Abstract
Aberrant changes in microRNA expression contribute to lymphomagenesis. Bromodomain and extra-terminal domain inhibitors such as OTX015 (MK-8628, birabresib) have demonstrated preclinical and clinical activity in hematologic tumors. MicroRNA profiling of diffuse large B-cell lymphoma cells treated with OTX015 revealed changes in the expression levels of a limited number of microRNAs, including miR-92a-1-5p, miR-21-3p, miR-155-5p and miR-96-5p. Analysis of publicly available chromatin immunoprecipitation sequencing data of diffuse large B-cell lymphoma cells treated with bromodomain and extra-terminal domain (BET) inhibitors showed that the BET family member BRD4 bound to the upstream regulatory regions of multiple microRNA genes and that this binding decreased following BET inhibition. Alignment of our microRNA profiling data with the BRD4 chromatin immunoprecipitation sequencing data revealed that microRNAs downregulated by OTX015 also exhibited reduced BRD4 binding in their promoter regions following treatment with another bromodomain and extra-terminal domain inhibitor, JQ1, indicating that BRD4 contributes directly to microRNA expression in lymphoma. Treatment with bromodomain and extra-terminal domain inhibitors also decreased the expression of the arginine methyltransferase PRMT5, which plays a crucial role in B-cell transformation and negatively modulates the transcription of miR-96-5p. The data presented here indicate that in addition to previously observed effects on the expression of coding genes, bromodomain and extra-terminal domain inhibitors also modulate the expression of microRNAs involved in lymphomagenesis.
Introduction
The important role of non-coding elements of the genome, specifically microRNAs (miRNAs), in mediating cellular transformation was first demonstrated in chronic lymphocytic leukemia.1 Since then, numerous miRNAs have been shown to function as tumor suppressors or oncogenes in both hematologic and solid tumors.2–8 miRNAs are short sequences of 19 – 25 nucleotides that function as part of an RNA-induced silencing complex (RISC).9 In humans, they function primarily by destabilizing messenger RNA (mRNA) and inhibiting the translation of mRNA into protein. This is achieved through binding of the 5ʹ seed region of a miRNA to its recognition sequence in the 3ʹ untranslated region of its target gene.10 A single miRNA can recognize multiple target genes and, conversely, different miRNAs can target an individual gene.11 Thus, in the context of cancer, miRNAs can intricately and markedly influence individual driver genes and entire signaling pathways crucial to the survival of cancer cells. Furthermore, a number of miRNAs have been shown to participate in a feedback loop with the protein product of their target gene.11
Diffuse large B-cell lymphoma (DLBCL) is an aggressive lymphoma that accounts for approximately 35-40% of all lymphoma cases.12 DLBCL frequently harbors mutations in chromatin-modifying enzymes indicating that perturbation of epigenetic regulation is an important trigger for B-cell transformation.13,14 A class of epigenetic drugs that has recently shown promising results in pre-clinical and clinical settings, and particularly in DLBCL, inhibits members of the bromodomain and extra-terminal domain (BET) protein family.15–25 In mammals, the BET family comprises four proteins, BRD2, BRD3, BRD4 and BRDT, which all share two highly conserved N-terminal bromodomains (BRD) and a C-terminal extra-terminal (ET) domain. BET proteins specifically bind to acetylated lysine residues via their dual BRD motifs, acting as epigenetic readers of acetyl-lysine marks. They therefore constitute an important component of the write-read-erase model via which epigenetic information is interpreted by cells.17 BET inhibitors act by preventing the interaction of BRD4 with acetylated histones.26 Here we show direct and indirect regulation of miRNA expression in DLBCL by a BET inhibitor.
Methods
Cell lines and molecules
Established human cell lines derived from DLBCL were cultured according to recommended conditions. Two germinal-center B-cell type DLBCL (GCB-DLBCL) cell lines, DOHH-2 and OCI-LY-1, were cultured in Roswell Park Memorial Institute medium and Iscove’s Modified Dulbecco’s Medium, respectively. The activated B-cell–like DLBCL (ABC-DLBCL) cell lines SU-DHL-2 and HBL-1 were cultured in Roswell Park Memorial Institute medium. Cell lines were obtained as previously described,27 and their identity was authenticated by short tandem repeat DNA profiling (IDEXX BioResearch, Ludwigsburg, Germany). All media were supplemented with fetal bovine serum (10%; DOHH-2 and OCI-LY-1 or 20%; SU-DHL-2 and HBL-1), penicillin-streptomycin-neomycin (5,000 units penicillin, 5 mg streptomycin and 10 mg neomycin/mL, Sigma) and L-glutamine (1%). OTX015 (MK-8628, birabresib) was provided by Oncoethix (Lausanne, Switzerland).
In vivo xenograft model
The xenograft model used here has been described elsewhere.15 Total RNA, previously extracted from these tumors, was used to analyze OTX015-mediated modulation of miRNA expression in vivo.
Western blotting analysis
Protein extractions, sodium dodecylsulfate polyacrylamide gel electrophoresis and immunoblotting were performed as previously described.15 The antibodies used were anti-PRMT5 (A1520; NeoBiolab), anti-GAPDH (9131; Cell Signaling) and anti-BRD4 (A301-985A; Bethyl).
One-step quantitative reverse transcription - polymerase chain reaction
Total RNA was extracted from cells treated with dimethyl sulfoxide (DMSO) or OTX015 using TRIzol (Thermo Scientific, Lausanne, Switzerland). One-step quantitative reverse transcription - polymerase chain reaction (qRT-PCR) was performed as previously described15 using 20 ng of RNA for each reaction. Forward and reverse primers used for quantification of PRMT5 mRNA were, respectively, 5ʹ-TCTCATGGTTTCCCATCCTC-3ʹ and 5ʹ-ACACAGATGGTTTGGCCTTC-3ʹ. Quantification of GAPDH expression served as an endogenous control. GAPDH primer sequences were, 5ʹ-CGACCACTTTGTCAAGCTCA-3ʹ (forward) and 5ʹ-CCCTGTTGCTGTAGCCAAAT-3ʹ (reverse). Expression of GAPDH was verified to be stable between the analyzed groups.
MicroRNA expression profiling
Total RNA was extracted as previously described.15 miRNA expression profiling was performed on RNA from DLBCL cell lines treated with DMSO or OTX015 using the Agilent Human microRNA microarray v. 3 or Nanostring nCounter Human V3A miRNA Expression Assay Kits. Profiling was done on RNA extracted from untreated lymphoma cell lines27,28 using the Nanostring nCounter Human V2. All samples were processed as previously described.29,30 Profiling data are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) database under the GEO project number GSE99208.
