t(4;11) MLL-AF4 pro-B acute lymphoblastic leukemia (ALL) is an aggressive hematologic malignancy that accounts for 50–85% of infant ALL cases.1 Retrospective analysis of Guthrie cards and twin concordance studies both confirmed the pre-natal origin of this disease.2 Patients show an accumulation of immature pro-B cells in the bone marrow, followed by a rapid and uncontrolled proliferation of leukemia blasts that hijack the immune system and invade peripheral organs such as the spleen, liver and central nervous system. The chromosomal translocation results in the fusion of the N-terminal part of MLL with almost the entire AF4 gene, which disrupts the epigenetic signature of hematopoietic cells.3 This induces a stem cell-like expression signature (e.g. HOXA cluster, MEIS1, RUNX1) as well as a pro-survival and proliferation phenotype (characterized by the upregulation of BCL2, MCL1, CDK6).4–8
The molecular signature at diagnosis has been well characterized, but there is no information on the initial changes during the first stages of leukemogenesis due to the challenge posed by the pre-natal origin of the disease. We recently described a pre-leukemia model of t(4;11) MLL-AF4 infant leukemia, which uses an Mll-AF4 invertor line and a VE-Cadherin-driven Cre recombinase to target the expression of Mll-AF4 to all definitive hematopoietic cells formed during embryonic development.9 While this model does not progress to the same rapid, acute leukemia phenotype observed in human patients, possibly due to species differences,10 it provides unique access to the prenatal pre-leukemic state in vivo. Mll-AF4 expression was shown to lead to increased engraftment and self-renewal potential of E14 fetal liver (FL) cells, as well as a high B-lymphoid clonogenic potential; however, the precise contribution of individual cell types was not addressed in detail.
Here, we separated the hematopoietic compartment into three stem/progenitor fractions, hematopoietic stem cells/multipotent progenitors (HSCs/MPPs), lymphoid-primed multipotent progenitors (LMPPs) and Lin−ckit+/common lymphoid progenitor cells (LK/CLPs), with a sorting strategy adapted to the fetal context (Online Supplementary Figure S1A),11 and used transplantation assays and gene expression analysis to further characterize the cell-of-origin of t(4;11) MLL-AF4 pro-B ALL. Details on materials and methods can be found in the Online Supplementary Appendix. Analysis of cell cycle distribution showed that the HSC/MPP population was more highly represented in the G0-G1 phase compared to LMPP and LK/CLP (Online Supplementary Figure S1B), and less in the G2/M phase (Online Supplementary Figure S1C). Mll-AF4 did not alter the cell cycle distribution of HSC/MPPs, LMPPs or LK/CLPs, suggesting that proliferation is not hijacked during early stages of leukemogenesis.
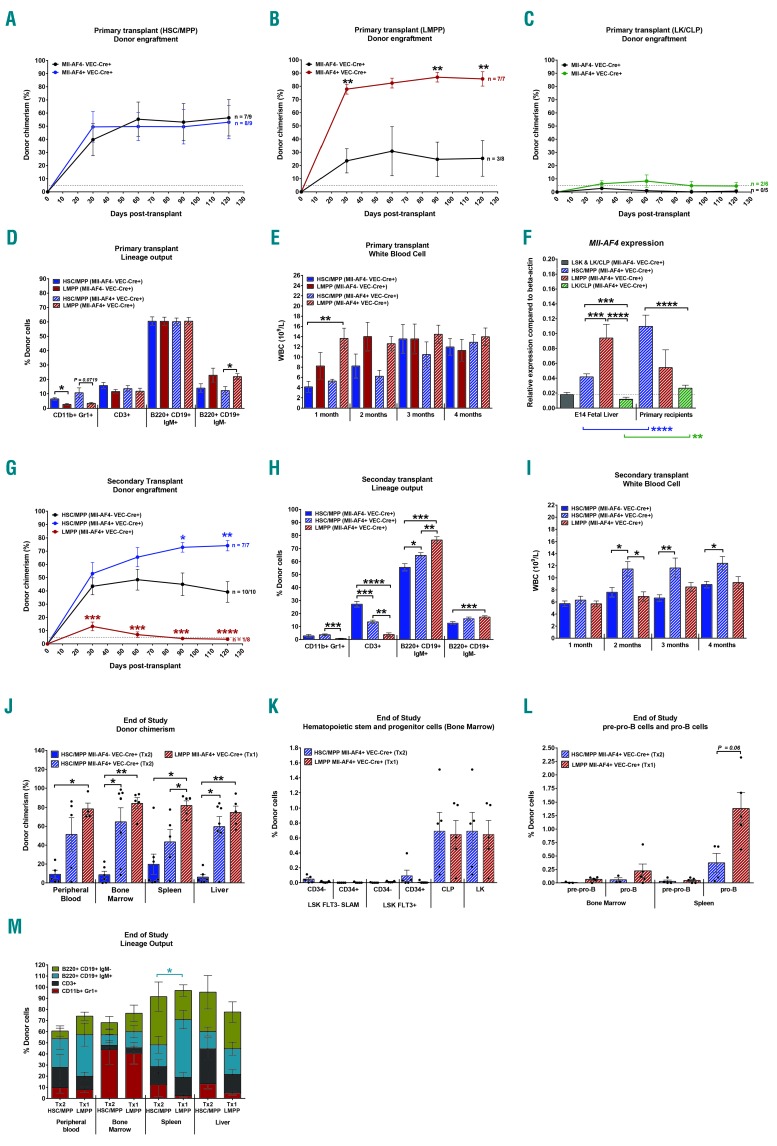
