Abstract
The bone marrow microenvironment regulates normal and malignant hematopoiesis, but the underlying molecular mechanisms remain poorly defined. Using a chimeric mice model, we demonstrate that Twist1 deletion in the bone marrow microenvironment results in alteration of multiple niche cells as well as downregulated expression of major hematopoietic stem cell supportive factors. The perturbed microenvironment reduces hematopoietic stem cell homing and retention, impairs hematopoietic stem cell self-renewal and induces myeloid skewing. Nevertheless, it accelerates the progression of MLL-AF9 leukemia, which is partially mediated by Jagged-2-dependent Notch signaling. Our data provide the first demonstration of a pivotal role of TWIST1 in favoring normal hematopoiesis and hampering leukemia development. They also bring new insights into the role of the bone marrow niche in driving the development of acute myeloid leukemia, and suggest possible new avenues, exploiting the niche, to improve leukemia treatments.
Introduction
Hematopoietic stem cells (HSC) reside in a special bone marrow (BM) niche, which regulates their localization, self-renewal and differentiation. Studies have identified several major cell types of the niche, including mesenchymal stem cells (MSC), osteolineage cells (OLC), adipocytes and vascular endothelial cells (EC).1–5 Besides the key cellular components, some growth and survival factors are also indispensable components of the niche, including C-X-C motif chemokine 12 ligand (CXCL12), vascular cell adhesion molecule1 (VCAM1),6,7 stem cell factor (SCF)4 and osteopontin.8 A sophisticated network of interactions between these multiple BM cells, extracellular factors and adhesion molecules is essential to regulate different HSC properties during homeostasis and keep normal hematopoiesis in check.
Acute myeloid leukemia (AML) has been considered for decades to be a disorder intrinsic to hematopoietic cells; however, evidence is accumulating that the microenvironment exerts more than a mere bystander effect. Leukemic cells can remodel the niche into a permissive environment favoring leukemic stem cell (LSC) expansion over normal HSC maintenance.9 Recently, emerging evidence even points to a role for the BM niche as a driver of disease maintenance/progression. Krause et al. showed that osteoblast-specific activation of the parathyroid hormone receptor enhances MLL-AF9 oncogene-induced AML in mouse transplantation models.10 To date, there are still few studies concerning the role of the bone marrow niche in initiating and maintaining AML and relevant mechanisms remain elusive.
TWIST1 is a highly conserved transcription factor belonging to the basic helix-loop-helix family and is implicated in diverse developmental systems.11–13 Studies have revealed that TWIST1 is a key regulator of MSC self-renewal, enhances their life-span, inhibits MSC osteo/chondrogenic differentiation and promotes adipogenic differentiation.14–16 Twist1 haploisufficiency leads to Saethre-Chotzen syndrome, which is characterized by alterations in osteogenic precursor cell proliferation, differentiation and survival.17 Recent studies have demonstrated that TWIST1 promotes angiogenesis by inducing EC proliferation and migration, and deregulation of this mechanism mediates pathological angiogenesis.18,19 Arthur et al. showed that overexpression of Twist1 in MSC enhances the capacity to maintain human CD34+ cells in long-term culture-initiating cell assays through increasing Cxcl12 expression.20 However, the effects of TWIST1 on multiple niche elements and its modulation of normal HSC maintenance and leukemia progression in vivo have not been functionally characterized so far.
To explore this issue, we generated a murine model of a Twist1-deficient microenvironment. We showed that the major niche cellular components and factors changed remarkably upon Twist1 deletion, causing severe dysfunction of normal HSC. Nevertheless, these alterations of the BM microenvironment promoted MLL-AF9 oncogene-induced AML progression in mouse transplantation models, not only pointing to TWIST1 as an instructive signal modulating the stem cell niche, but also emphasizing the importance of the niche for AML development.
Methods
Mice
Twist1flox/flox mice were purchased from Mutant Mouse Regional Resource Centers. ER-Cre mice were a gift from Professor Weiping Yuan. C57BL/6 and B6.SJL mice were purchased from the animal facility of State Key Laboratory of Experimental Hematology. Twist1flox/flox mice were crossed with ER-Cre mice to generate ER-Cre;Twist1fl/fl and ER-Cre;Twist1+/+ mice. Eight- to 12-week old mice were used. Cre expression was induced by daily intraperitoneal injection of tamoxifen (75 mg/kg of total body weight in corn oil; Sigma-Aldrich, St. Louis, MO, USA) for 5 days. All animal procedures complied with the animal care guidelines approved by the Institutional Animal Care and Use Committees of the State Key Laboratory of Experimental Hematology.
Transplantation assays
For non-competitive BM transplantation, to create the chimeras described in Online Supplementary Figure S1A, 2x106 whole BM cells from B6.SJL (CD45.1) mice were transplanted into ER-Cre;Twist1+/+ or ER-Cre;Twist1fl/fl (CD45.2) recipients that were lethally irradiated (9.5 Gy from a Cesium source, 4-24 h before transplantation). Sixteen weeks later, tamoxifen was injected to induce Twist1 deletion. For competitive transplantation, 300 BM long-term HSC (CD45.1) from tamoxifen-treated ER-Cre;Twist1+/+ or ER-Cre;Twist1fl/fl chimeric mice were mixed with 2×105 congenic BM support cells and injected into lethally irradiated CD45.2 recipients. For the MLL-AF9 AML model, 5x105 GFP+ leukemic cells were transplanted into Twist1-deleted or control chimeric recipients.
Flow cytometry analysis and cell sorting
The BM cell suspensions were flushed from femora and tibiae. Spleen cells were pestled by the plug of a 10 mL syringe. The cells were then filtered through a 74 mm nylon mesh. For flow cytometric analysis of stromal cells, BM was flushed using phosphate-buffered saline with 2% bovine serum, the bones were minced with scissors, then the plugs were digested in 1 mg/mL collagenase I (OLC) or IV (MSC and EC) (Sigma-Aldrich) dissolved in Hank’s balanced salt solution with 10% fetal bovine serum for 90 min (collagenase I) or 30 min (collagenase IV) at 37°C. The dissociated cells were collected and kept on ice. Cells were incubated with conjugated antibodies. Stained cells were analyzed with FACS LSR II or sorted with a FACS Aria II instrument (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed by FlowJo software.
Statistical analysis
The significance of differences between two groups was determined using unpaired two-tailed Student t tests. Data are presented as means ± standard deviations. Overall survival curves were plotted according to the Kaplan-Meier method with the log-rank test applied for comparisons. *P<0.05, **P<0.01, ***P<0.001.
Details of other experimental procedures are given in the Online Supplementary Methods.