MicroRNA quantification with TaqMan microRNA assays
Profiling results for selected miRNAs were validated using the following TaqMan MicroRNA Assays (Applied Biosystems): hsa-miR-96-5p, assay ID: 000186; hsa-miR-92a-1-5p, assay ID: 002137, hsa-miR-21-3p, assay ID: 002438; hsa-miR-155-5p, assay ID: 002623; RNU6B, assay ID: 001093. Reverse transcription and quantitative PCR were performed using the TaqMan MicroRNA Reverse Transcription Kit and the TaqMan Universal PCR Master Mix according to the manufacturer’s instructions. Briefly, for each sample, 10 ng of total RNA was used for reverse transcription and 1.33 μL of the reverse transcription product was used in triplicate wells for the quantitative PCR (qPCR). All qPCR reactions were performed on an Applied Biosystems StepOnePlus System. Amplification of RNU6B served as a normalizing control for RNA quantity. Data were analyzed using the ΔΔCt method to obtain relative quantities. Expression of RNU6B was verified as stable between the analyzed groups.
Data mining
miRNA expression data obtained from each profiling platform were analyzed independently. For Agilent arrays, the hybridization signal values for the multiple probes were obtained using the Agilent Feature Extraction Software 10.7.3 (Agilent Technologies). For the Nanostring nCounter, raw expression data were log-transformed and normalized by the quantile method after application of manufacturer-supplied correction factors. For both platforms, differentially expressed miRNAs were defined using R/Bioconductor with the linear model for microarray data analysis (limma) with a contrast matrix for the comparisons of interest on the datasets filtered to exclude features below the detection threshold (defined for each sample by a cut-off corresponding to twice the standard deviation of negative control probes plus the means) in at least half of the samples. The transcripts bearing an absolute log-fold change greater than 0.2 and a P-value less than 0.05 at any experimental time point were defined as differentially expressed. Overlapping among lists was performed using the VENNY on-line tool.31 Experimentally validated transcript targets of the miRNAs were obtained using the MicroRNA Target Filter in Ingenuity Pathway Analysis (Qiagen). Functional annotation of the targets was performed with the Gene Set Enrichment Analysis tool for overlap analysis using the hallmarks and the c2.cp of the Molecular Signatures Database 5.232 and hypergeometric P-values after correction for multiple hypothesis testing according to Benjamini and Hochberg.
Publicly available chromatin immunoprecipitation (ChIP) sequencing datasets obtained by ChIP followed by high-throughput DNA sequencing were downloaded and re-analyzed. They comprised datasets obtained in the ABC-DLBCL cell line HBL-1 (SRP043524)23 and in the GCB-DLBCL cell line OCI-LY-1 (SRP022129),22 both treated with the BET inhibitor JQ1 or DMSO alone. Sequence reads obtained from ChIP fragments were aligned to the human reference genome hg19 using Bowtie, allowing up to one mismatch per fragment length. Redundant reads were removed and only reads uniquely mapping to the reference genome were used for further analysis. The peaks that were genomic regions enriched by ChIP, relative to the background reads, were detected using HOMER (v2.6), a suite of tools for Motif Discovery and next-generation sequencing analysis, with a default option (false discovery rate = 0.001 and Poisson P-value cut-off = 1e-04). Differential peaks were defined as having at least a four-fold difference in enrichment within a 200 bp region between the two conditions (DMSO versus JQ1) and a Poisson enrichment P-value less than 1e-04. All discovered putative peaks were ranked by their normalized tag counts (number of tags found at the peak, normalized to 10×106 total mapped tags) and annotated with the annotatePeaks.pl subroutine. We defined miRNA promoters using FANTOM533 and the precursors of microRNAs downloaded from miRBase (v20)34 to annotate the BRD4 ChIP sequencing datasets. We defined enriched regions located within 5 kb regions of predicted promoters and pre-miRNAs as candidate BRD4 binding sites. For global ChIP sequencing visualization, we used ngs.plot (https://code.google.com/p/ngsplot/) for inspection of both average and ‘laid out’ coverages as curves or heatmaps.
Chromatin immunoprecipitation
Cells (SU-DHL-2 and HBL-1) were cross-linked with 1% formaldehyde. Crosslinking was quenched with 125 mM glycine. Cells were washed with ice-cold phosphate-buffered saline containing 1 × HALT protease inhibitor (Thermo Scientific, Lausanne, Switzerland) and resuspended in sodium dodecylsulfate lysis buffer (ChIP Assay Kit, Millipore, Schaffhausen, Switzerland) before sonication using the Bioruptor Plus. For each immunoprecipitation reaction, chromatin from 1×106 cells was incubated overnight with anti-PRMT5 (A1520; NeoBiolab), anti-BRD4 (A301-985A; Bethyl) or 3 μg of the negative control antibody, anti-IgG (Millipore). Immune complexes were collected by incubation with 20 μL magnetic protein G beads at 4°C for 1.5 h. Protein G-bound complexes were sequentially washed with Low Salt Wash Buffer, High Salt Wash Buffer, LiCl Wash Buffer and twice with TE Buffer (ChIP Assay Kit, Millipore). Protein/DNA complexes were eluted using 1% sodium dodecylsulfate and 0.1 M NaHCO3. Following reversal of crosslinks (65°C overnight), samples were treated with RNAse A and then Proteinase K. DNA samples were purified using the QIAquick PCR purification kit (Qiagen, Hombrechtikon, Switzerland). Chromatin samples to which no antibody had been added were processed in parallel as input references. For qPCR analysis of ChIP samples, triplicate wells containing 1 μL of purified ChIP DNA plus PCR master mix were prepared. Reactions were performed on a StepOnePlus Real-Time PCR system (Applied Biosystems). Standard curves were constructed using sonicated and purified chromatin. ChIP-qPCR was performed using primers specific for the upstream regulatory regions of PRMT5 and miR-96-5p. Primer sequences were as follows: PRMT5 forward; 5ʹ-AGCGCGAGGAGAAAGATG-3ʹ, PRMT5 reverse; 5ʹ-CTATTTCGGGGACGCAATTC-3ʹ, miR-96 forward; 5ʹ-AGCTGGGAGACCTTGCTTC-3ʹ, miR-96 reverse; 5ʹ-TCACCCCTCCTAACCCAAAT-3ʹ.