All three fractions were transplanted to assess their engraftment, self-renewal and differentiation potentials. Mll-AF4 did not affect the engraftment of HSC/MPPs or LK/CLPs, but led to a significantly higher engraftment of the LMPP fraction (Figure 1A-C). We had previously found that HSC/MPPs and LMPPs from E14 FL could form B-lymphoid colonies with a pro-B phenotype when Mll-AF4 is expressed.9 We therefore assessed the lineage output in the peripheral blood of the primary recipients. While there was no difference in T (CD3+) and mature B-cell (B220+CD19+IgM+) production, LMPPs had a lower myeloid (CD11b+Gr1+) and higher immature B-cell (B220+CD19+IgM−) output compared with HSC/MPPs (Figure 1D). This skewing was, however, independent of Mll-AF4 expression and therefore represents an intrinsic property of LMPPs. Mll-AF4+ LMPP primary recipients had the highest white blood cell count one month after transplant, suggesting a faster contribution to the hematopoietic system (Figure 1E); however, this difference diminished over time. The expression of Mll-AF4 is significantly higher in E14 fetal liver LMPPs compared to HSC/MPPs and LK/CLPs, which may offer an explanation for their enhanced engraftment in primary recipients (Figure 1F, left set of graphs).
Figure 1.
In the presence of Mll-AF4, E14 fetal liver (FL) lymphoid-primed multipotent progenitor cells (LMPPs) display higher engraftment potential and a B-lymphoid bias. (A-C) Primary transplant of 1000 hematopoietic stem cells/multipotent progenitor cells (HSC/MPPs) (A), 750 LMPPs (B) and 1000 lineage-ckit+/common lymphoid progenitor cells (LK/CLPs) (C). Total donor chimerism in peripheral blood is shown and dotted line represents 5% threshold for considering mice as being repopulated. Repopulated mice/total injected shown next to the curve. (D) Donor chimerism in individual lineages of primary recipients four months after transplant. (E) White blood cell count in the peripheral blood of primary recipients. (F) Quantitative PCR of Mll-AF4 in fresh fetal liver HSC/MPP, LMPP and LK/CLP and sorted cells from primary recipients. (G) Secondary transplant of HSC/MPP and LMPP-derived bone marrow cells from primary recipients. (H) Donor chimerism in individual lineages of secondary recipients four months after transplant. (I) White blood cell count in the peripheral blood of secondary recipients. (J) Donor chimerism in the peripheral blood, bone marrow, spleen and liver at end of study. (K) Donor-derived HSCs, LMPPs, CLP and LK in the bone marrow at end of study. (L) Donor-derived precursor-progenitor lymphoid B cell (pre-pro-B) and progenitor B (pro-B) cells in the bone marrow and spleen at end of study. (M) Donor chimerism in individual lineages at end of study. A non-parametric Mann-Whitney test was used to compare datasets with a significance cut-off of P<0.05 (*), P<0.01 (**), P<0.001 (***) or P 0.0001 (****).