Results
Microenvironmental Twist1 deficiency leads to decreased numbers of mesenchymal stem cells and mature osteoblasts, an increased proportion of endothelial cells, and altered expression of cell factor genes
To explore the role of TWIST1 in the BM niche and its regulation of HSC, we generated microenvironment Twist1-deleted and control chimeric mice according to the method described by Schreck and Saez.21,22 In brief, 2×106 BM cells from B6.SJL wild-type (WT) mice (CD45.1) were transplanted into ER-Cre;Twist1fl/fl and ER-Cre;Twist1+/+ recipients (CD45.2) (Online Supplementary Figure S1A). Sixteen weeks later, about 90% of the cells in the peripheral blood of recipients were donor-derived cells (Online Supplementary Figure S1B). Tamoxifen was then injected intraperitoneally for 5 days to induce Twist1 deletion. Two weeks after the last injection, mRNA detection demonstrated that Twist1 had been knocked out in all the MSC, OLC, and EC isolated from Twist1Δ/Δ mice with similar knockout levels (Online Supplementary Figure S1C), while the expression of Twist2 was almost unchanged (data not shown).
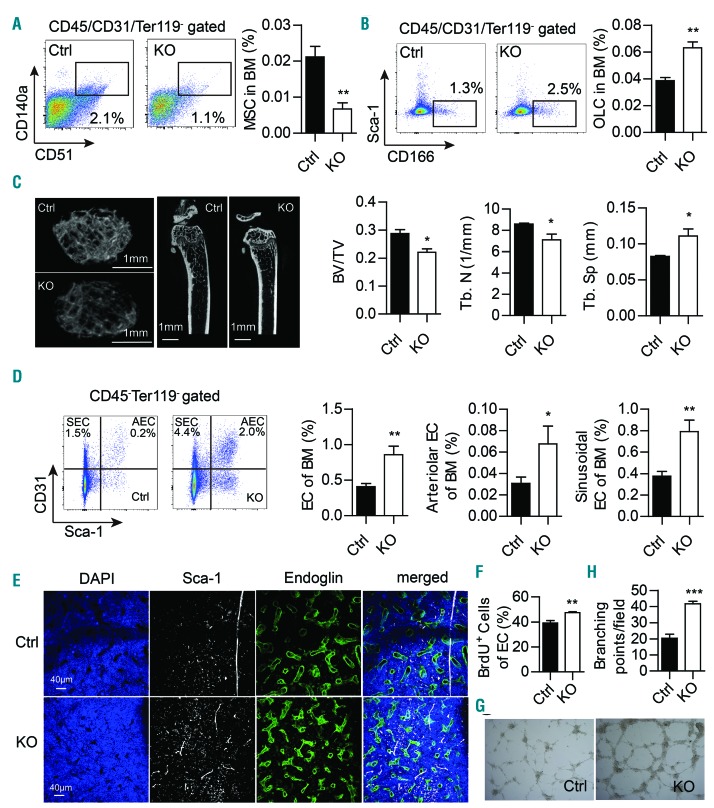
To define components of the Twist1-deleted BM microenvironment that may be altered, stromal populations and extracellular factors were assessed in Twist1-deleted and control chimeric mice. We observed that conditional deletion of Twist1 led to a significant decrease in the number of MSC (CD140a+CD51+CD45/Ter119/CD31-)23 in the BM compared with that in control mice, as determined by flow cytometry (Figure 1A). The decrease in MSC number was further confirmed by a fibroblastic colony-forming unit assay (Online Supplementary Figure S2A). Furthermore, Twist1-deleted MSC showed a decrease in proliferative cells and an increase in apoptotic cells (Online Supplementary Figure S2B,C), indicating the mechanism underlying the reduced number of MSC.
Figure 1.
Twist1 deficiency in the bone marrow microenvironment leads to decreased frequency of mesenchymal stem cells and mature osteoblasts, and an increased proportion of endothelial cells. (A) Flow cytometry (FACS) analysis of bone marrow (BM) msesenchymal stem cells (MSC, CD140a+CD51+CD45/Ter119/CD31−) in chimeric control (Ctrl) and knockout (KO) mice. Representative FACS profiles are shown on the left, and cell frequency is shown on the right (n=4, three independent experiments). (B) FACS analysis of BM osteolineage cells (OLC, Sca-1−CD166+CD45/Ter119/CD31−) in chimeric Ctrl and KO mice. Representative FACS profiles are shown on the left, and cell frequency is shown on the right (n=5, three independent experiments). (C) Micro-computed tomography analysis of the trabecular bone of chimeric Ctrl and KO mice. Representative images are shown on the left. Scale bars, 1 mm. Trabecular bone volume/total volume (BT/BV), trabecular number (Tb. N) and trabecular spacing (Tb. Sp) in the femoral metaphysis are shown on the right (n=4, two independent experiments). (D) FACS analysis of BM endothelial cells (EC) in chimeric Ctrl and KO mice. Representative FACS profiles of sinusoidal EC (SEC, CD45−Ter119−CD31+Sca-1−) and arteriolar EC (AEC, CD45−Ter119−CD31+Sca-1+) are shown on the left. Frequencies of BM total EC (CD45−Ter119−CD31+), AEC and SEC are shown on the right (n=6, two independent experiments). (E) Immunofluorescent images of the BM microvasculature in the femoral diaphysis of animals of each genotype are shown after staining for Sca-1 (white, arteries), Endoglin (green, sinusoids) and 4’,6-diamidi-no-2-phenylindole (DAPI, blue), as detailed in the Methods. Scale bars, 40 mm. (n=3, two independent experiments). (F) Proliferation analysis of EC in chimeric Ctrl and KO mice (n=4, two independent experiments). (G) In vitro tube formation assay with EC from chimeric Ctrl and KO mice. (H) Quantification of the tube formation assay (n=3, two independent experiments). Column plots show the mean ± standard deviation. *P<0.05; **P<0.01, ***P<0.001 (Student t test).
Twist1 deficiency resulted in a marked increase in the frequency of Sca-1−/CD166+ stromal cells (Figure 1B), which include immature and mature OLC.24 Meanwhile, the expression of osteoblastic differentiation genes Runx2, Ogn and Gpnmb14,23 was significantly upregulated in Twist1-deleted MSC (Online Supplementary Figure S2D). To assess the ability of MSC to differentiate into the osteoblastic lineage, we induced osteoblast differentiation in MSC and found that Twist1 deletion clearly increased alkaline phosphatase activity and matrix mineralization (Online Supplementary Figure S2E). These results establish that Twist1 deficiency enhanced MSC commitment toward osteoblasts. However, expression of the mature osteoblast marker, Bglap, was downregulated (Online Supplementary Figure S2F). In addition, micro-computed tomography analysis also revealed a significant decrease in mature osteoblasts in Twist1-deleted mice, which was reflected by a reduction of trabecular volume (Figure 1C). Collectively, Twist1 deletion promotes MSC to differentiate toward the osteoblast lineage with a block of mature osteoblast differentiation.
