bradyzoites are found within cysts, a modified parasitophorous vacuole. This parasite has a large family of SRS (surface antigen-1 related sequence) proteins which are reported to be involved in attachment of these organisms to their mammalian host cells and in immune subversion during latent infection. We have identified a novel mucin-domain containing SRS protein, using a glycoepitope-specific antibody, which recognizes the cyst wall. SRS13 has two SRS domains and between these domains is a threonine-rich tandem repeat mucin like domain that is similar to the mucin like domain seen in another cyst wall specific SRS protein CST1 (SRS44). SRS13 is upregulated in bradyzoites and O-GalNAc glycosylated by ppGalNAc-T2 and T3. Similar to the cyst wall protein CST1, SRS13 localizes to the cyst wall, but unlike CST1, SRS13 is dispensable for normal cyst wall formation. Together, these findings elucidate the role of a second SRS mucin domain protein, SRS13, in bradyzoite biology and expands the previously reported functions of the SRS protein family.
Keywords: Toxoplasma gondii, O-GalNAc Glycosylation, SRS, Cyst, Mucin
INTRODUCTION
Toxoplasma gondii is an obligate intracellular protozoan parasite that infects up to a third of world’s population (Tenter et al. 2000). The predilection of this parasite for the central nervous system, causing necrotizing encephalitis, and for the eye, causing chorioretinitis, constitutes its major threat to patients. T. gondii can cause lethal encephalitis in immune compromised individuals, such as patients who have received organ transplants and those with advanced HIV infection. Infection can also cause congenital disease, when a seronegative woman is infected during pregnancy, resulting in blindness and intellectual disabilities in children infected in utero. The asexual cycle of T. gondii consists of two life cycle stages: rapidly replicating tachyzoites that cause acute infection and the slower growing bradyzoites that are found in tissue cysts and are responsible for chronic or latent infection. While the asexual stages of these parasites cause infection in numerous homeothermic vertebrates, replication as a sexual cycle occurs only in the enterocytes of the Felidae. Transmission of T. gondii is due to ingestion of tissue cysts containing bradyzoites, the latent asexual stage found in animal tissues, or ingestion of oocysts containing sporozoites, the sexual stage found in cat feces.
A family of GPI-anchored proteins termed SRS (Surface antigen glycoprotein 1 Related Sequence) proteins covers the surface of T. gondii. These proteins have a SRS fold containing conserved 4 or 6 cysteine pairs that form intra-molecular disulfide bonds ((He et al. 2002) IPR007226/ PF04092). Typical SRS proteins contain either one or two SRS domains, form homodimers, and have a C-terminus glycosylphosphatidylinositol (GPI) anchor involved in tethering these proteins on the plasma membrane. Discrete repertories of SRS proteins are developmentally expressed and are specific to each stage of differentiation. There are 144 SRS genes in the T. gondii ME49 genome, of which 35 are pseudo genes (Wasmuth et al. 2012). These SRS proteins can be classified into eight subfamilies based on their sequence similarity (Wasmuth et al. 2012). Various SRS proteins have been demonstrated to have important biological functions such as adhesion to the host cell surface (SAG3/ SRS57), virulence (SAG1 / SRS29B), and immune modulation (SRS9 / SRS16B) (Dzierszinski et al. 2000; Van et al. 2007). Previously, our group demonstrated that a unique large SRS protein CST1 (SRS44), a mucin-like domain containing glycoprotein with thirteen SRS domains that belongs to SRS Family 2, is necessary for the construction of an intact cyst wall and confers structural rigidity to the parasite cyst wall (Tomita et al. 2013). We also determined that the mucin domain of CST1 is glycosylated by the polypeptide N- acetylgalactosaminyltransferase isoforms ppGalNAc-T2 and T3 in a hierarchical manner, that these ppGalNAc-Ts are responsible for the initiation of mucin type O-linked GalNAc glycosylation, and that glycosylation of this mucin domain is required for cyst wall structural integrerty (Tomita et al. 2017). Curiously, the monoclonal antibodies that bind to the mucin domain of CST1 recognized another glycoprotein identified as an additional band at ~150 kDa in addition to the CST1(Tomita et al. 2013), which we have identified is SRS13. In this paper, we present data on the characterization of SRS13 as a mucin domain containing SRS protein belonging to SRS Family 2 that localizes to T. gondii cyst wall and matrix.
MATERIALS AND METHODS
T. gondii cell culture and strains
Human foreskin fibroblasts (HFF: ATCC-CRL-2522) were maintained in 10% fetal bovine serum pH 7 Dulbecco’s modified Eagle’s medium (DMEM) with penicillin-streptomycin at 5% C02. Confluent monolayers were infected with Type II PruΔku80 strain of T. gondii, which is widely used for genetic studies (Fox et al. 2011). For in vitro bradyzoite differentiation, parasites strains were grown in differentiation medium (DMEM medium adjusted to pH8.2 with 50 mM HEPES and supplemented with 1% fetal bovine serum and penicillin-streptomycin) for three days at atmospheric C02.