Results
BET inhibition modulates the expression of a subset of microRNAs
We have previously shown that the BET inhibitor OTX015 modulates the expression of multiple coding transcripts in DLBCL cells.15 Here we assessed the effect of OTX015 on global miRNA expression. GCB-DLBCL OCI-LY-1 and ABC-DLBCL HBL-1 cells were treated with 500 nM OTX015 for 4 and 24 h. Total RNA isolated from vehicle- and OTX015-treated cells was interrogated with the Nanostring nCounter. Fourteen miRNAs were modulated (5 downregulated, 9 upregulated) by the BET inhibitor in OCI-LY-1 cells and 11 (5 downregulated, 6 upregulated) in HBL-1 cells (Table 1).
Table 1.
miRNAs modulated by the BET inhibitor OTX015 in four DLBCL cell lines.
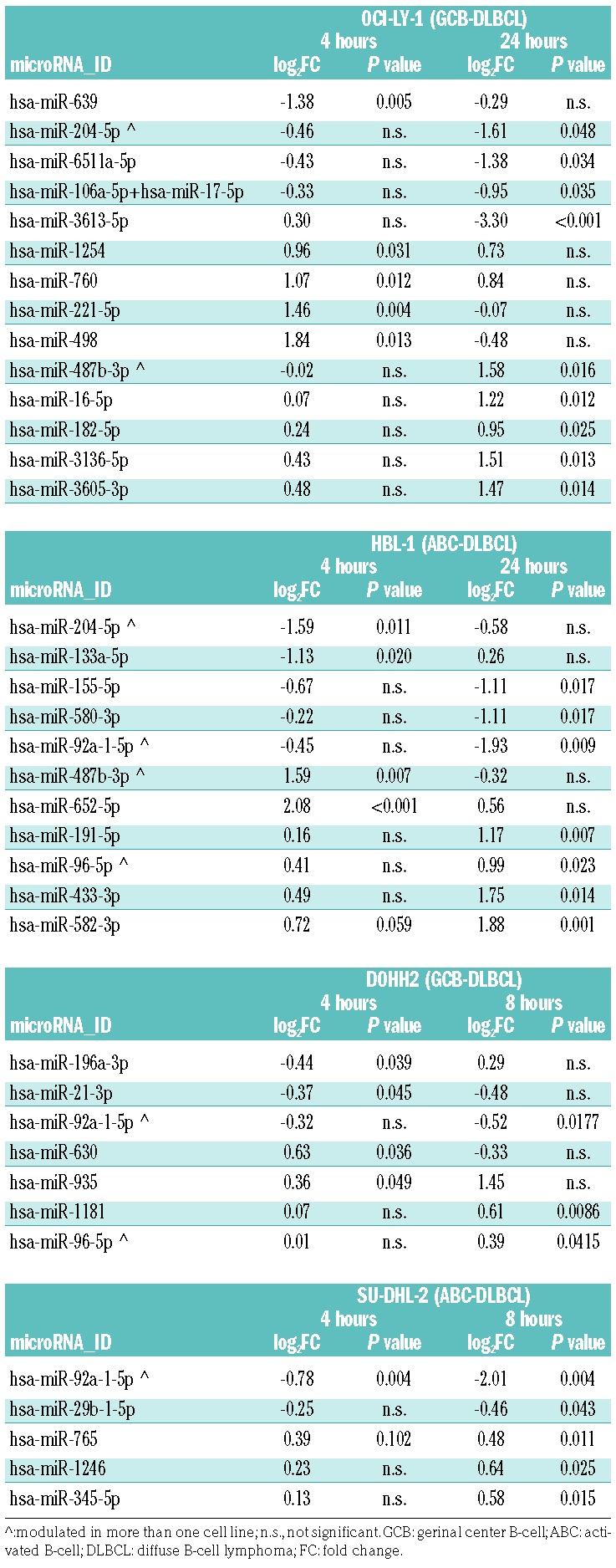
Additionally, we used the Agilent Human miRNA microarray v.3. platform to perform miRNA profiling on two more DLBCL cell lines, DOHH-2 (GCB-DLBCL) and SU-DHL-2 (ABC-DLBCL), the same cell lines we had previously used for mRNA profiling of OTX015-treated cells.15 In this case, seven miRNAs (3 downregulated, 4 upregulated) were affected by OTX015 in the GCB-DLBCL, and five (2 downregulated, 3 upregulated) in the ABC-DLBCL cell line (Table 1).
A few miRNAs were affected in more than one cell line, although we could not determine clear subtype-specific differences in miRNA modulation since only one GCB-and one ABC-DLBCL cell line were interrogated on each profiling platform. The oncogenic miR-92a-1-5p,35 belonging to the miR-17-92 cluster, was downregulated in three of four cell lines (2 ABC-DLBCL, 1 GCB-DLBCL). miR-204-5p, involved in BRAF resistance in melanoma,36 was downregulated and miR-487b-3p, expressed at lower levels in DLBCL versus follicular lymphoma,37 was upregulated in both cell lines analyzed with the Nanostring platform. The tumor suppressor miR-96-5p38,39 was upregulated in HBL-1 and DOHH-2 cells. Besides these, among the miRNAs modulated by the BET inhibitor there were others known to be involved in lymphomagenesis. The oncogenic miRNAs hsa-miR-21-3p40–44 and miR-15545,46 were downregulated, while, besides miR-96-5p, another miRNA with a tumor suppressor function, miR-16-5p,47 was also upregulated by the BET inhibitor. qRT-PCR was used to validate the expression of two lymphoma oncomiRNAs modulated by BET inhibition: miR-155-5p and miR-92a-1-5p (Online Supplementary Figure S1). The latter also appeared significantly downregulated after in vivo treatment of SU-DHL-2 xenografts (Online Supplementary Figure S2).
microRNAs modulated by BET inhibition control important pathways in diffuse large B-cell lymphoma
Functional annotation analysis identified the p53 pathway, apoptosis, MYC-targets, cell cycle regulation, B-cell receptor signaling, interleukin-6 signaling, the STAT3 pathway, PI3K and nuclear factor-κB signaling among the biological processes significantly associated with the miRNAs that exhibited expression changes in HBL-1 and OCI-LY-1 DLBCL cells treated with OTX015 (Online Supplementary Table S1). The same pathways were predicted to be affected based on the modulated miRNAs in DOHH-2 and SU-DHL-2 cells treated with OTX015 (Online Supplementary Table S1). These signaling pathways and processes were similar to those we previously observed when analyzing the gene expression profiles of OTX015-treated DLBCL cells.15
BRD4 binds to the upstream regulatory regions of multiple miRNAs
To further study the role of BET proteins in miRNA regulation, we took advantage of two publicly available ChIP-sequencing datasets obtained in the ABC-DLBCL cell line HBL-1 (SRP043524)23 and in the GCB-DLBCL cell line OCI-LY-1 (SRP022129),22 both treated with the BET inhibitor JQ1 or DMSO alone. Analysis of these datasets revealed that half of the regions bound by BRD4 were in intronic and intergenic regions where miRNAs are often located.48 We detected 794 miRNAs with at least one BRD4-binding event within their regulatory regions in ABC-DLBCL HBL-1 cells and 757 in the GCB-DLBCL OCI-LY-1 cell line (Online Supplementary Table S2).