We then assessed the self-renewal potential of HSC/MPPs and LMPPs in secondary transplantations. Mll-AF4 expression increased the repopulation of secondary recipients with HSC/MPPs compared with the Mll-AF4- control (Figure 1G), although engraftment levels were not significantly different from those observed with Mll-AF4+ HSC/MPPs in primary recipients (Online Supplementary Figure S1D). Mll-AF4+ LMPPs, on the other hand, only showed limited self-renewal (Figure 1G), suggesting that Mll-AF4 can only enhance self-renewal in cells that already possess this property. Furthermore, we detected an upregulation of Mll-AF4 expression following transplantation in HSC/MPPs sorted from primary recipients, while it remained unchanged in LMPPs, which may also contribute to the higher self-renewal potential of Mll-AF4+ HSC/MPPs (Figure 1F, right set of graphs). Both populations displayed skewing towards the B lineage in the presence of Mll-AF4 in secondary recipients, which, in the case of LMPPs, resulted in an almost entirely B-lymphoid-biased output (Figure 1H). The white blood cell count was significantly higher in Mll-AF4+ HSC/MPP secondary recipients from two months after transplantation (Figure 1I).
We did a post-mortem analysis of HSC/MPP secondary recipients and LMPP primary recipients (6-15 months old). As shown above, hematopoietic progenitors such as LMPPs, can only engraft primary recipients, whereas HSCs can serially engraft. Therefore, we compared LMPP primary recipients and HSC/MPP secondary recipients to assess differences in hematopoietic output established by HSCs and LMPPs. Mll-AF4+ HSC/MPPs and LMPPs showed a significantly higher engraftment in the bone marrow and liver compared to Mll-AF4− HSC/MPPs (Figure 1J). Furthermore, LMPP primary recipients had more donor cells in the spleen compared to Mll-AF4+ HSC/MPP recipients. While there were no significant differences in the stem/progenitor compartment in the bone marrow (Figure 1K), Mll-AF4+ LMPP primary recipients displayed a trend towards more pro-B cells in the spleen compared to Mll-AF4+ HSC/MPP secondary recipients (Figure 1L), which resulted in a higher proportion of mature B220+CD19+IgM+ cells (Figure 1M).
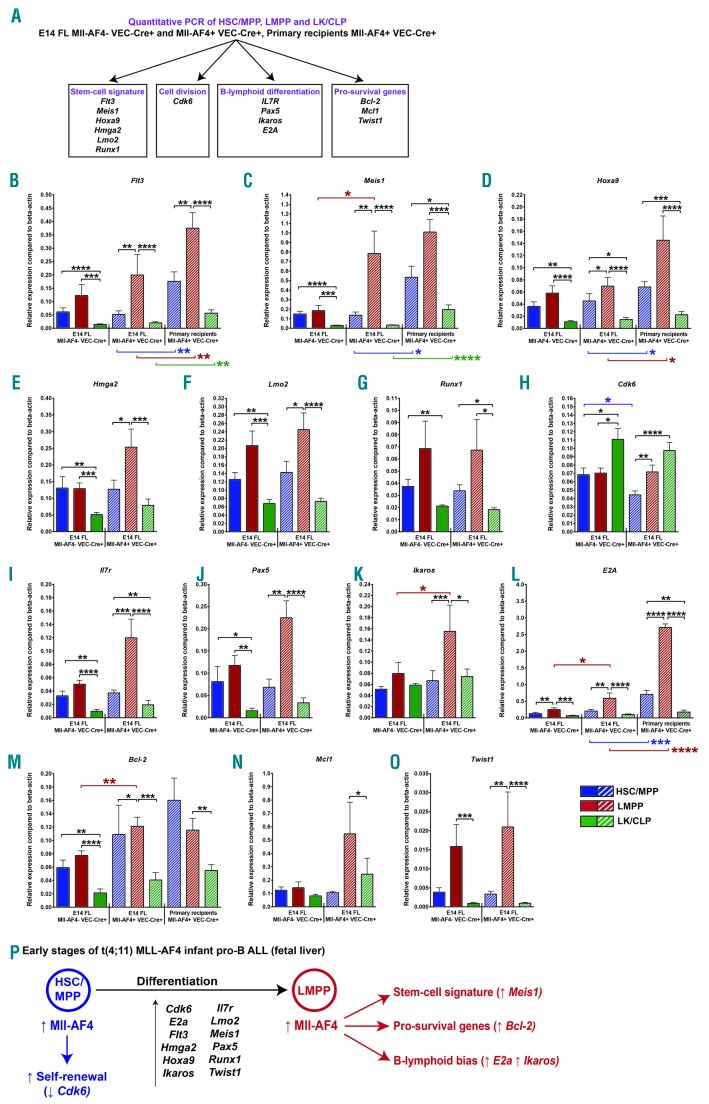
To explain the cell type-specific effects, we assessed the expression of 14 genes associated with t(4;11) MLL-AF4 pro-B ALL and, more specifically, linked to HSC signature, B-lymphoid differentiation, cell division and pro-survival phenotypes: Flt3,12 Meis1,4 Hoxa9,4 Hmga2,13 Lmo2,14 Runx1,5 Cdk6,8,15 Il7r,16 Pax5,16 Ikaros,16 E2a,16 Bcl-2,7 Mcl1,7 and Twist16 (Figure 2A). Flt3 is up-regulated in LMPPs regardless of Mll-AF4 status (Figure 2B), while Meis1 is up-regulated specifically in response to Mll-AF4 expression in LMPPs (Figure 2C). The expression of Hoxa9 in Mll-AF4+ LMPPs is significantly higher compared with Mll-AF4+ HSC/MPPs and/or Mll-AF4+ LK/CLPs (Figure 2D). Hmga2, Lmo2 and Runx1 expression in Mll-AF4+ LMPPs is significantly higher compared with Mll-AF4+ HSC/MPPs and/or Mll-AF4+ LK/CLPs (Figure 2E-G). Cdk6 expression was higher in LK/CLPs, and decreased in HSC/MPPs upon Mll-AF4 expression (Figure 2H). This can partly explain the enhanced self-renewal of Mll-AF4+ HSC/MPPs compared to Mll-AF4− HSC/MPPs (Figure 1G). The expression of IL7r and Pax5 was generally higher in LMPPs, but was not significantly affected by Mll-AF4 expression, although there was a clear trend (Figure 2I and J), whereas Ikaros and E2a were strongly up-regulated in Mll-AF4+ LMPPs compared to Mll-AF4− LMPPs and Mll-AF4+ HSC/MPPs and LK/CLPs (Figure 2K and L). This likely explains the strong B-lymphoid bias observed in the transplant recipients (Figure 1H). We observed a significant upregulation of Bcl-2 in Mll-AF4+ LMPPs compared to Mll-AF4− LMPPs (Figure 2M), which is a direct transcriptional target of MLL-AF4.7 Mcl1 expression was relatively stable (Figure 2N), but Twist1 was up-regulated in Mll-AF4+ LMPP compared to HSC/MPP and LK/CLP Mll-AF4+ cells (Figure 2O). We assessed the expression of Flt3, Meis1, Hoxa9, E2a and Bcl-2 in sorted Mll-AF4+ HSC/MPPs, LMPPs and LK/CLPs from primary recipients to measure expression changes induced by transplantation stress and a change in microenvironment: FL versus bone marrow (Figure 2B-D, L and M). The relative expression pattern amongst the three populations was similar in freshly sorted FL cells and sorted cells from primary recipients. However, there is a general upregulation of Flt3, Meis1, Hoxa9 and E2a in cells from primary recipients (Figure 2B-D, and L), which may explain the shorter disease latency following transplantation observed previously.9 The strong upregulation of E2a also likely explains the strong B-lymphoid bias observed in LMPP recipients (Figure 1H). This study suggests that the FL LMPP sets the stage for the transformation process of t(4;11) MLL-AF4 infant pro-B ALL through the higher expression of Flt3, Hoxa9, Lmo2, Runx1, Il7r, Pax5, E2a and Twist1 (Figure 2P). The activation of Mll-AF4 increases the expression of E2a, Ikaros, Meis1 and Bcl-2, leading to a strong B-lymphoid bias and a survival advantage. This study is a step forward towards understanding the molecular mechanisms of infant leukemogenesis, and also further supports our previous proposition that the FL LMPP is the cell-of-origin of t(4;11) Mll-AF4 pro-B ALL.