Identification of the SalmonE reactive glycoprotein by mass spectrometry
Purification of the monoclonal antibody SalmonE binding protein and identification was performed as previously described (Tomita et al. 2013). Briefly, the monoclonal antibody SalmonE was crosslinked to the Protein L agarose beads with disuccinimidyl suberate. Human foreskin fibroblasts infected with a ME49 strain of T. gondii for three days in pH 8.1 medium with 10% FBS at 0.5% C02 were lysed with 1% TritonX-100 in PBS. It was incubated with SalmonE-beads for 2 hours at 4 °C. The beads were extensively washed with 1% TritonX-100 PBS, and the eluate was separated on SDS-PAGE. The gel was stained with Coomassie Brilliant Blue and visible bands were excised. The protein(s) in each band was reduced and alkylated using TCEP and iodoacetamide then digested with trypsin in 25mM ammonium bicarbonate/0.01% ProteaseMax at 50°C for 1 hour. The resulting digest was cleaned with C18 ziptip and the peptides eluted onto a MALDI plate with a saturated solution of α-cyanohydroxycinnamic acid in 70% acetonitrile/0.1% trifluoroacetic acid. MS/MS analysis of the digested sample was carried out using the AB Sciex 4800 MALDI-TOF-TOF (Applied Biosystems), operated at 20kV accelerating voltage in the reflector positive ion mode. The MS/MS data generated were converted to mgf files and searched against EPICDB (Madrid-Aliste et al. 2009) using the in-house Mascot Protein Search engine (Matrix Science) for protein identification.
Generation of SRS13 deficient mutant parasite
Type II Prugniaud strain capable of high efficiency homologous recombination (PruΔku80) (Fox et al. 2011) was used as the background strain for the creation of Δsrs13 strain. The SRS13 gene was deleted by a use of a double-homologous recombination vector (Fig. 1c). Briefly, a selectable marker hypoxanthine-xanthine-guanine phosphoribosyltransferase [HXGPRT] cassette flanked by 1kb upstream and downstream genomic DNA sequence of the SRS13 gene were concatenated into pRS416 yeast shuttle vector using the yeast strain ATCC#90845. This construct deletes the entire SRS13 gene from the locus. The parasites were transfected with the linearized Δsrs13 vector and selected in the presence of 25 μg/ml mycophenolic acid and 50 μg/ml xanthine. Parasites were cloned using limiting dilution in 96-well plates. Integration of Δsrs13 vector at the SRS13 locus was verified by PCR (Fig. 1c). The supplementary file contains a description of the Δsrs13 vector, wild type SRS13 locus, and Δsrs13 locus with nucleotide sequences and annotations including gene models and primer sequences in GenBank format (Supplementary File).
Figure 1.
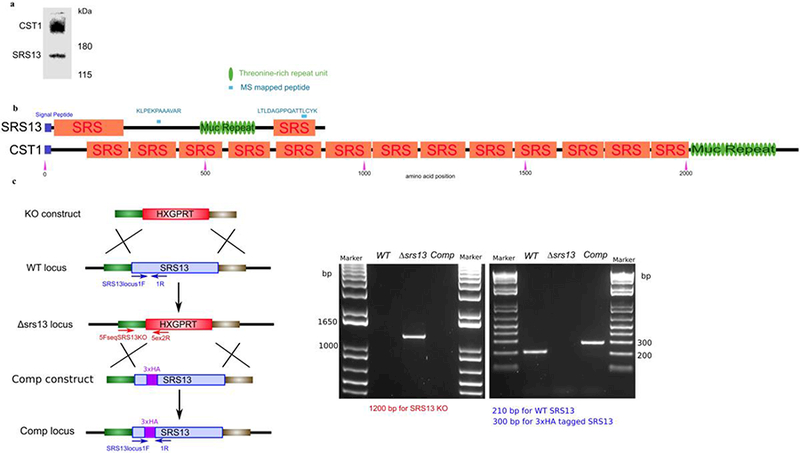
a. The O-GalNAc glycoepitope-specific monoclonal antibody SalmonE binds to CST1 as well as SRS13. Immunoblot of in vitro bradyzoite T. gondii lysate (3d in differentiation medium) probed with monoclonal antibody SalmonE demonstrating that this antibody binds not only to CST1 (>180 kDa) but also another glycoprotein SRS13 (150kDa).
b. Schematic of SRS13 protein and CST1 domain configurations. The two peptides that identified by mass spectrometry are indicated as blue lines. The purple boxes represent signal peptides, orange boxes represent SRS domain, and green ovals represent threonine-rich tandem repeat units. . All the protein features are in scale.
c. Deletion and complementation of the SRS13 gene. Schematic of SRS13 deletion and complementation (left) describing the homologous recombination. The SRS13 gene was replaced with a selectable HXGPRT cassette forming the Δsrs13 locus; the HXGPRT cassette was replaced with N- terminal 3xHA tagged SRS13 forming the complemented Δsrs13:SRS13 locus. Verification of the SRS13 deletion was performed by diagnostic PCR (left agarose gel) with one primer from the 5’ UTR of SRS13 and another from the HXGPRT cassette. Verification of complementation was performed using primer sets flanking the 3xHA sequence on 5’ end of SRS13 gene. The sequence of plasmids and SRS13 loci with primer annotations are in the electronic supplemental material.
Generation of HA-tagged parasites
To visualize the SRS13 gene, it was epitope-tagged with 3xHA either at its N-terminus after the predicted signal peptide sequence or at its C-terminus immediately before the stop codon (Supplementary files provided information on the vector sequence, N- and C- tagged loci, and primers in Genbank format). Parasites (PruΔku80) were transiently transfected with a Cas9 plasmid (Shen et al. 2014) with a pyrimethamine resistance marker and gRNA specific to the either N- or C-terminus of SRS13 gene. Parasites were co-transfected with PCR fragments of 3xHA with 40-nucleotide homologous arms immediately adjacent to the cleavage site targeted by the gRNA. The transfected parasites were then cultured in the presence of 1 μM pyrimethamine for three days and then cloned by limiting dilution in a 96-well plate. The presence of 3xHA sequence was verified using IFA.