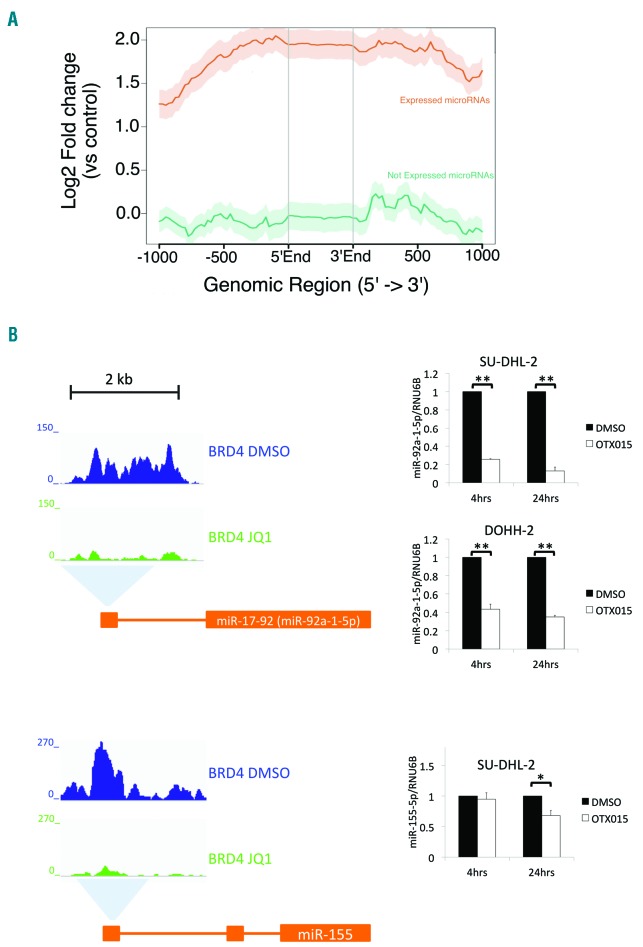
To determine whether BRD4 binding was associated with the expression of miRNA genes we profiled miRNA expression levels in a panel of 35 lymphoma cell lines using the Nanostring nCounter (Online Supplementary Table S3). Comparison of miRNA expression levels and BRD4 binding sites demonstrated that BRD4 peaks were more prevalent in the proximity of expressed miRNAs than non-expressed miRNAs (P<0.001) and were positively correlated with miRNA expression levels (Figure 1A). When we compared BRD4 binding in the presence or absence of the BET inhibitor JQ1, we identified 707 miRNAs with decreased BET bromodomain binding after exposure to the BET inhibitor in ABC-DLBCL HBL-1 cells and 348 in GCB-DLBCL OCI-LY-1 cells (Online Supplementary Table S2). Downregulation of miR-92a-1-5p and miR-155p expression following BET inhibitor-mediated reduction of BRD4 binding was also confirmed by qRT-PCR analysis (Figure 1B).
Figure 1.
BRD4 binds to the regulatory regions of microRNAs. (A) The genomic regions within ± 1 kb of miRNA promoters that are bound by BRD4 obtained using ngs.plot. Lines represent the average expression profiles of “expressed” (red line) and “not expressed” (green line) miRNA. (B) Analysis of publicly available chromatin immunoprecipitation sequencing data of diffuse large B-cell lymphoma (DLBCL) cells showed that BET inhibitor treatment reduces BRD4 binding at the 5ʹ regulatory regions of the miR-17-92 cluster, which contains miR-92a-1-5p, and the miR-155 host gene (left panel). An activated B-cell (ABC)-DLBCL cell line (SU-DHL-2) and a germinal center B-cell (GCB)-DLBCL cell line (DOHH-2) were treated with OTX015 for 4 and 24 h before TaqMan quantitative reverse transcription polymerase chain reaction analysis of miR-92a-1-5p and miR-155-5p expression (right panel). miR-155-5p expression is only shown for SU-DHL-2 as it is an ABC-DLBCL specific oncomiR. Expression of RNU6B was used for normalization. For each timepoint, the mean fold-change relative to the dimethyl sulfoxide (DMSO) control is shown. Charts show the mean of at least three independent experiments. *P<0.05; **P<0.01. Error bars denote the standard error.
BET inhibition mediates upregulation of miR-96-5p by downregulating PRMT5 expression
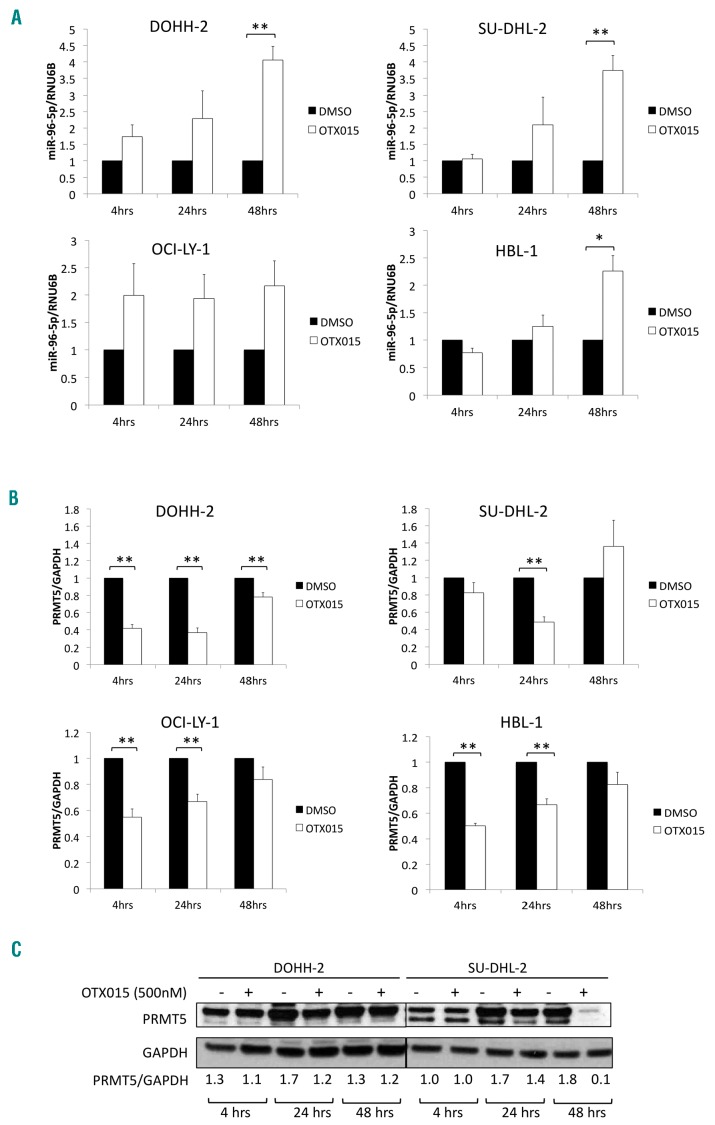
The observed upregulation of the tumor suppressor miR-96-5p after exposure of DLBCL cell lines to OTX015 could not be explained by a direct BRD4-mediated effect of the BET inhibitor on the miRNA promoter. Thus, the miRNA profiling results were further validated by qRT-PCR in two GCB-DLBCL cell lines (DOHH-2, OCI-LY-1) and two ABC-DLBCL cell lines (SU-DHL-2, HBL-1) treated with OTX015 for 4, 24 and 48 h. For DOHH-2, SU-DHL-2 and HBL-1 cells, there was a time-dependent upregulation of miR-96-5p. For OCI-LY-1 cells, miR-96-5p was similarly upregulated at all three time points (Figure 2A).
