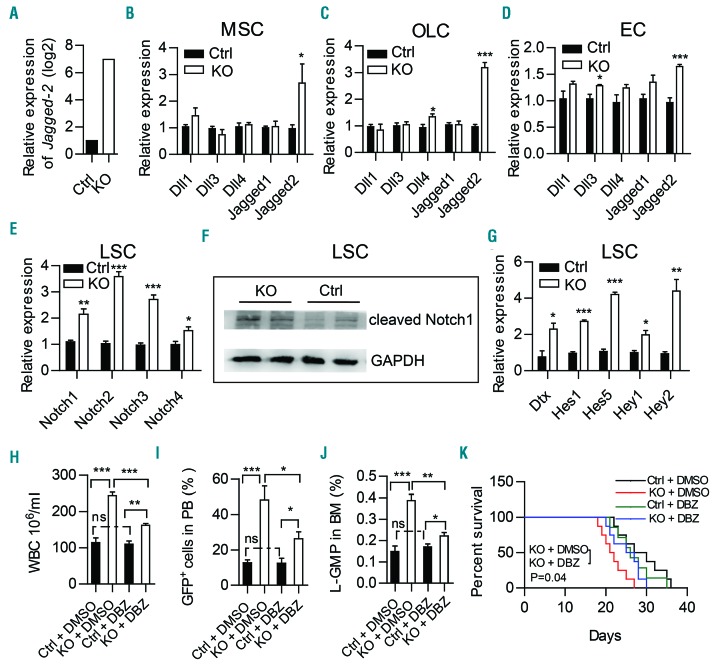
Figure 6.
Notch signaling is activated in leukemic stem cells and inhibition of Notch signaling partially rescues MLL-AF9-induced acute myeloid leukemia progression in Twist1-deficient mice. Expression of Jagged-2 in RNA-sequencing analysis of stromal cells (CD45−Ter119−) from chimeric control (Ctrl) and knockout (KO) mice. (B-D) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of Notch ligands (Dll1, Dll3, Dll4, Jagged-1 and Jagged-2) in freshly sorted mesenchymal stem cells (MSC) (B), osteolineage cells (OLC) (C) and endothelial cells (EC) (D) from chimeric Ctrl and KO mice. (E) qRT-PCR analysis of Notch receptors (Notch1-4) in freshly sorted GFP+c-Kit+Gr-1− from chimeric Ctrl and KO mice. (F) Western blot showed a significant increase of cleaved Notch1 expression in leukemic stem cells (LSC) from chimeric KO mice compared to Ctrl mice. (G) qRT-PCR analysis of downstream genes (Dtx, Hes1, Hes5, Hey1 and Hey2) regulated by the Notch pathway in freshly sorted GFP+c-Kit+Gr-1− from chimeric Ctrl and KO mice. (B-E, G) Data represent the mean ± standard deviation from three independent experiments.*P<0.05, **P<0.01, ***P<0.001 (Student t test). (H-J) GFP+ leukemic cells were transplanted into chimeric Ctrl and KO recipient mice. Five days later, the mice were treated daily with vehicle (dimethylsulfoxide, DMSO) or γ-secretase inhibitor (DBZ) (2 mmol per kg body weight) for 10 days. The counts of white blood cells (WBC) (H), leukemic cells in peripheral blood (PB) (I), and L-GMP (IL-7R−Lin−GFP+c-KithiCD34+CD16/32hi) cells in bone marrow (BM) (J) are shown (n=4, two independent experiments). Column plots show the mean ± standard deviation. *P<0.05; **P<0.01, Student t test). (K) Survival curve of chimeric Ctrl and KO recipients treated with DMSO or DBZ (n=7-8, log-rank test).