Complementation of SRS13 gene
Δsrs13 parasites were complemented with an N-terminal 3xHA tagged copy of SRS13 gene at the endogenous locus. Δsrs13 parasites were transiently transfected with Cas9 plasmid with a pyrimethamine cassette and gRNA specific to the HXGPRT sequence that is located at Δsrs13 locus. The parasites were co-transfected with a linearized donor plasmid that contains 1kbp homologous sequence upstream and downstream of the SRS13 gene as well as a N-terminal 3xHA tagged copy of SRS13 gene amplified from the epitope tagged parasite genomic DNA. The transfected parasites were cultured in the presence of 1 μM pyrimethamine for three days then in the presence of 6-thioxanthine for three additional days. Parasites were then cloned by limiting dilution in a 96-well plate and the presence of the SRS13 gene was verified in cloned parasites using PCR.
Immunoblotting
HFF cells were infected with T. gondii in normal DMEM medium with 10% FBS at 5% C02 or in differentiation medium at atmospheric C02 for three days. Harvested parasites were passed through 27G needle three times and then filtered through a 5μm polycarbonate membrane to remove host cells. Parasites were lysed in 1% TritonX-100 in PBS containing cOmplete™ EDTA-free protease inhibitor (ThermoFisher), and the lysates were cleared by centrifugation. The supernatants were then boiled with sample buffer, separated by 4–15% SDS-PAGE, and transferred to PVDF membrane. The membrane was incubated with SalmonE (1:500), rabbit anti-TgALD1 (1:5000) or DBA lectin (at 10 μg/ml, EY Laboratories) for 1 hour at room temperature. After washing three times with PBS, the membranes were incubated with HRP conjugated anti-mouse antibody, anti-rabbit antibody, or streptavidin-HRP at 1:2000 and subsequently the signal was detected using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher).
Immunocytochemistry
HFF cells cultured on cover-glasses were infected with T. gondii in differentiation medium at atmospheric CO2 for five days. The cells were then fixed with 4% paraformaldehyde in PBS for 30 minutes and permeablized with 0.2% TritonX100 in 3% BSA in PBS for 20 minutes. Following that treatment, the cells were washed with PBS three times and then blocked with 3% BSA in PBS at 4°C overnight. Cover-glasses with cultured, fixed, permeabilized cells were then incubated with rat anti-HA monoclonal antibody (3F10 clone, Roche) at 1:200 in PBS with 1% BSA for 90 minutes at 37°C and washed with PBS three times. Afterward, an anti-rat Alexa Fluor 594 antibody at a 1:500 dilution was incubated with the cover-glasses for 90 minutes at 37°C, and they were washed with PBS three times. Following this washing procedure, each cover-glass was mounted on a slide using Prolong Gold Antifade containing DAPI (ThermoFisher). Each coverglass was then examined using a confocal fluorescence microscope (SP5, Leica).
Parasite Growth and Differentiation Assays
A growth assay was performed by enumerating the number of parasites per vacuole at 36 hours after infection in either tachyzoite or bradyzoite conditions. Briefly, parasites were passed through 27G needle six times then filtered through 5 μm PVDF filter. HFF monolayers grown on cover glasses in 24- well tissue culture plates were infected (2000 parasites/well) in either pH 7 or differentiation medium and then incubated in 5% or atmospheric CO2 respectively for 2 hours. Noninvading parasites were removed by washing with PBS once and the cells were then cultured in either pH 7 medium or differentiation medium. At 36 and 72 hours after infection, the cells were fixed and permeabilized as described above.To evaluate growth, 36-hour samples were incubated with anti-Toxoplasma polyclonal rabbit antibody at 1:5000 dilution and with anti-GRA1 monoclonal mouse antibody at 1:1000. To evaluate differentiation, 72-hour samples were incubated with anti-GFP rabbit monoclonal antibody G10362 (Thermo Fisher) at 1:1000 and anti-Toxoplasma polyclonal mouse antibody at 1:1000 dilutions in PBS with 1% BSA for 90 minutes at 37°C. Following incubation with antibodies samples (for either the growth or differentiation assay) were then washed three times with PBS and incubated with Alexa Flour 594 anti-mouse antibody and Alexa Flour 488 anti-rabbit antibody both at 1:2000 for 90 minutes at 37°C. After three washes with PBS, the cover glasses were mounted on glass slides with Prolong Gold with DAPI. For the growth assay with 36-hour samples, numbers of parasites per vacuole were visually enumerated using a fluorescent microscope by counting at least 100 vacuoles per line with technical triplicates. Statistical significance was determined using multiple t test of each parasites per vacuole category with the Holm-Sidak method with alpha = 0.05 (Graphpad Prism 7.0). For the differentiation assay, vacuoles were counted and categorized into either bradyzoite or tachyzoite containing vacuoles based on the expression of GFP in bradyzoites by the LDH2 promoter.
Transmission electron microscopy
The BALB/cdm1 mice were infected with 2000 parasites per mouse intraperitoneally with either WT or Δsrs13 strain for four weeks in the presence of sulfamerazine at 30mg/L in drinking water. After 4 weeks the brains of the mice were removed, homogenized and cysts purified by isopycnic centrifugation as previously described (Tomita et al. 2013). The isolated cysts were fixed with 2.5% glutaraldehyde, 2% paraformaldehyde in 0.1M sodium cacodylate buffer, postfixed in 1% osmium tetroxide followed by 2% uranyl acetate, dehydrated through a graded series of ethanol and embedded in LX112 resin (LADD Research Industries, Burlington VT). Ultrathin sections were cut on a Reichert Ultracut UCT, stained with uranyl acetate followed by lead citrate and viewed on a JOEL 1200EX transmission electron microscope at 80kv.
Ethical approval
All animal experiments were conducted according to the U.S.A. Plublic Health Service Policy on Human Care and Use of Laboratory Animals. Animals were maintained in an AAALAC-approved facility and the Institutional Care Committee (IAUC) of the Albert Einstein College of Medicine, Bronx, New York (Animal Protocol 20150908; Animal Welfare Assurance number A3312–01), approved all protocols.