Figure 2.
OTX015 modulates microRNA-96-5p expression in diffuse large B-cell lymphoma models. (A) OTX015 upregulates miR-96-5p in a time-dependent manner. Two germinal center B-cell (GCB)-diffuse large B-cell lymphoma (DLBCL) cell lines (DOHH-2, OCI-LY-1) and two activated B-cell (ABC)-DLBCL cell lines (SU-DHL-2, HBL-1) were treated with dimethyl sulfoxide (DMSO) or 500 nM OTX015 for 4, 24, and 48 h. Expression of miR-96-5p was determined by TaqMan quantitative reverse transcription polymerase chain reaction (qRT-PCR). Expression of RNU6B was used for normalization. For each time-point, the mean fold-change relative to the DMSO control is shown. (B) OTX015 treatment of DLBCL cells downregulates PRMT5. Two GCB-DLBCL (DOHH-2, OCI-LY-1) and two ABC-DLBCL (SU-DHL-2, HBL-1) cell lines were treated with DMSO or 500 nM OTX015 for 4, 24, and 48 h. Expression of PRMT5 was determined by qRT-PCR. GAPDH expression was used for normalization. For each time-point, the mean fold-change relative to the DMSO control is shown. (C) OTX015 reduces PRMT5 protein levels in DOHH-2 and SU-DHL-2 cells treated with DMSO or 500 nM OTX015. GAPDH was used as a loading control. PRMT5 signals were quantified using ImageJ (http://rsb-web.nih.gov/ij/) and normalized to GAPDH signals. Representative images of two independent Western blot analyses are shown. The graphs show the mean of three independent experiments. **P<0.01. Error bars denote the standard error.
In lymphomas, miR-96-5p expression is regulated as part of a negative feedback loop with the protein arginine methyltransferase, PRMT5.39 Overexpression of PRMT5 mediates transcriptional repression of this miRNA via symmetric dimethylation of histones H3 and H4 in the promoter of miR-96-5p. Conversely, binding of miR-96-5p to the 3ʹ untranslated region of PRMT5 inhibits its translation.39,49 We hypothesized that the upregulation of miR-96-5p observed in OTX015-treated DLBCL cells could be due to a perturbation of its downregulation by PRMT5. To assess this, we performed qRT-PCR analysis of PRMT5 in DLBCL cells treated with OTX015 for 4, 24 and 48 h. PRMT5 mRNA was markedly downregulated at 4 and 24 h. At 48 h PRMT5 levels were similar in DMSO- and OTX015-treated cells for all four cell lines (Figure 2B). At the protein level (Figure 2C), moderate downregulation of PRMT5 was evident at 24 h in DOHH-2 cells treated with OTX015. In SU-DHL-2 cells, PRMT5 was moderately downregulated at 24 h and was negligible at 48 h. These results indicate that OTX015 could downregulate the levels of expression of both PRMT5 RNA and protein in DLBCL cells. Hence upregulation of miR-96-5p following OTX015 treatment was associated with downregulation of PRMT5 protein, particularly in SU-DHL-2 cells.