Figure 2.
The lymphoid-primed multipotent progenitor cell (LMPP) population displays an MLL-AF4 gene expression signature. (A) Quantitative PCR strategy in fresh fetal liver cells (n = 6-7) and cells derived from primary recipients (n = 3). Quantitative PCR of (B) Flt3, (C) Meis1, (D) Hoxa9, (E) Hmga2, (F) Lmo2, (G) Runx1, (H) Cdk6, (I) Il7r, (J) Pax5, (K) Ikaros, (L) E2a, (M) Bcl-2, (N) Mcl1, (O) Twist1. (P) Early stages of t(4;11) MLL-AF4 infant leukemia based on the pre-leukemia mouse model. A non-parametric Mann-Whitney test was used to compare datasets with a significance cut-off of P<0.05 (*), P<0.01 (**), P<0.001 (***) or P<0.0001(****).
Supplementary Material
Acknowledgments
The authors would like to thank Fiona Rossi and Claire Cryer from the SCRM FACS Facility for cell sorting services and flow cytometry advice and would like to acknowledge the support of Andrew Dyer, Hollie McGrath, Linda Dunn-Campbell and John Agnew from the SCRM Animal Facility in animal experimentation. Core facilities at the Edinburgh MRC Centre for Regenerative Medicine are supported by center grant MR/K017047/1.
Footnotes
Funding: this study was supported by a Bloodwise Bennett Senior Fellowship (KO) and a grant from the Kay Kendall Leukaemia Fund and Cancer Research UK (KO).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Biondi A, Cimino G, Pieters R, Pui C-H. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96(1):24–33. [PubMed] [Google Scholar]
- 2.Gale KB, Ford AM, Repp R, et al. Backtracking leukemia to birth: Identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci USA. 1997;94(25):13950–13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 Methylation Profiles Define Murine and Human MLL-AF4 Leukemias. Cancer Cell. 2008;14(5):355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL Amino Terminal Sequences with Menin Is Required for Transformation. Cancer Res. 2007;67(15):7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson Adam C, Ballabio E, Geng H, et al. RUNX1 Is a Key Target in t(4;11) Leukemias that Contributes to Gene Activation through an AF4-MLL Complex Interaction. Cell Rep. 2013;3(1):116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22(24):3403–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benito Juliana M, Godfrey L, Kojima K, et al. MLL-Rearranged Acute Lymphoblastic Leukemias Activate BCL-2 through H3K79 Methylation and Are Sensitive to the BCL-2-Specific Antagonist ABT-199. Cell Rep. 2015;13(12):2715–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Linden M, Willekes M, van Roon E, et al. MLL fusion-driven activation of CDK6 potentiates proliferation in MLL-rearranged infant ALL. Cell Cycle. 2014;13(5):834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett Neil A, Malouf C, Kapeni C, et al. Mll-AF4 Confers enhanced self-renewal and lymphoid potential during a restricted window in development. Cell Rep. 2016;16(4):1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S, Luo Roger T, Ptasinska A, et al. Instructive Role of MLL-Fusion Proteins Revealed by a Model of t(4;11) Pro-B Acute Lymphoblastic Leukemia. Cancer Cell. 2016;30(5):737–749. [DOI] [PubMed] [Google Scholar]
- 11.Böiers C, Carrelha J, Lutteropp M, et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 2013;13(5):535–548. [DOI] [PubMed] [Google Scholar]
- 12.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106(1):254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, Eguchi-Ishimae M, Yagi C, et al. HMGA2 as a potential molecular target in KMT2A-AFF1-positive infant acute lymphoblastic leukaemia. Br J Haematol. 2015;171(5):818–829. [DOI] [PubMed] [Google Scholar]
- 14.Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci USA. 1998;95(7):3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurenti E, Frelin C, Xie S, et al. CDK6 Levels Regulate Quiescence Exit in Human Hematopoietic Stem Cells. Cell Stem Cell. 2015;16(3):302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy RR, Kincade PW, Dorshkind K. The Protean Nature of Cells in the B Lymphocyte Lineage. Immunity. 2007;26(6):703–714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.