RESULTS
Identification of SRS13
The monoclonal antibody SalmonE that binds to the mucin domain of CST1 in an O-linked GalNAc glycosylation dependent manner (Tomita et al. 2013) also binds to an additional 150 kDa band (Figure 1a). To identify this unknown protein, immunoprecipitation was performed using monoclonal antibody SalmonE on lysates obtained from an in vitro differentiation-induced parasite culture (Figure S1a). Mass spectrometric analysis of the 150 kDa band identified two peaks at m/z of 1748 (peptide: LTLDAGPPQATTLCYK) and 1250 (peptide: KLPEKPAAAVAR) (Figure S1b, S1c, and S1d). Based on the T. gondii genome sequence data available at ToxoDB.org, both peptides were mapped to a SRS-domain containing protein SRS13 (TGME49 22370).
SRS13 contains a signal peptide and two SRS domains of which one at N-terminus is a degraded domain (Figure 1b). Between the two SRS domains, there is a 169 amino acid long stretch of mucin-like threonine- rich tandem repeats. In this mucin-like domain there are 13 repeats of a 13 amino-acid long consensus sequence of KTQPETTTVAPPA (based on the HMM-based algorithm HHrepID (Biegert & Soding 2008)). According to the available T. gondii gene model, SRS13 is predicted to be a 92 kDa protein; however on SDS-PAGE it migrates as a 150 kDa band. This is probably due to extensive post-translational modification of this protein by O-linked glycosylation in its mucin-like domain. A similar finding of higher migration was seen with CST1 (SRS44), the only other SRS protein that contains a mucin-like domain depicted in Fig. 1b.
SRS13 is dispensable for the in vitro growth
To study the function of the SRS 13 protein, the SRS13 gene was deleted from the genome of the type II Prugniaud Δku80 strain of T. gondii using homologous recombination (Fig. 1c) as previously described (Tomita et al. 2013). The SRS13 deficient parasite (Δsrs13) was complemented by inserting an HA-tagged copy of SRS13 at the endogenous Δsrs13 locus. Deletion and complementation of SRS13 gene was confirmed by PCR (Fig. 1c). To assess the effect of SRS13 deletion in the parasite growth, the numbers of parasites per vacuole were measured in both tachyzoite and bradyzoite conditions at 36 hours after infection (Fig. 2a). There was no statistically significant difference between the WT and Δsrs13 parasite growth both in tachyzoite and bradyzoite conditions. In comparison, the growth of Δcst1 parasites were significantly reduced compared with WT (multiple t test, p < 0.05) both in tachyzoite and bradyzoite conditions. This demonstrates that SRS13 deletion does not influence the in vitro growth under either tachyzoite or bradyzoite induction conditions.
Figure 2.
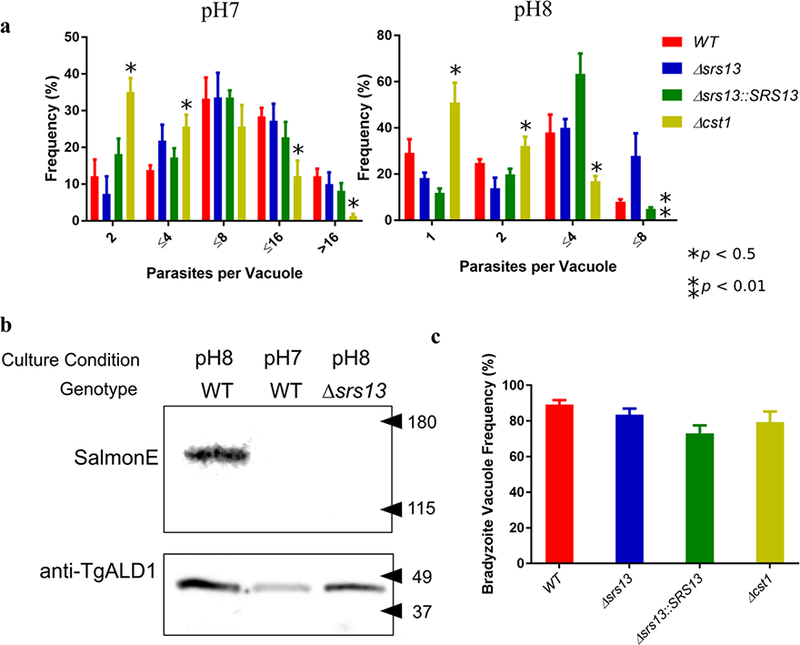
a. SRS13 does not influence parasite growth under either tachyzoite or bradyzoite conditions. The number of parasites per vacuole were counted at 36 hours after infection either in tachyzoite medium (pH7) or bradyzoite medium (pH8). At least 100 vacuoles were counted for each strain of parasite. Bars represent SD values; * represents statistical significance (*p <0.5, **p <0.01, multiple t test).
b. Immunoblot image of parasite lysate with SalmonE. This demonstrates that SRS13 (150 kDa band) protein is upregulated in bradyzoites (pH8 medium). TgALD1 was used for parasite loading control. Figure 1a is a part of this membrane.
c. SRS13 does not influence in vitro differentiation efficiency. The graph shows the rate of bradyzoite specific GFP positive vacuoles in differentiation medium at 72 hours after infection. The bars represent SD values. There was no statistically significant difference between WT and Δsrs13.
SRS13 is upregulated in bradyzoites
Many of SRS proteins are stage specifically expressed (Kim and Boothroyd 2005). To assess if the SRS13 protein is also stage specifically expressed, both bradyzoite and tachyzoite parasite lysates were immunoblotted with monoclonal antibody SalmonE and SRS13 was found to be highly upregulated in bradyzoites compared to tachyzoites (Figure 2b). This result is congruent with the 66.8 fold upregulation of SRS13 mRNA expression in bradyzoites compared with tachyzoites seen in the RNAseq data (Behnke et al. 2008) available at ToxoBD.org.