BRD4 binds to the 5ʹ regulatory region of PRMT5 in diffuse large B-cell lymphoma cells and treatment with a BET inhibitor reduces BRD4 binding
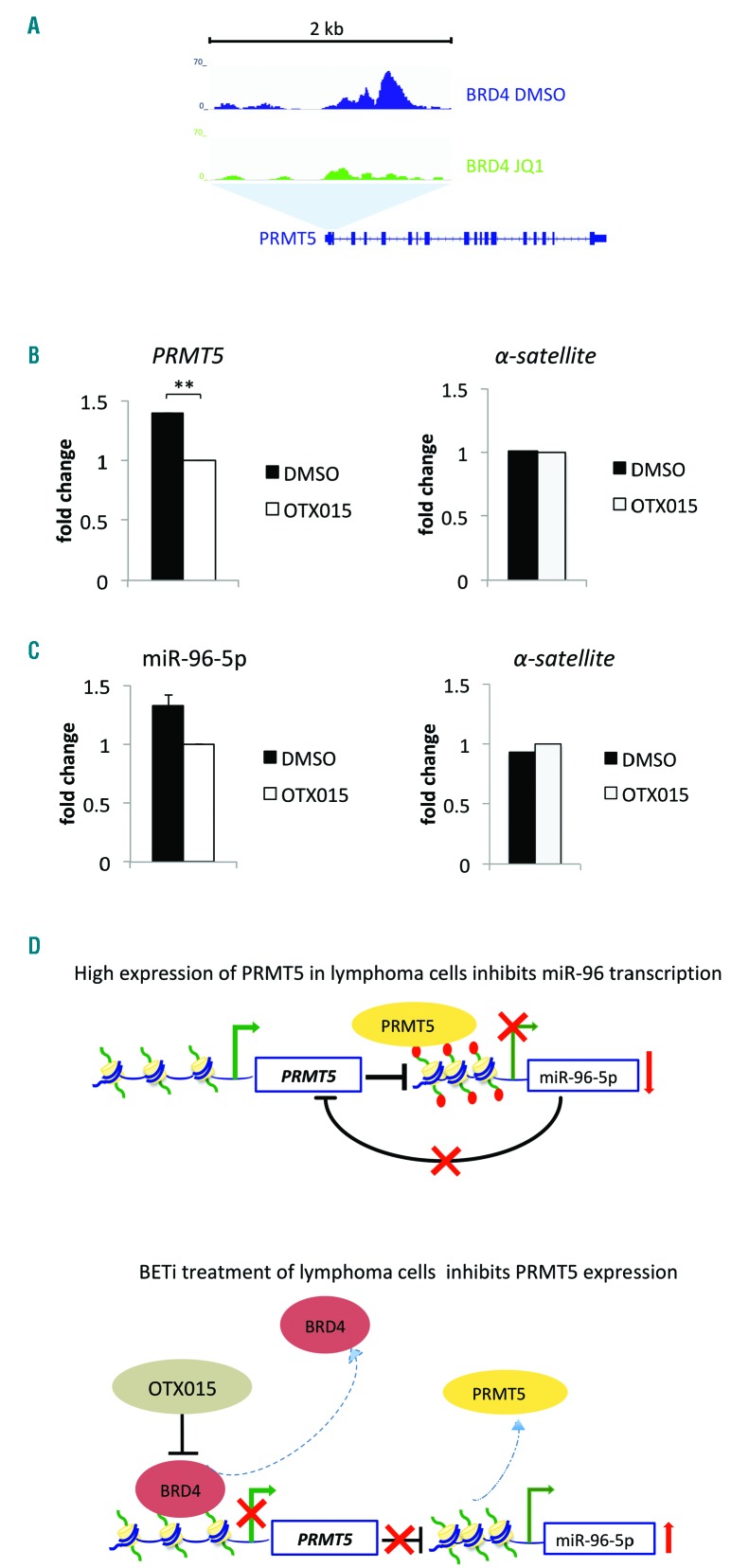
As OTX015 mediates transcriptional repression by displacing BRD4 from chromatin,15,21 we hypothesized that PRMT5 was transcriptionally regulated by BRD4. To assess this, we re-analyzed the two public ChIP sequencing datasets of DLBCL cells treated with the BET inhibitor JQ1,22,23 which has a similar mechanism of action to OTX015 and exhibits an overlapping profile of targeted genes.15,22 This revealed that BRD4 bound to the 5ʹ region of PRMT5 and that this binding was reduced following treatment with the BET inhibitor (Figure 3A). In agreement with the public ChIP sequencing data of JQ1-treated DLBCL cells, when we performed ChIP-qPCR analysis of SU-DHL-2 cells treated with OTX015, we observed decreased binding of BRD4 to the 5ʹ region of PRMT5 (Figure 3B). The decrease in PRMT5 expression following OTX015 treatment was, therefore, likely due to a reduced association of BRD4 to the 5ʹ regulatory region of PRMT5. Additionally, DNAse hypersensitivity sites and H3K27 acetylation, both marks of active transcription, were enriched at the BRD4 binding site in PRMT5 (data not shown). As PRMT5 and miR-96-5p regulate each other in a negative feedback loop, we tested for binding of PRMT5 to the promoter of miR-96-5p. OTX015 treatment led to reduced recruitment of PRMT5 to the miR-96-5p promoter (Figure 3C) indicating that upregulation of miR-96-5p in BET inhibitor-treated cells was mediated through downregulation of PRMT5 (Figure 3D).
Figure 3.
OTX015 reduces binding of BRD4 to PRMT5 and diminishes recruitment of PRMT5 to the microRNA-96-5p promoter. (A) Analysis of publicly available chromatin immunoprecipitation (ChIP) sequencing data of diffuse large B-cell lymphoma (DLBCL) cells treated with the BET inhibitor JQ1 showed that BRD4 binds to the 5ʹ regulatory region of PRMT5 and that BET inhibitor treatment reduces BRD4 recruitment to PRMT5. (B,C) ChIP was performed for DLBCL cells treated with dimethyl sulfoxide (DMSO) or 500 nM OTX015 for 48 h. Anti-BRD4, anti-PRMT5 and anti-IgG (negative control) antibodies were used for immunoprecipitations. (B) Chromatin pulled down with anti-BRD4 and anti-IgG in DMSO- and OTX015-treated SU-DHL-2 cells was amplified with primers specific for the 5ʹ regulatory region of PRMT5 identified by the analysis in (A). (C) Chromatin pulled down with anti-PRMT5 and anti-IgG antibodies in DMSO- and OTX015-treated HBL-1 cells was amplified with primers specific for the promoter of miR-96-5p. Amplification of the same immunoprecipitated chromatin samples was performed with primers specific for the chromosome 4 human alpha satellite sequence as an additional negative control (representative results from one of two biological replicates are shown). The graphs show the mean fold-difference between DMSO- and OTX015-treated cells after normalization to input and IgG background subtraction. ChIP-quantitative polymerase chain reaction experiments were repeated twice in triplicate. (D) Proposed model for the upregulation of miR-96-5p expression following treatment of DLBCL cells with OTX015. Upper panel; previous work by others has shown that PRMT5 is overexpressed in lymphoma cells in which it mediates transcriptional repression of miR-96 and that overexpression of miR-96 negatively regulates PRMT5 translation.39,49 Lower panel; here we showed that BRD4 binds to the upstream regulatory region of PRMT5. Treatment of DLBCL cells with a BET inhibitor reduced BRD4 occupancy at the PRMT5 locus and also reduced the expression of PRMT5 mRNA and protein. Additionally, the BET inhibitor diminished the occupancy of PRMT5 at the miR-96 promoter and increased miR-96 expression. **P<0.01. Error bars denote the standard error.
Discussion
The present study shows that the BET inhibitor OTX015 modulates the expression of miRNAs in DLBCL cells. The regulation may occur directly, due to the binding of BRD4 to the regulatory regions of specific miRNAs, or indirectly as demonstrated for miR-96-5p, a miRNA with important functions in the proliferation and survival of B-cell malignancies.49 The ability of OTX015 to alter miRNA expression demonstrates that the effects of BET inhibition on the transcriptome extend beyond coding genes to comprise also non-coding regions of the genome. miRNAs function by regulating the expression of genes at the transcript level, where they can mediate mRNA degradation, reduce mRNA stability or prevent translation of mRNA into protein.