Emerging data demonstrate the role of vascular EC in HSC maintenance, and arterioles and sinusoids exhibit dif ferent properties in relation to HSC distribution and quiescence.4,25,26 Ciuculescu et al. reported that Rac deletion in MSC leads to an inverted ratio of marrow arterioles and sinusoid vessels and impaired hematopoiesis.27 We observed that Twist1 deletion resulted in increased CD45-Ter119−CD31+, CD45−Ter119−CD31+Sca1+ and CD45−Ter119- CD31+Sca1− populations enriched for total EC, arteriolar EC and sinusoidal EC,28 respectively (Figure 1D). The increase of arteriolar and sinusoidal EC was further confirmed by the observation that arteries (Sca-1-staining) and sinusoids (Endoglin-staining) were both significantly increased in Twist1-deleted mice compared with control mice by immunofluorescence of femoral sections (Figure 1E). The increase of EC may be the result of cell proliferation as determined by increased bromodeoxyuridine incorporation into CD45−Ter119−CD31+ stromal cells in Twist1-deleted mice (Figure 1F). We next performed a tube formation assay to determine the effect of TWIST1 on new blood vessel development. Consistent with increased microvessels in knockout mice in vivo, capillary tube formation of Twist1-deleted EC was also increased on matrigel (Figure 1G,H), indicating that Twist1 deletion promotes angiogenesis.
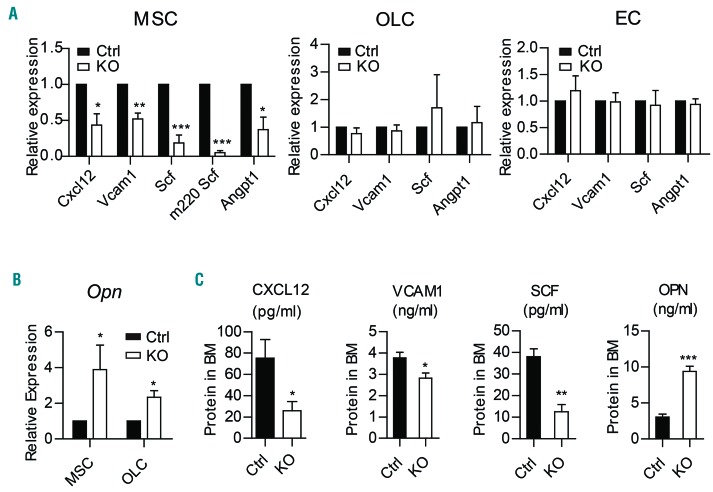
To further evaluate the impact of Twist1 deficiency on cell factors, we performed quantitative real-time polymerase chain reaction to analyze the expression of key niche factors. The results showed significant decreases in the expression of Cxcl12, Vcam1, Angiopoietin-1 (Angpt1) and Scf, particularly the membrane-bound isoform Scf (m220 Scf), which was found to be extremely important for HSC maintenance, in MSC (5×104 cells) from Twist1-deleted mice as compared to control mice (Figure 2A). The expression of Opn, which negatively regulates the HSC pool, was obviously increased in both MSC and OLC (Figure 2B). Enzyme-linked immunoassay demonstrated the reduced protein levels of CXCL12, VCAM1, SCF and elevated level of osteopontin (Figure 2C) in BM supernatants of Twist1-deleted mice as compared to those in control mice.
Figure 2.
Twist1 deletion in the bone marrow microenvironment changes expression of niche factors. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of the expression of C-X-C motif chemokine ligand 12 (Cxcl12), Vascular cell adhesion molecule 1 (Vcam1), Stem cell factor (Scf) and Angiopoietin-1 (Angpt1) in freshly sorted mesenchymal stem cells (MSC), osteolineage cells (OLC) and endothelial cells (EC) from chimeric control (Ctrl) and knockout (KO) mice (n=4). (B) qRT-PCR analysis of the expression of Osteopontin (Opn) in MSC and OLC from chimeric Ctrl and KO mice (n=4). (C) Enzyme-linked immunosorbent assay analysis of BM protein concentrations of CXCL12, VCAM1, SCF and OPN in chimeric Ctrl and KO mice (n=5-8). Column plots show the mean ± standard deviation from three independent experiments. *P<0.05; **P<0.01; ***P<0.001 (Student t test).
Collectively, Twist1 deletion leads to significant alterations in various key niche components, demonstrating its functional importance in the BM microenvironment, and implying its potential regulatory role in HSC maintenance.
Microenvironmental Twist1 deficiency impairs the homing and retention of hematopoietic stem cells but promotes their mobilization
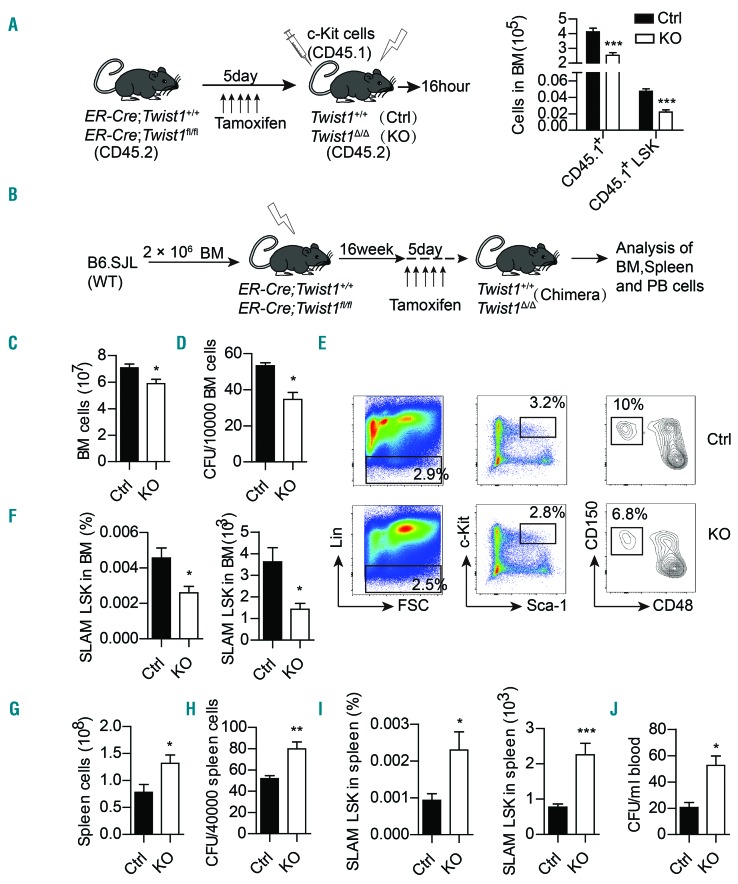
Most HSC are retained in the BM niche in a quiescent, nonmotile mode by adhesion to stromal cells, which are essential for normal hematopoiesis and for protection from myelotoxic injury. Twist1 deletion resulted in decreased expression of BM CXCL12 and VCAM1, which are critical for HSC retention, homing and mobilization,29,30 implying that TWIST1 may have a functional impact on the migration of HSC. We first evaluated homing of normal HSC to the BM of Twist1-deleted mice. Freshly isolated c-Kit+ cells from B6.SJL (CD45.1) mice were injected into lethally irradiated Twist1-deleted or control mice. Sixteen hours after transplantation, the absolute number of CD45.1+ and CD45.1+ LSK (Lin-Sca-1+c-Kit+) cells was significantly decreased in Twist1-deleted mice compared to the number in controls (Figure 3A), indicating that Twist1 deficiency impairs homing of hematopoietic stem/progenitor cells (HSPC) to the BM microenvironment.
Figure 3.