SRS13 is not required for bradyzoite differentiation
Since the SRS13 was highly upregulated in bradyzoite culture conditions, we sought to evaluate the effect of the SRS13 deletion on the differentiation of the parasites from tachyzoites to bradyzoites. Figure 2c demonstrates the in vitro differentiation rate of parasites at three days after infection in bradyzoite differentiation medium. The differentiation of a given vacuole was determined by the GFP expressed under the control of bradyzoite specific promoter LDH2. The figure demonstrates that lack of SRS13 does not influence in vitro bradyzoite differentiation.
SRS13 O-GalNAc glycosylation is dependent on ppGalNAc-T2 and T3
Our previous study demonstrated that monoclonal antibody SalmonE binds to the mucin domain of CST1 in O-GalNAc glycosyltransferase ppGalNAc-T2 and T3 dependent manner and that this binding also disappears after ß-elimination, which removes all O-linked glycans (Tomita et al. 2017). Therefore, binding of SalmonE to SRS13 was assessed in ppGalNAc-T deficient mutants. Figure 3A shows that SRS13 could be detected by SalmonE in wild type and Δt1 T. gondii, but was absent in the Δt2, Δt3, and Δsrs13 strains of T. gondii. This implies that the glycans on the mucin domain of SRS13, which are detected by SalmonE, are dependent on the presence of ppGalNAc-T2 and T3.
Figure 3.
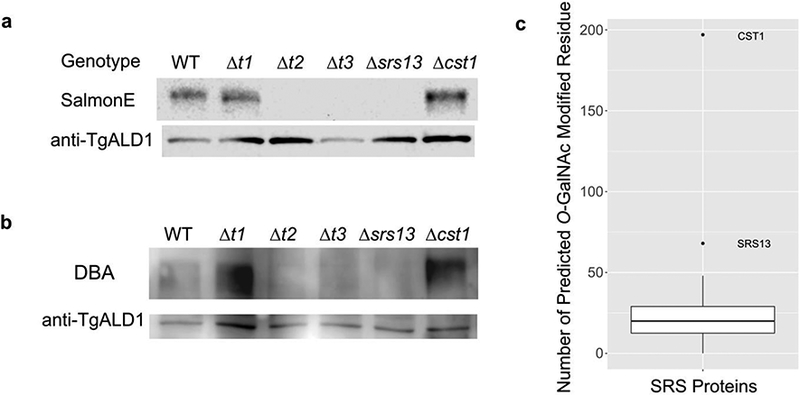
a. SRS13 is O-GalNAc glycosylated in ppGalNAc-T2 and -T3 dependent manner. Immunoblot of various ppGalNAc-T deficient parasite lysates probed with glycoepitope-specific SalmonE. There is a lack of SRS13 signal in the Δt2 and Δt3 parasites. TgALD1 was used as a loading control.
b. DBA lectin overlay of ppGalNAc-T deficient parasite lysates. This demonstrates a lack of DBA binding to SRS13 in Δt2 and Δt3 parasites.
C. SRS13 and CST1 are only two SRS proteins that have mucin-like domains. This box-and-whisker plot shows the number of predicted O-GalNAc modified residues for each SRS protein. Both SRS13 and CST1 have far larger number of predicted glycosylation sites (i,e they are outside the 1.5 x inter-quartile range) indicating that they are unique among the SRS protein.
DBA lectin, which has binding specificity to glycan structure GalNAcα1–3GalNAc, binds to the mucin domain of CST1 in a ppGalNAc-T2 and T3 dependent manner and its binding disappears after ß- elimination (Tomita et al. 2017). To investigate whether DBA lectin also binds to SRS13, parasite lysates were probed with DBA lectin (Figure 3b). We demonstrated that the DBA lectin binds to the SRS13 and its reactivity was lost in the ppGalNAc-T2 and T3 deletion mutants (Δt2 and Δt3). This indicates that the glycans on SRS13 that are detected by DBA lectin require T2 and T3 glycosyltransferase. Taken together, these results persuasively suggest that the SRS13 mucin domain is O-GalNAc glycosylated by ppGalNAc-T2 and T3.
To assess if the occurrence of mucin domains is common in SRS proteins an in silico prediction of O- GalNAc glycosylation potential was performed for all SRS proteins using NetOGlyc 4.0 (Steentoft et al. 2013). Figure 3c is the box-and-whisker plot displaying the distribution of all SRS proteins based on the number of predicted O-GalNAc modified residues. SRS13 and CST1 are the only outliers (>1.5 Inter Quartile Range), out of all SRS proteins, indicating that they are the uniquely equipped with mucin domains and that these domains are likely highly O-GalNAc glycosylated.
SRS13 localizes to the bradyzoite matrix and cyst wall
To assess the localization of SRS13, an endogenous copy of the SRS13 gene was tagged with a 3x hemagglutinin (3xHA) epitope sequence at either amino-terminus (after the signal peptide) or at the carboxyl-terminus (Fig. 4a). Figure 4b is an immunofluorescence image of in vitro HA-tagged parasites cultured at pH 7 (tachyzoite conditions) and pH 8 (bradyzoite conditions). Under tachyzoite culture conditions, SRS13 was not seen by IFA, which is consistent with the immunoblot and RNAseq data on the expression of SRS13. Under bradyzoite culture conditions, SRS13 expression was robust and by IFA SRS13 localized to the cyst wall and matrix. The observed expression pattern of SRS13 to the cyst wall co-localized with the CST1 detected by a polyclonal anti-CSTl raised to a peptide portion of CST1. This verifies that SRS13 is a bradyzoite specific cyst wall and matrix protein.