By analyzing publicly available ChIP sequencing data22,23 in combination with our miRNA profiling data of baseline and BET inhibitor-treated lymphoma cells, we determined that a subset of miRNAs were bound by BRD4 and that this binding decreased after BET inhibitor treatment. For a number of these miRNAs reduced binding of BRD4 after BET inhibition was associated with reduced expression. Our finding that BRD4 directly binds to the regulatory regions of miRNA genes to regulate their expression in lymphomas complements the recent report describing that components of the miRNA processing machinery, namely DGCR8 and Drosha, are localized to super-enhancers of miRNAs in a tissue-specific manner and that association of these proteins with super-enhancers is reduced following treatment with the BET inhibitor JQ1.50 Indeed, these observations indicate that the targeted effects of BET inhibition on specific genes in different cellular contexts also likely comprises non-coding transcripts that are specifically modulated in different transformed cell types. With respect to this, we observed that the promoters of two established lymphoma oncomiRNAs, miR-155-5p and miR-92a-1-5p, were bound by BRD4. When lymphoma cells were treated with a BET inhibitor, BRD4 was diminished at these sites and this was associated with downregulation of miRNA expression. miR-155-5p is upregulated in activated B cells and ABC-DLBCL,45,46,51 often because of amplification of its locus.45 Its high expression is associated with poor outcome and resistance to R-CHOP therapy and its knockdown compromises the viability of ABC-DLBCL cells.43,45 Of interest, BET inhibition decreased miR-155-5p expression only in the two ABC-DLBCL cell lines, while the GCB-DLBCL cell line showed upregulation of miR-155-5p suggesting that different mechanisms may regulate the transcription of this miRNA gene in different DLBCL subtypes.
miR-92a-1-5p is one of the six members of the miR-17-92 cluster located on chromosome 13q31.3 and is overexpressed in different lymphoma subtypes including DLBCL.35 It is a transcriptional target of the MYC oncoprotein,35 which is itself rapidly and robustly downregulated by treatment with a BET inhibitor.15 Downregulation of miR-92a-1-5p was already very pronounced after 4 h of BET inhibitor treatment. Additionally, miR-92a-1-5p was the only miRNA commonly identified by the two different platforms that we utilized for miRNA profiling. The miR-17-92 cluster is involved in activation of the PI3K/AKT/mTOR pathway, lymphoma pathogenesis and chemoresistance.52,53 The miR-17-92 cluster can acquire super-enhancers during neoplastic transformation50,54 and BRD4 exhibits a preference for binding at super-enhancers.22 This provides further support for our observation of direct regulation of the miR-17-92 cluster by BRD4.
miR-21-3p was also downregulated by BET inhibitor treatment. This miRNA is overexpressed in B-cell lymphomas.40–42 It inhibits translation of the tumor suppressor PTEN43 and knockdown of miR-21 increases the sensitivity of lymphoma cells to CHOP treatment.44
When we performed functional annotation analysis of the OTX015-modulated miRNAs, we identified the same signaling pathways and processes that we had previously identified from gene expression profiles of OTX015-treated DLBCL cells,15 indicating that changes in miRNA expression likely contribute to modulating some of the transcripts and pathways that have been previously identified in cells treated with a BET inhibitor.15,22,23
In lymphoma cells, which overexpress PRMT5, the negative feedback loop comprising miR-96-5p and PRMT5 is usually poised in favor of PRMT5.39 PRMT5 catalyzes the symmetric methylation of arginine residues on histones H3 and H4, giving rise to the repressive epigenetic marks H3R8me2S and H4R3me2S. Inhibition of PRMT5 expression, either pharmacologically or by the use of antisense oligonucleotides, severely compromises lymphoma cell viability and induces apoptosis.39,49 In the DLBCL cells we treated with OTX015, PRMT5 protein was negligible after 48 h of treatment in the ABC-DLBCL cell line SU-DHL-2, which we previously showed undergoes pronounced apoptosis in response to OTX015.15 DOHH-2 cells, in which we did not observe apoptosis following OTX015 treatment,15 exhibited moderate PRMT5 downregulation. The less marked downregulation of PRMT5 protein in DOHH-2 cells treated with OTX015 was in contrast to its pronounced downregulation at the transcript level. It was nevertheless associated with a pronounced increase in miR-96-5p levels indicating that, for this cell line, factors other than PRMT5 downregulation may contribute to releasing miR-96-5p from transcriptional repression. Inhibition of PRMT5 has been shown to release miR-96-5p from transcriptional repression in lymphoma cells.39 We observed that BET inhibitor-mediated inhibition of PRMT5 was associated with decreased binding of BRD4 to the 5ʹ regulatory region of PRMT5 and that this resulted in reduced occupancy of PRMT5 at the miR-96-5p promoter. The disruption of the negative feedback loop comprising PRMT5 and miR-96-5p by OTX015 shows how the anti-tumor effects of BET inhibition are further amplified by modulation of secondary targets, which might themselves also contribute to further suppress direct targets of BET inhibitors. With respect to this, overexpression of miR-96-5p downregulates phosphorylated STAT3 (p-STAT3) without affecting levels of total STAT3 in T-cell anaplastic large-cell lymphoma cells.38 We have previously shown that OTX015 treatment decreases p-STAT3 in ABC-DLBCL cells.15 It is therefore possible that in addition to the direct effects of OTX015 on the expression of genes involved in JAK/STAT signaling,15 the overexpression of miR-96-5p contributes to maintaining p-STAT3 repressed.
Our study provides the first evidence of BET inhibitor-mediated modulation of miRNAs in lymphomas. This modulation can occur by inhibiting the interaction of BRD4 with genes whose products regulate miRNA expression, or through the direct inhibition of BRD4 at miRNA regulatory regions, or, as recently suggested, by interfering with the processing of pri-miRNA to pre-miRNAs.50 Unlike coding transcripts, miRNAs are highly stable in blood and as such, levels of circulating miRNAs have been used for diagnosis and screening in a number of diseases. In lymphomas, the overexpression of specific miRNAs in plasma and serum samples has been shown to be an accurate biomarker for diagnosis, prognosis and response to therapy.45 The circulating miRNAs that have been identified as biomarkers in lymphoma are among those that we have identified as regulated by BET inhibition (miR-92, miR-21, miR-155). The assessment of circulating miRNAs could, therefore, be used as a robust and non-invasive way to monitor response to BET inhibitor treatment.