Decreased hematopoietic stem/progenitor cell homing and retention in bone marrow and increased hematopoietic stem/progenitor cell mobilization to spleen and peripheral blood in Twist1-deficient mice. (A) Experimental scheme of the hematopoietic stem/progenitor cell (HSPC) homing assay (left), and absolute number of CD45.1+ and CD45.1+LSK (Lin-Sca-1+c-Kit+) cells homing to the bone marrow (BM) (right) (n=5, two independent experiments). (B) Experimental scheme for analysis of HSPC retention and mobilization. (C-F) Analysis of BM cells of chimeric control (Ctrl) and knockout (KO) mice. Total BM cells (C) of femora and tibiae, number of BM progenitor cells (D) measured by colony-forming cell (CFC) assay in methylcellulose, and frequency and number of SLAM LSK cells (CD150+CD48−LSK) (E-F) are shown (n=4-6, three independent experiments). (G-J) Analysis of peripheral blood cells of chimeric Ctrl and KO mice. Number of total cells (G), number of colony-forming units (CFU) (H), frequency and number (I) of SLAM LSK cells in spleen, and number of CFU (J) in peripheral blood are shown (n=4-6, three independent experiments). Column plots show the mean ± standard deviation. *P<0.05; **P<0.01; ***P<0.001 (Student t test).
We next determined the contribution of TWIST1 to HSC retention and mobilization by assessing total cells and HSPC in the BM, spleen and peripheral blood of Twist1-deleted mice and control mice (Figure 3B). The results revealed that Twist1 deletion led to decreases in the numbers of total cells (Figure 3C), colony-forming units (Figure 3D), and HSC-enriched SLAM LSK (CD150+CD48-LSK) in the BM (Figure 3E,F) but significant increases in those in the spleen and circulation (Figure 3G-J).
Granulocyte colony-stimulating factor (G-CSF) is a hematopoietic cytokine known as the prototypic mobilizing agent.31 To determine whether Twist1 deletion promot ed secretion of endogenous G-CSF, we examined G-CSF levels in the BM supernatants of Twist1-deleted and control mice by enzyme-linked immunosorbent assay. We found that G-CSF protein expression was significantly elevated in Twist1-deficient mice as compared to that in controls. Furthermore, quantitative real-time polymerase chain reaction results revealed that among MSC, OLC, EC and macrophagocytes, which are all producers of G-CSF, OLC and macrophagocytes were the major source of G-CSF (Online Supplementary Figure S3A,B). We next assessed HSPC mobilization after infusion of exogenous G-CSF and found that Twist1 deletion significantly increased exogenous G-CSF-induced mobilization of total cells and colony-forming cells to the blood and spleen, and SLAM LSK to the spleen (Online Supplementary Figure S3C-H).
Taken together, these results demonstrate a functional role of TWIST1 in HSC homing, retention, baseline mobilization and stress mobilization in response to G-CSF.
Twist1 deficiency impairs hematopoietic stem cell quiescence and self-renewal, and induces enhanced early myeloid lineage differentiation
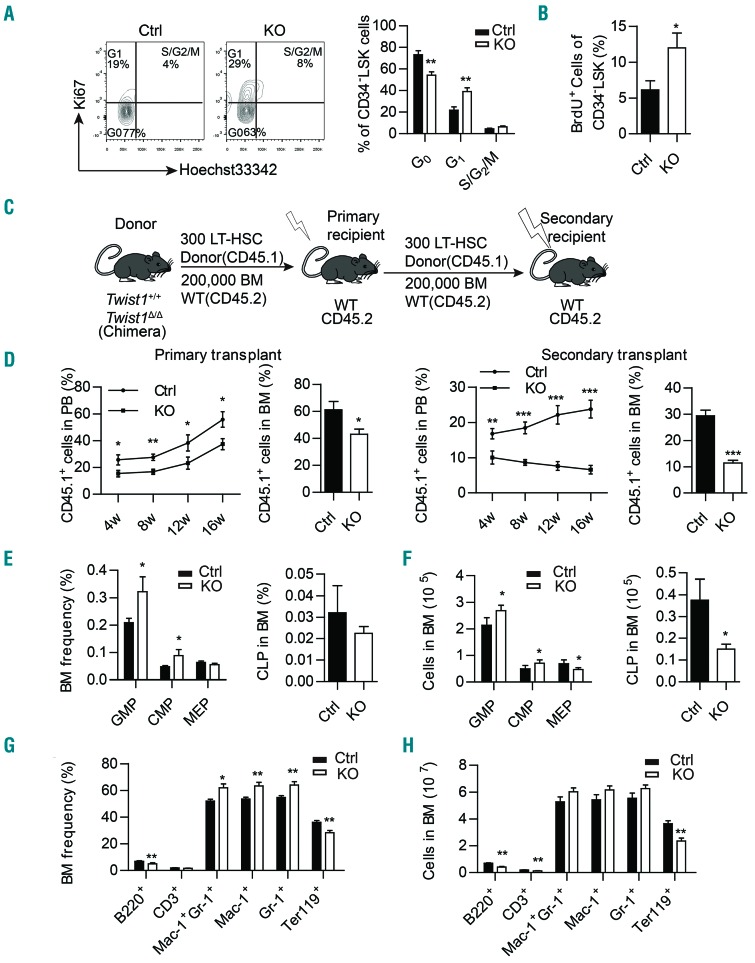
Quiescence, self-renewal and committed differentiation are important properties of HSC, which could be controlled by stromal cells, extracellular matrix, cytokines and chemokines.32,33 Our study revealed that Twist1 deletion altered multiple stromal cells and the level of expression of HSC supportive factors, so we next investigated whether these HSC features were consequently changed. Immunophenotypic analysis demonstrated that Twist1 deletion resulted in a significant decrease in the number of long-term HSC (CD34−Flt3−LSK) in the BM (Online Supplementary Figure S4A). Ki67 staining revealed a significant decrease in the percentage of HSC (CD34−LSK) in G0 phase in Twist1-deleted mice, together with an increase in G1 phase (Figure 4A). Bromodeoxyuridine labeling further identified a higher frequency of proliferating cells in CD34-LSK cells from Twist1-deleted mice compared to control mice (Figure 4B). These data suggest that TWIST1 in the microenvironment plays an important role in maintaining HSC quiescence, and loss of Twist1 drives aberrant proliferation of HSC.
Figure 4.