Figure 4.
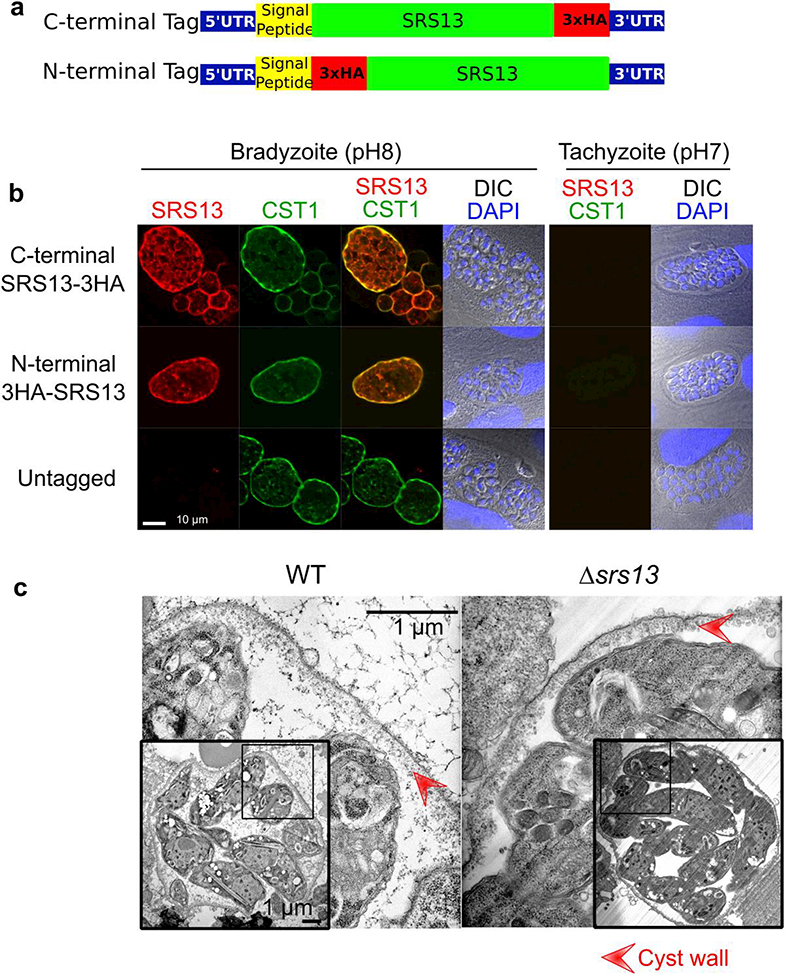
a. Schematic for the HA tagging strategies. For C-terminal tagging a 3xHA sequence was added immediately before the stop codon. For N-terminal tagging a 3xHA sequence was added immediately after the predicted signal peptide sequence..
b. SRS13 is localized to cyst wall and bradyzoite matrix but not in tachyzoite. Immunocytochemical image of in vitro T. gondii cultured in either bradyzoite condition (pH8 for 5 days) or tachyzoite condition (pH7 for 3 days) probed with anti-HA antibody (red), rabbit polyclonal anti-CST1 antibody (green), and DAPI (blue).
c. SRS13 is not required for the formation of in vivo cyst wall. Transmission electron (TEM) micrograph of in vivo mouse brain cysts in low and high magnifications are shown. The lower magnification TEM image of whole cysts are in the insert panels. Red arrowheads indicate the cyst wall.
SRS13 is dispensable for formation of the cyst wall
There is a significant parallel between SRS13 and CST1; in that both have Family-2 SRS domains, are O- GalNAc glycosylated at mucin-like domains, and localize to cyst wall. Our previous study demonstrated that an absence of the cyst wall mucin protein CST1 disrupted cyst wall formation causing an absence of the amorphous material found in the cyst wall and increased cyst fragility (Tomita et al. 2017). To investigate the possibility that SRS13 is also necessary for formation of in vivo cyst wall, BALB/cDM1 mice were infected with either WT or Δsrs13 T. gondii to produce brain cysts. No difference in virulence (e.g. no difference in mortality, data not shown) was seen in mice infected with either strain of T. gondii. Four weeks after the infection brain cysts were purified from both WT and Δsrs13 T. gondii -infected mouse brains. Transmission electron microscopy of the WT brain cysts demonstrated the typical granular cyst wall layer (Figure 4c, red arrowhead), which is absent in Δcst1 cysts (Tomita et al. 2017). The Δsrs13 parasites had a well-developed cyst wall with a typical granular cyst wall layer (Figure 4c) similar to WT parasites. This indicates that unlike CST1, SRS13 is dispensable for the formation of a proper cyst wall. No significant breakage of Δsrs13 T. gondii was observed during their purification, suggesting that SRS13 did not affect cyst wall fragility in constrast to CST1 deletion that caused a significant increase in cyst wall fragility (Tomita et al. 2017).
Phylogenetic analysis of SRS13
CST1(SRS44) orthologues are found in the genomes of the majority of tissue cyst forming coccidians. In contrast, examination of the tissue-cyst forming coccidians for SRS13 orthologs demonstrates that SRS13 is absent in most of the coccidia, e.g. Sarcocystis neurona lacks a SRS13 ortholog. Of the coccidia, an SRS13 ortholog was only found in Neospora caninum and Hammondia hammondi. The ortholog of SRS13 in H. hammondi is almost identical to that of T. gondii except that H. hammondi has 14 threonine-rich tandem repeats (consensus: QPEKP[L/V]ATTTAPP) instead of the 13 seen in T. gondii (consensus: KTQPETTTVAPPA). Phylogenetically, H. hammondi is very closely related to T. gondii. The SRS13 ortholog in N. caninum has six tandem repeats with the consensus of DAPQPGPPQP, indicated that this repeat region is not O-GalNAc glycosylated, instead of the threonine-rich (mucin-like) tandem repeats present in both T. gondii and H. hammondi. Glycosylation of SRS13 is, therefore, probably limited to T. gondii and H. hammondi.