In conclusion, our observations contribute to a better understanding of the targeted effects of BET inhibitors, revealing a novel aspect of the activity of this class of compounds in lymphomas.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/12/2049
References
- 1.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. [DOI] [PubMed] [Google Scholar]
- 3.Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15): 6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. [DOI] [PubMed] [Google Scholar]
- 5.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. [DOI] [PubMed] [Google Scholar]
- 6.Kluiver J, Poppema S, de Jong D, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243–249. [DOI] [PubMed] [Google Scholar]
- 7.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hezaveh K, Kloetgen A, Bernhart SH, et al. Alterations of microRNA and microRNA-regulated messenger RNA expression in germinal center B-cell lymphomas determined by integrative sequencing analysis. Haematologica. 2016;101(11):1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC. Micro RNAs are complementary to 3ʹ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. [DOI] [PubMed] [Google Scholar]
- 11.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. [DOI] [PubMed] [Google Scholar]
- 12.Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;66(6):443–459. [DOI] [PubMed] [Google Scholar]
- 13.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boi M, Gaudio E, Bonetti P, et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res. 2015;21(7):1628–1638. [DOI] [PubMed] [Google Scholar]
- 16.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13(5):337–356. [DOI] [PubMed] [Google Scholar]
- 17.Stathis A, Bertoni F. BET Proteins as targets for anticancer treatment. Cancer Discov. 2018;8(1):24–36. [DOI] [PubMed] [Google Scholar]
- 18.Stathis A, Zucca E, Bekradda M, et al. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 2016;6(5):492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amorim S, Stathis A, Gleeson M, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3(4):e196–204. [DOI] [PubMed] [Google Scholar]
- 20.Berthon C, Raffoux E, Thomas X, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3(4):e186–195. [DOI] [PubMed] [Google Scholar]
- 21.Henssen A, Althoff K, Odersky A, et al. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clin Cancer Res. 2016;22(10):2470–2481. [DOI] [PubMed] [Google Scholar]
- 22.Chapuy B, McKeown MR, Lin CY, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceribelli M, Kelly PN, Shaffer AL, et al. Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci USA. 2014;111(31):11365–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riveiro ME, Astorgues-Xerri L, Vazquez R, et al. OTX015 (MK-8628), a novel BET inhibitor, exhibits antitumor activity in non-small cell and small cell lung cancer models harboring different oncogenic mutations. Oncotarget. 2016;7(51):84675–84687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez R, Riveiro ME, Astorgues-Xerri L, et al. The bromodomain inhibitor OTX015 (MK-8628) exerts anti-tumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget. 2017;8(5):7598–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noel JK, Iwata K, Ooike S, Sugahara K, Nakamura H, Daibata M. Development of the BET bromodomain inhibitor OTX015. Mol Cancer Ther. 2013;12(11 Suppl): C244. [Google Scholar]
- 27.Chila R, Basana A, Lupi M, et al. Combined inhibition of Chk1 and Wee1 as a new therapeutic strategy for mantle cell lymphoma. Oncotarget. 2015;6(5):3394–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarantelli C, Gaudio E, Arribas AJ, et al. PQR309 is a novel dual PI3K/mTOR inhibitor with preclinical antitumor activity in lymphomas as a single agent and in combination therapy. Clin Cancer Res. 2018;24(1):120–129. [DOI] [PubMed] [Google Scholar]
- 29.Valeri N, Braconi C, Gasparini P, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dal Bo M, D’Agaro T, Gobessi S, et al. The SIRT1/TP53 axis is activated upon B-cell receptor triggering via miR-132 up-regulation in chronic lymphocytic leukemia cells. Oncotarget. 2015;6(22):19102–19117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007. [cited; Available from: http://bioin-fogp.cnb.csic.es/tools/venny/index.html.
- 32.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lizio M, Harshbarger J, Shimoji H, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dal Bo M, Bomben R, Hernandez L, Gattei V. The MYC/miR-17-92 axis in lymphopro-liferative disorders: a common pathway with therapeutic potential. Oncotarget. 2015;6(23):19381–19392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Martinez M, Benito-Jardon L, Alonso L, Koetz-Ploch L, Hernando E, Teixido J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018;78(4):1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Culpin RE, Proctor SJ, Angus B, Crosier S, Anderson JJ, Mainou-Fowler T. A 9 series microRNA signature differentiates between germinal centre and activated B-cell-like diffuse large B-cell lymphoma cell lines. Int J Oncol. 2010;37(2):367–376. [DOI] [PubMed] [Google Scholar]
- 38.Vishwamitra D, Li Y, Wilson D, et al. MicroRNA 96 is a post-transcriptional suppressor of anaplastic lymphoma kinase expression. Am J Pathol. 2012;180(5):1772–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26(15):3558–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arribas AJ, Gomez-Abad C, Sanchez-Beato M, et al. Splenic marginal zone lymphoma: comprehensive analysis of gene expression and miRNA profiling. Mod Pathol. 2013;26(7):889–901. [DOI] [PubMed] [Google Scholar]
- 41.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117(11):3140–3146. [DOI] [PubMed] [Google Scholar]
- 42.Junker F, Chabloz A, Koch U, Radtke F. Dicer1 imparts essential survival cues in Notch-driven T-ALL via miR-21-mediated tumor suppressor Pdcd4 repression. Blood. 2015;126(8):993–1004. [DOI] [PubMed] [Google Scholar]
- 43.Go H, Jang JY, Kim PJ, et al. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6(17):15035–15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai H, Wei J, Deng C, Yang X, Wang C, Xu R. MicroRNA-21 regulates the sensitivity of diffuse large B-cell lymphoma cells to the CHOP chemotherapy regimen. Int J Hematol. 2013;97(2):223–231. [DOI] [PubMed] [Google Scholar]
- 45.Iqbal J, Shen Y, Huang X, et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood. 2015;125(7):1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121(5):1156–1161. [DOI] [PubMed] [Google Scholar]
- 47.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 49.Alinari L, Mahasenan KV, Yan F, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125(16):2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative MicroRNA network analysis. Cell. 2017;168(6):1000–1014.e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmadvand M, Eskandari M, Pashaiefar H, et al. Over expression of circulating miR-155 predicts prognosis in diffuse large B-cell lymphoma. Leuk Res. 2018;70:45–48. [DOI] [PubMed] [Google Scholar]
- 52.Rao E, Jiang C, Ji M, et al. The miRNA-17-92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26(5):1064–1072. [DOI] [PubMed] [Google Scholar]
- 53.Benhamou D, Labi V, Novak R, et al. A c-Myc/miR17-92/Pten axis controls PI3K-mediated positive and negative selection in B cell development and reconstitutes CD19 deficiency. Cell Rep. 2016;16(2):419–431. [DOI] [PubMed] [Google Scholar]
- 54.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.