Twist1 deletion causes impaired quiescence, self-renewal of hematopoietic stem cells and myeloid skewing. (A) Percentage of the cell cycle distribution of CD34−LSK (Lin−Sca-1+c-Kit+) cells in chimeric control (Ctrl) and knockout (KO) mice. Representative flow activated cell sorting profiles are shown on the left, and cell frequency is shown on the right (n=4-5, two independent experiments). (B) Proliferation analysis of CD34−LSK cells in chimeric Ctrl and KO mice (n=4-5, two independent experiments). (C) Schematic overview of the serial transplantation assay. (D) Percentages of donor-derived peripheral blood (PB) cells and bone marrow (BM) cells after the primary and secondary competitive transplants (n=5, two independent experiments). (E-F) Frequency (E) and number (F) of common myeloid progenitors (CMP, CD34+CD16/32−Lin−Sca-1−c-Kit+), granulocyte/macrophage progenitors (GMP, CD34+CD16/32+Lin−Sca-1−c-Kit+), megakaryocyte/erythroid progenitors (MEP, CD34−CD16/32−Lin−Sca-1−c-Kit+) and common lymphoid progenitors (CLP, Lin−Sca-1lowc-KitlowIL7R+) in chimeric Ctrl and KO mice (n=4-5, three independent experiments). (G-H) Frequency (G) and number (H) of B cells (B220+), T cells (CD3+), myeloid cells (Mac-1+ and Gr-1+) and erythrocytes (Ter119+) in chimeric Ctrl and KO mice (n=4-5, three independent experiments). Column plots show the mean ± standard deviation. *P<0.05; **P<0.01; ***P< 0.001 (Student t test).
To clarify the role of TWIST1 in HSC self-renewal, we conducted serial transplantation assays (Figure 4C). Three hundred long-term HSC (CD34−Flt3−LSK, CD45.1) from Twist1-deleted or control chimeric mice were transplanted into lethally irradiated recipients (CD45.2), together with 2×105 CD45.2+ support BM cells. Then secondary transplantation assays were performed 16 weeks later. Three hundred long-term HSC (CD45.1+) from the primary recipients were transplanted into lethally irradiated secondary recipients, together with 2×105 CD45.2+ support BM cells. Donor cells from Twist1-deleted mice showed lower engraftment capacity than control cells out to 16 weeks in both primary and secondary transplantation (Figure 4D). From the above, it can be concluded that Twist1 deficiency in niche cells impairs HSC self-renewal capacity.
We then performed FACS analysis to evaluate the differentiation capacity of HSC, and found increases in the numbers of common myeloid progenitors (CD34+CD16/32−Lin−Sca-1−c-Kit+, 1.5-fold, P=0.047) and granulocyte/macrophage progenitors (GMP: CD34+CD16/32+Lin−Sca-1−c-Kit+, 1.3-fold, P=0.022) in the BM of Twist1-deleted mice in comparison with those in controls, accompanied by decreases in the numbers of megakaryocyte/erythroid progenitors (MEP, CD34-CD16/32−Lin−Sca-1−c-Kit+, 1.5-fold, P=0.028) and common lymphoid progenitors (Lin−Sca−1lowc-KitlowIL7R+, 2.5-fold, P=0.046) (Figure 4E,F). In accordance with the change of progenitors, the proportion of mature myeloid cells (Mac1+, 1.2-fold, P=0.007; Gr-1+, 1.2-fold, P=0.008; Mac-1+Gr-1+, 1.2-fold, P=0.011) was also significantly increased, accompanied by reduced proportions of B lymphoid cells (B220+, 1.4-fold, P=0.004) and erythrocytes (Ter119+, 1.3- fold, P=0.008) in the BM of Twist1-deleted mice (Figure 4G,H). These results suggest that Twist1 deletion in niche cells promotes HSC differentiation into the myeloid lineage. We observed that apart from the increase of HSPC in the spleen of Twist1-deleted mice, the numbers of mature myeloid cells and erythrocytes were also increased, while the number of lymphoid cells remained unchanged (Online Supplementary Figure S4B-D), indicating the occurrence of extramedullary hematopoiesis in the spleen of Twist1-deleted mice.
Taken together, these data suggest that TWIST1 in the BM microenvironment plays a critical role in HSC maintenance, and Twist1 deletion impairs all the fundamental features of HSC.
Twist1 deficiency promotes progression of MLL-AF9-induced acute myeloid leukemia
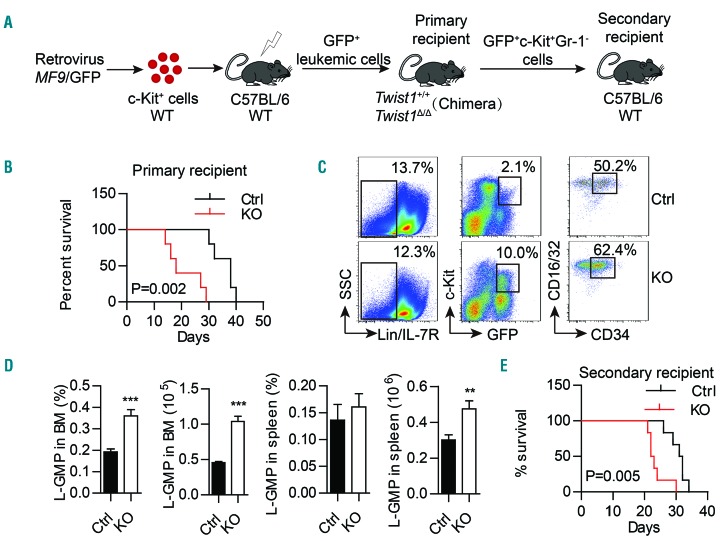
It has been reported that the MLL-AF9 AML model exhibited multiple alterations in the niche compartments, including decreased frequencies of MSC and osteoblasts, an increased number of vascular EC, and downregulated expression of Vcam1, Cxcl12, Angpt1, and Scf, together with upregulation of Opn.34 Intriguingly, these phenotypes are quite similar to the niche alterations in our Twist1-deleted mice. Considering the emerging importance of the BM niche for leukemia maintenance and progression, we were tempted to speculate that the altered niche in Twist1-deleted mice may play a role in the development of MLL-AF9 AML. To validate this hypothesis, we transduced WT BM c-Kit+ cells with retrovirus expressing MLL-AF9, and injected these cells into lethally irradiated WT recipient mice, in which the disease was rapidly induced with massive BM and spleen infiltration of GFP+ leukemic cells. We next injected 5×105 GFP+ spleen cells from these mice into Twist1-deleted and control chimeric recipient mice (Figure 5A). Notably, the overall survival of Twist1-deleted recipients was significantly shorter than that of control recipients (Figure 5B), and Twist1-deleted recipient mice exhibited a greater infiltration of total cells and leukemic cells in the BM, peripheral blood and spleen than that of control mice (Online Supplementary Figure S5A-G).
Figure 5.
Twist1 deletion in the bone marrow microenvironment promotes the progression of acute myeloid leukemia. (A) Experimental scheme of the MLL-AF9 acute myeloid leukemia model and leukemic stem cell (LSC, GFP+c-Kit+Gr-1−) transplantation. (B) Kaplan–Meier survival curve of chimeric control (Ctrl) and knockout (KO) recipient mice (n=5, three independent experiments, log-rank test). (C) Representative flow cytometry profiles of L-GMP (IL-7R−Lin−GFP+c-KithiCD34+CD16/32hi). (D) Frequency and absolute number of L-GMP in the bone marrow (BM) and spleen of Ctrl and KO recipients (n=5, three independent experiments. Column plots show the mean ± standard deviation. **P<0.01; ***P<0.001, Student t test). (E) Kaplan–Meier survival curve of mice transplanted with LSC from chimeric Ctrl and KO mice (n=6, two independent experiments, log-rank test).