DISCUSSION
Toxoplasma gondii, unlike most coccidian parasites, can transition from bradyzoite to tachyzoite and vice- versa, bypassing its definitive host (i.e. cats) and permitting clonal propagation in its intermediate hosts. During chronic (i.e. latent) infection, whenever transformation to tachyzoites occurs any tachyzoites that develop are removed by the immune system. In contrast, cysts (containing bradyzoites) are sequestered from the immune system. The dynamic equilibrium between encysted (bradyzoite) and replicating (tachyzoite) life cycle stages leads to recurrent antigenic stimulation as well as the persistent antibody titers observed in latent infection. In the absence of an effective immune response released organisms that differentiate into tachyzoites cause acute infection. Tissue cysts, therefore, serve as a reservoir for the reactivation of toxoplasmosis when the host becomes immunocompromised by conditions such as AIDS, organ transplantation, or due to the impaired immune response that occurs when pathogens are acquired in utero.
CST1 and SRS13 share many similarities including a threonine-rich tandem repeat (mucin domain), Family 2 SRS domain, reactivity to the glycoepitope-specific monoclonal antibody SalmonE and to DBA lectin in a ppGalNAc-T2 and -T3 dependent manner, and upregulation of gene expression and protein levels in the bradyzoite stage. Therefore, we speculated that SRS13 would like CST1 be a componenet of the cyst wall. Our investigation demonstrated that, similar to CST1, SRS13 is also found in the cyst wall and matrix, but unlike CST1, it is not an essential component for assembly of the cyst wall. Typical SRS proteins, including those specifically expressed in bradyzoite are tethered on the parasite plasma membrane through C-terminal GPI-anchors. So far, CST1 and SRS13 are the only SRS proteins that are not associated with plasma membrane, but are instead secreted into cyst matrix and then associate with the cyst wall.
In this paper, we have investigated the role of ppGalNAc-T2 and T3 in O-GalNAc glycosylation of SRS13. Previous studies in a cell free system suggested that T2 is an initiator glycosyl transferase that modifies a naked peptide (Stwora-Wojczyk et al. 2004b); while T3 requires pre-O-GalNAc glycosylation of its target peptide and then transfers GalNAc to adjascent accepter residues (Stwora-Wojczyk et al. 2004a). This hierarchical O-GalNAc glycosylation was confirmed in T.gondii using the CST1 mucin domain as a model substrate for T2 and T3 glycosyltransferases (Tomita et al. 2017). Based on this data and the observations in the current paper, it is likely that the single unit of mucin-like domain (threonin-rich tandem repeat) of SRS13 is initially glycosylated by T2. Subsequently, T3 detects the initial O-GalNAc modified by T2 and transfers GalNAc to adjascent threonines to fully occupy the single repeat unit. Other glycosyl transferases then extend the O-GalNAc modification utilizing other sugars to form more complex glycans. In the absence of the “initiator” T2, the “filling-in” T3 cannot modify the mucin domain of SRS13 resulting in the observed lack of DBA lectin binding. Without the “filling-in” T3, the mucin domain is sparsely O-GalNAc glycosylated by T2 and this is not sufficient for detection by the DBA lectin. The lack, however, of O- GalNAc glycosylation does not necessarily mean that this expressed SRS13 is devoid of any glycosylation. A variety of proteins are modified with other glycans in T. gondii including N-linked glycans, O-GlcNAc, O-fucose (Luk et al. 2008; Perez-Cervera et al. 2011; Bandini et al. 2016), and these post translations glycomodifications could also occur on SRS13.
One question that can be asked is: What is the function of the mucin domains in these two SRS proteins, SRS13 and CST1? Mucins are known to provide protection against proteolytic digestion and maintain hydration. In mammals, secreted mucins form polymer networks resistant to endogenous proteases. For example, a network of mucins protect and lubricate the gastrointestinal tract (reviewed in (Johansson et al. 2013)). To this end, it is possible that these cyst wall mucins of T. gondii could help to maintain the hydration of parasites in the cysts when the host animal dies and being desiccated. The cyst wall mucins could be also providing a physical barrier against the proteolytic enzymes released from the surrounding dead cells. These physical properties could conceivably give enough time for the parasites to survive in a dead animal until another potential host such as scavenging animals eats them. If cyst wall mucins were forming a physical barrier, it would still allow active endocytosis for nutrition acquisition. A recent study has shown that cysts are not in a stationary state, but in rather dynamic equilibrium. A subset of bradyzoites within cysts are actively dividing (Sinai et al. 2016) and the cyst membrane has been demonstrated to rapidly endocytose nutrients (Acquarone et al. 2017).