Previous studies using MLL-AF9 AML models have established that LSC are enriched in the leukemic GMP populations (IL-7R−Lin−GFP+c-KithiCD34+CD16/32hi)34,35 or c-Kit+Gr-1−.36 We found that the infiltration of leukemic GMP cells was significantly increased in the BM and periphery of Twist1-deleted recipient mice compared to control mice (Figure 5C,D), and so was that of GFP+c-Kit+Gr-1− cells (Online Supplementary Figure S6A-C). Additionally, cell cycle analysis showed that the proportion of LSC (GFP+c-Kit+Gr-1−) in the G0 phase was decreased and that in the G1 or S/G2/M phases was increased in both BM and spleen from Twist1-deleted mice, but rates of LSC apoptosis in BM and spleen did not differ between Twist1-deleted and control mice (Online Supplementary Figure S6D-G). We then performed secondary transplants using LSC (Figure 5A), injecting 5000 GFP+c-Kit+Gr-1− cells isolated from Twist1-deleted and control mice into WT recipient mice. We found that the survival of the secondary recipients of LSC grafts from leukemic Twist1-deleted donors was significantly reduced compared to that of recipients of control LSC grafts (Figure 5E).
Taken together, these results demonstrate that the altered niche in Twist1-deleted mice accelerates the pro gression of MLL-AF9-induced AML by increasing the expansion and leukemogenic capacity of LSC.
Microenvironmental Twist1 deletion promotes acute myeloid leukemia development partially through the Notch signaling pathway
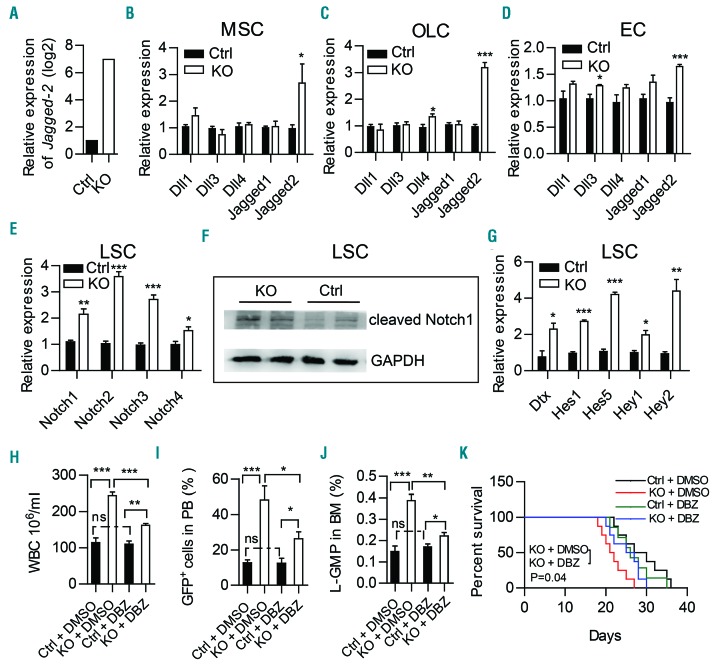
To understand the mechanism underlying TWIST1 function in the development of AML, we performed RNA- sequencing analysis on BM stromal cells isolated from Twist1-deleted or control chimeric mice. There were 6948 differentially expressed genes (4764 downregulated and 2184 upregulated; fold change of >2 and P value <0.05). Analysis of leukemia-related signaling revealed a marked increase of Jagged-2 expression in stromal cells from Twist1-deleted mice (Figure 6A). Previous studies have demonstrated that TWIST1 regulates the Notch pathway in prospective coronal suture mesenchyme and osteoprogenitors.37,38 Moreover, aberrant Notch signaling is a common mechanism in niche-induced AML and pre-leukemic conditions.39–42 To investigate whether Twist1 deficiency promotes the development of MLL-AF9 AML through Notch signaling, we determined the expression of all Notch ligands (Dll1, Dll3, Dll4, Jagged-1, Jagged-2) in MSC, OLC and EC of Twist1-deleted mice. The results revealed that Jagged-2 was significantly upregulated in all these cells (Figure 6B-D). Additionally, the levels of expression of all four Notch receptors (Notch1-4), cleaved Notch1 and the Notch targets Dtx, Hes1, Hes5, Hey1, and Hey2 were significantly upregulated in LSC from Twist1-deleted mice compared to those of controls (Figure 6E-G), indicating increased Notch signaling in this population. Furthermore, pharmacological inhibition of Notch signaling with a γ-secretase inhibitor (DBZ) (Figure 6H-K) or blockade of Notch with dominant-negative MAML1 (DNMAML1) (Online Supplementary Figure S7) partially rescued leukemic cell infiltration and LSC engraftment, and prolonged the overall survival of Twist1-deleted recipients. These data suggest that a Twist1-deleted microenvironment contributes to MLL-AF9 AML development at least in part via Notch signaling.
Figure 6.
Notch signaling is activated in leukemic stem cells and inhibition of Notch signaling partially rescues MLL-AF9-induced acute myeloid leukemia progression in Twist1-deficient mice. Expression of Jagged-2 in RNA-sequencing analysis of stromal cells (CD45−Ter119−) from chimeric control (Ctrl) and knockout (KO) mice. (B-D) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of Notch ligands (Dll1, Dll3, Dll4, Jagged-1 and Jagged-2) in freshly sorted mesenchymal stem cells (MSC) (B), osteolineage cells (OLC) (C) and endothelial cells (EC) (D) from chimeric Ctrl and KO mice. (E) qRT-PCR analysis of Notch receptors (Notch1-4) in freshly sorted GFP+c-Kit+Gr-1− from chimeric Ctrl and KO mice. (F) Western blot showed a significant increase of cleaved Notch1 expression in leukemic stem cells (LSC) from chimeric KO mice compared to Ctrl mice. (G) qRT-PCR analysis of downstream genes (Dtx, Hes1, Hes5, Hey1 and Hey2) regulated by the Notch pathway in freshly sorted GFP+c-Kit+Gr-1− from chimeric Ctrl and KO mice. (B-E, G) Data represent the mean ± standard deviation from three independent experiments.*P<0.05, **P<0.01, ***P<0.001 (Student t test). (H-J) GFP+ leukemic cells were transplanted into chimeric Ctrl and KO recipient mice. Five days later, the mice were treated daily with vehicle (dimethylsulfoxide, DMSO) or γ-secretase inhibitor (DBZ) (2 mmol per kg body weight) for 10 days. The counts of white blood cells (WBC) (H), leukemic cells in peripheral blood (PB) (I), and L-GMP (IL-7R−Lin−GFP+c-KithiCD34+CD16/32hi) cells in bone marrow (BM) (J) are shown (n=4, two independent experiments). Column plots show the mean ± standard deviation. *P<0.05; **P<0.01, Student t test). (K) Survival curve of chimeric Ctrl and KO recipients treated with DMSO or DBZ (n=7-8, log-rank test).