The cyst wall is the critical structure for survival, reactivation and transmission of T. gondii. The biogenesis, composition, and functions of the cyst wall need to be fully defined to provide the information required to develop a comprehensive model of this structure, which is essential for understanding latency and chronic infection in T. gondii. Furthermore, understanding T. gondii developmental biology and formation of the cyst wall will provide the necessary underpinning for control strategies such as vaccine development as well as the development of therapeutic agents that could eliminate latency preventing reactivation toxoplasmosis, which cannot be achieved by current medications for T. gondii.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH) grants National Institute of Allergy and Infectious diseases (NIAID) AI095094 (LMW) and GM GM102779 (TT).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- Acquarone M, Ferreira- da- Silva M da F, Guimarães EV, Barbosa HS (2017) Toxoplasma gondii Tissue Cyst: Cyst Wall Incorporation Activity and Matrix Cytoskeleton Proteins Paving the Way to Nutrient Acquisition. In: Toxoplasmosis. InTech [Google Scholar]
- Bandini G, Haserick JR, Motari E, Ouologuem DT, Lourido S, Roos DS, Costello CE, Robbins PW, Samuelson J (2016) O -fucosylated glycoproteins form assemblies in close proximity to the nuclear pore complexes of Toxoplasma gondii. Proc Natl Acad Sci 113:11567–11572. doi: 10.1073/pnas.1613653113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Radke JB, Smith AT, Sullivan WJ, White MW (2008) The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol 68:1502–18. doi: 10.1111/j.1365-2958.2008.06249.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierszinski F, Mortuaire M, Cesbron-Delauw MF, Tomavo S (2000) Targeted disruption of the glycosylphosphatidylinositol-anchored surface antigen SAG3 gene in Toxoplasma gondii decreases host cell adhesion and drastically reduces virulence in mice. Mol Microbiol 37:574–82 [DOI] [PubMed] [Google Scholar]
- Fox B a, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, Cesbron-Delauw M-F, Weiss LM, Bzik DJ (2011) Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot Cell 10:1193–206. doi: 10.1128/EC.00297-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Grigg ME, Boothroyd JC, Garcia KC (2002) Structure of the immunodominant surface antigen from the Toxoplasma gondii SRS superfamily. Nat Struct Biol 9:606–11. doi: 10.1038/nsb819 [DOI] [PubMed] [Google Scholar]
- Johansson MEV, Sjövall H, Hansson GC (2013) The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 10:352–61. doi: 10.1038/nrgastro.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-K, Boothroyd JC (2005) Stage-specific expression of surface antigens by Toxoplasma gondii as a mechanism to facilitate parasite persistence. J Immunol (Baltimore, Md 1950) 174:8038–8048 [DOI] [PubMed] [Google Scholar]
- Luk FCY, Johnson TM, Beckers CJ (2008) N-linked glycosylation of proteins in the protozoan parasite Toxoplasma gondii. Mol Biochem Parasitol 157:169–178. doi: 10.1016/J.MOLBIOPARA.2007.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid-Aliste CJ, Dybas JM, Angeletti RH, Weiss LM, Kim K, Simon II, Fiser A, Hogue Angeletti R (2009) EPIC-DB: a proteomics database for studying Apicomplexan organisms. BMC Genomics 10:38. doi: 10.1186/1471-2164-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cervera Y, Harichaux G, Schmidt J, Debierre-Grockiego F, Dehennaut V, Bieker U, Meurice E, Lefebvre T, Schwarz RT (2011) Direct evidence of O-GlcNAcylation in the apicomplexan Toxoplasma gondii: a biochemical and bioinformatic study. Amino Acids 40:847–56. doi: 10.1007/S00726-010-0702-4 [DOI] [PubMed] [Google Scholar]
- Shen B, Brown KM, Lee TD, Sibley LD (2014) Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5:e01114–14. doi: 10.1128/mBio.01114-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai AP, Watts EA, Dhara A, Murphy RD, Gentry MS, Patwardhan A (2016) Reexamining Chronic Toxoplasma gondii Infection: Surprising Activity for a “Dormant” Parasite. Curr Clin Microbiol Reports 3:175–185. doi: 10.1007/s40588-016-0045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-BG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32:1478–88. doi: 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stwora-Wojczyk MM, Dzierszinski F, Roos DS, Spitalnik SL, Wojczyk BS (2004. a) Functional characterization of a novel Toxoplasma gondii glycosyltransferase: UDP-N-acetyl-D- galactosamine:polypeptide N-acetylgalactosaminyltransferase-T3. Arch Biochem Biophys 426:231–40. doi: 10.1016/j.abb.2004.02.013 [DOI] [PubMed] [Google Scholar]
- Stwora-Wojczyk MM, Kissinger JC, Spitalnik SL, Wojczyk BS (2004. b) O-glycosylation in Toxoplasma gondii: Identification and analysis of a family of UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferases. Int J Parasitol 34:309–322. doi: 10.1016/j.ijpara.2003.11.016 [DOI] [PubMed] [Google Scholar]
- Tentera M, Heckerotha R, Weiss LM (2000) Toxoplasma gondii: from animals to humans. Int J Parasitol 30:1217–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Bzik DJ, Ma YF, Fox B a., Markillie LM, Taylor RC, Kim K, Weiss LM (2013) The Toxoplasma gondii Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence. PLoS Pathog 9:1–15. doi: 10.1371/journal.ppat.1003823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Sugi T, Yakubu R, Tu V, Ma Y, Weiss LM (2017) Making Home Sweet and Sturdy: Toxoplasma gondii ppGalNAc-Ts Glycosylate in Hierarchical Order and Confer Cyst Wall Rigidity. MBio 8:e02048–16. doi: 10.1128/mBio.02048-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van TT, Kim S-K, Camps M, Boothroyd JC, Knoll LJ (2007) The BSR4 protein is up-regulated in Toxoplasma gondii bradyzoites, however the dominant surface antigen recognised by the P36 monoclonal antibody is SRS9. Int J Parasitol 37:877–885. doi: 10.1016/j.ijpara.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Wasmuth JD, Pszenny V, Haile S, Jansen EM, Gast AT, Sher A, Boyle JP, Boulanger MJ, Parkinson J, Grigg ME (2012) Integrated Bioinformatic and Targeted Deletion Analyses of the SRS Gene Superfamily Identify SRS29C as a Negative Regulator of Toxoplasma Virulence. MBio 3:. doi: 10.1128/mBio.00321-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