Discussion
In the current study, we demonstrated that excision of the Twist1 gene from the BM microenvironment resulted in a significant decrease in the numbers of MSC and mature osteoblasts, and an increase in the number of EC. The expression of CXCL12, VCAM1 and SCF was reduced, while that of osteopontin was increased. These changes led to a marked impairment of HSC localization, self-renewal, quiescence and differentiation. By transplanting MLL-AF9 cells into the Twist1-deleted and control chimeric mice, we verified that Twist1 deletion resulted in accelerated development of leukemia, at least partially through Notch signaling (Figure 7). These results reveal the essential role of TWIST1 in supporting normal hematopoiesis and perturbing AML development.
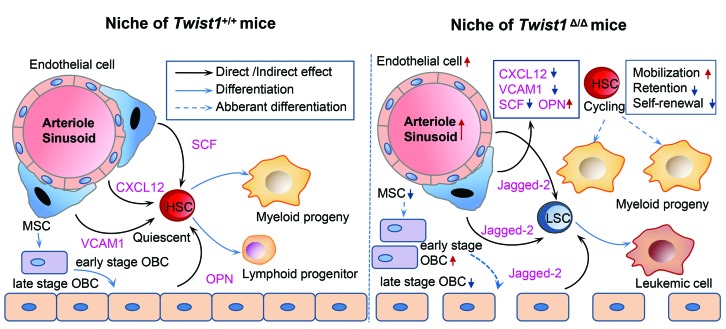
Figure 7.
Overview of the alterations in the bone marrow niche of Twist1-deleted mice. Simplified scheme of the normal hematopoietic stem cell (HSC) niche and its alterations in the Twist1-deleted mice. The left panel shows that HSC are around arterioles, sinusoids and endosteum where factors such as C-X-C motif chemokine ligand 12 (CXCL12), vascular cell adhesion molecule1 (VCAM1), stem cell factor (SCF) and osteopontin (OPN) secreted by mesenchymal stem cells (MSC), endothelial cells and osteoblastic cells (OBC) influence their self-renewal, quiescence, retention and differentiation. The right panel summarizes alterations of the niche, HSC and leukemic stem cells (LSC) observed in Twist1-deleted mice.
In our model, Twist1 deletion in the BM microenvironment leads to an increased number of EC and microvessel density, suggesting the existence of an indirect and powerful mechanism for promoting angiogenesis in vivo. Ohki et al. reported that G-CSF can markedly increase vascular endothelial growth factor (VEGF) release from G-CSF-responsive myelomonocytic cells, which promote the co-recruitment of VEGFR1+ (VEGF receptor 1) cells contributing to neo-angiogenesis.43 Since we have found elevated G- CSF secretion in the BM supernatant of Twist1-deleted mice, we then determined VEGF mRNA expression in BM cells and its protein concentration in BM supernatant. As expected, the results revealed a marked increase of VEGF production in Twist1-deleted mice compared with control mice (Online Supplementary Figure S8A,B). These observations provide a possible explanation for the EC alterations, i.e., Twist1 deficiency in the BM microenvironment leads to increased production of G-CSF, which in turn induces the secretion of VEGF, exerting a promotive effect on the proliferation of EC. This effect overrides the direct inhibitory role of Twist1 deletion on EC, and results in increased numbers of EC. As VEGF can be produced by various cell types, the specific mechanism needs further investigation.
Our understanding of niche contributions to AML has increased tremendously over the past decade. However, most studies have focused on how the leukemic cells actively shape their microenvironment to reinforce disease progression. There are a limited number of reports showing that certain niche alterations can act as a driver of AML initiation or progression, without having been educated by leukemic cells.40,44,45 Our present study demonstrates that environmental deletion of Twist1, a conserved transcriptional factor gene, results in diverse cellular and factor alterations common to the microenvironmental dysregulation exhibited by AML. These alterations appear to be predis posing or initiating factors for the evolution of AML and point to TWIST1 as an instructive signal that alters the function of the niche.
The opposing effects of TWIST1 on normal HSC and LSC found in this study are of value. In an effort to elucidate the underlying mechanism, we performed RNA-sequencing and quantitative real-time polymerase chain reaction analysis. We found that Twist1 deletion leads to increased expression of the Notch ligand Jagged-2 in all the OLC, EC and MSC. LSC from Twist1-deleted chimeric mice have robust expression of all Notch receptors and canonical downstream Notch target genes, suggesting the aberrant activation of Notch signaling. A previous report showed that Notch activation promotes expansion and self-renewal of LSC,40 consistent with our results obtained by deletion of Twist1. We also found activation of Notch receptors and target genes in normal HSC (CD34−LSK) after Twist1 deletion (Online Supplementary Figure S9). In contrast to the promoting role in LSC, Notch activation in HSC has been reported to cause loss of stem cell quiescence,46 which often correlates with impaired self-renewal capacity of HSC, in line with the observations in our mouse model. Besides the direct impact of activated Notch signaling on LSC and HSC, the augmented proliferation and infiltration of LSC compared to normal HSC could be favorable for their competition for the niche over HSC. Various studies have demonstrated that LSC could positively remodel the BM microenvironment to enhance support of LSC at the expense of HSC,47,48 and this remodeling may in turn further promote leukemia progression and impair normal hematopoiesis. In addition, the reduced expression of Cxcl12, Scf and Angpt1 in Twist1-deleted mice may also account for the opposing impact of Twist1 deletion on HSC and LSC, since compared with HSC, LSC are less factor-dependent.34,45,49
In consideration of the important role of TWIST1 in reg ulating MSC, osteoblasts and EC, and to exclude the interference of hematopoietic cells, which were found to express Twist1 in our previous work,50 we generated the chimeric mouse model, in which Twist1 was diffusely deleted in the BM microenvironment. The BM niche comprises multiple cell types, which not only closely connect but also communicate with each other via cell factors and adhesion molecules throughout the BM. Due to the complexity of the niche, an overall environmental knockout strategy will facilitate the detection of direct and indirect effects of TWIST1 on the niche components. Utilizing our model, we uncovered extensive cellular and factor alterations in the BM niche and the AML-like microenvironmental phenotype resulting from Twist1 deficiency, and demonstrated the essential role of TWIST1 in HSC maintenance and suppression of AML evolution. To refine the contribution of different cell populations, studies in which Twist1 is modified in specific stromal cell subsets are ongoing in our laboratory.
In conclusion, we used a Twist1-deficient chimera model to obtain, for the first time in vivo, direct evidence that TWIST1 in the microenvironment plays a key role in maintaining the hematopoietic phenotype and hampering leukemia progression. These findings provide new insights into the importance of the BM niche for AML development, and lay the foundation for tackling leukemia from a different angle to improve current treatments.
Supplementary Material
Acknowledgments
This work was supported by grants from The National Key Research and Development Program of China (2016YFA0100603), CAMS Initiative for Innovative Medicine (2016-I2M-1-017), National Natural Science Foundation of China (81470278, 81670158, 81600138, and 81700106), and Tianjin Municipal Science and Technology Commission grants (17JCZDJC35100 and 17JCQNJC10800).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/12/1969
References
- 1.Mendez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425(6960):836–841. [DOI] [PubMed] [Google Scholar]
- 3.Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–161. [DOI] [PubMed] [Google Scholar]
- 4.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–988. [DOI] [PubMed] [Google Scholar]
- 7.Greenbaum A, Hsu YM, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495(7440):227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stier S, Ko Y, Forkert R, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201(11):1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng H, Hao S, Liu Y, et al. Leukemic marrow infiltration reveals a novel role for Egr3 as a potent inhibitor of normal hematopoietic stem cell proliferation. Blood. 2015;126(11):1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause DS, Fulzele K, Catic A, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19(11):1513–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15(8):3439–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verzi MP, Anderson JP, Dodou E, et al. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol. 2002;249(1):174–190. [DOI] [PubMed] [Google Scholar]
- 13.Chen ZF, Behringer RR. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9(6):686–699. [DOI] [PubMed] [Google Scholar]
- 14.Isenmann S, Arthur A, Zannettino AC, et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27(10):2457–2468. [DOI] [PubMed] [Google Scholar]
- 15.Goodnough LH, Chang AT, Treloar C, Yang J, Scacheri PC, Atit RP. Twist1 mediates repression of chondrogenesis by beta-catenin to promote cranial bone progenitor specification. Development. 2012;139(23): 4428–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjiantoniou E, Iseki S, Uney JB, Phylactou LA. DNazyme-mediated cleavage of Twist transcripts and increase in cellular apoptosis. Biochem Biophys Res Commun. 2003;300(1):178–181. [DOI] [PubMed] [Google Scholar]
- 17.Howard TD, Paznekas WA, Green ED, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15(1):36–41. [DOI] [PubMed] [Google Scholar]
- 18.Mahmoud MM, Kim HR, Xing R, et al. TWIST1 integrates endothelial responses to flow in vascular dysfunction and atherosclerosis. Circ Res. 2016;119(3):450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mammoto T, Jiang A, Jiang E, Mammoto A. Role of Twist1 phosphorylation in angio-genesis and pulmonary fibrosis. Am J Respir Cell Mol Biol. 2016;55(5):633–644. [DOI] [PubMed] [Google Scholar]
- 20.Arthur A, Cakouros D, Cooper L, et al. Twist-1 enhances bone marrow mesenchymal stromal cell support of hematopoiesis by modulating CXCL12 expression. Stem Cells. 2016;34(2):504–509. [DOI] [PubMed] [Google Scholar]
- 21.Schreck C, Istvanffy R, Ziegenhain C, et al. Niche WNT5A regulates the actin cytoskeleton during regeneration of hematopoietic stem cells. J Exp Med. 2017;214(1):165–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saez B, Ferraro F, Yusuf RZ, et al. Inhibiting stromal cell heparan sulfate synthesis improves stem cell mobilization and enables engraftment without cytotoxic conditioning. Blood. 2014;124(19):2937–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinho S, Lacombe J, Hanoun M, et al. PDGFRalpha and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med. 2013;210(7):1351–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura Y, Arai F, Iwasaki H, et al. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood. 2010;116(9):1422–1432. [DOI] [PubMed] [Google Scholar]
- 25.Kunisaki Y, Bruns I, Scheiermann C, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502(7473):637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acar M, Kocherlakota KS, Murphy MM, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526(7571):126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciuculescu MF, Park SY, Canty K, Mathieu R, Silberstein LE, Williams DA. Perivascular deletion of murine Rac reverses the ratio of marrow arterioles and sinusoid vessels and alters hematopoiesis in vivo. Blood. 2015;125(20):3105–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balderman SR, Li AJ, Hoffman CM, et al. Targeting of the bone marrow microenvironment improves outcome in a murine model of myelodysplastic syndrome. Blood. 2016;127(5):616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratajczak MZ. A novel view of the adult bone marrow stem cell hierarchy and stem cell trafficking. Leukemia. 2015;29(4):776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19(2):257–267. [DOI] [PubMed] [Google Scholar]
- 31.Taichman RS, Emerson SG. Human osteoblasts support hematopoiesis through the production of granulocyte colony-stimulating factor. J Exp Med. 1994;179(5):1677–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayani H. A glance into somatic stem cell biology: basic principles, new concepts, and clinical relevance. Arch Med Res. 2003;34(1):3–15. [DOI] [PubMed] [Google Scholar]
- 33.Bowers M, Zhang B, Ho Y, Agarwal P, Chen CC, Bhatia R. Osteoblast ablation reduces normal long-term hematopoietic stem cell self-renewal but accelerates leukemia development. Blood. 2015;125(17):2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanoun M, Zhang D, Mizoguchi T, et al. Acute myelogenous leukemia-induced sympathetic neuropathy promotes malignancy in an altered hematopoietic stem cell niche. Cell Stem Cell. 2014;15(3):365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen HY, Ting MC, Maxson RE. Jagged1 functions downstream of Twist1 in the specification of the coronal suture and the formation of a boundary between osteogenic and non-osteogenic cells. Dev Biol. 2010;347(2):258–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamath BM, Stolle C, Bason L, et al. Craniosynostosis in Alagille syndrome. Am J Med Genet. 2002;112(2):176–180. [DOI] [PubMed] [Google Scholar]
- 39.Kode A, Mosialou I, Manavalan SJ, et al. Foxo1-dependent induction of acute myeloid leukemia by osteoblasts in mice. Leukemia. 2016;30(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kode A, Manavalan JS, Mosialou I, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506(7487):240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Zhang H, Rodriguez S, et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-kappaB-dependent manner. Cell Stem Cell. 2014;15(1):51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YW, Koo BK, Jeong HW, et al. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008;112(12):4628–4638. [DOI] [PubMed] [Google Scholar]
- 43.Ohki Y, Heissig B, Sato Y, et al. Granulocyte colony-stimulating factor promotes neovascularization by releasing vascular endothelial growth factor from neutrophils. FASEB J. 2005;19(14):2005–2007. [DOI] [PubMed] [Google Scholar]
- 44.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464(7290):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zambetti NA, Ping Z, Chen S, et al. Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre-leukemia. Cell Stem Cell. 2016;19(5):613–627. [DOI] [PubMed] [Google Scholar]
- 46.Chiang MY, Shestova O, Xu L, Aster JC, Pear WS. Divergent effects of supraphysiologic Notch signals on leukemia stem cells and hematopoietic stem cells. Blood. 2013;121(6):905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernasconi P, Farina M, Boni M, Dambruoso I, Calvello C. Therapeutically targeting SELF-reinforcing leukemic niches in acute myeloid leukemia: a worthy endeavor? Am J Hematol. 2016;91(5):507–517. [DOI] [PubMed] [Google Scholar]
- 48.Huang MM, Zhu J. The regulation of normal and leukemic hematopoietic stem cells by niches. Cancer Microenviron. 2012;5(3):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S, Zambetti NA, Bindels EM, et al. Massive parallel RNA sequencing of highly purified mesenchymal elements in low-risk MDS reveals tissue-context-dependent activation of inflammatory programs. Leukemia. 2016;30(9):1938–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong CY, Liu XY, Wang N, et al. Twist-1, a novel regulator of hematopoietic stem cell self-renewal and myeloid lineage development. Stem Cells. 2014;32(12):3173–3182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.